Abstract
Similarity measurement of lung nodules is a critical component in content-based image retrieval (CBIR), which can be useful in differentiating between benign and malignant lung nodules on computer tomography (CT). This paper proposes a new two-step CBIR scheme (TSCBIR) for computer-aided diagnosis of lung nodules. Two similarity metrics, semantic relevance and visual similarity, are introduced to measure the similarity of different nodules. The first step is to search for K most similar reference ROIs for each queried ROI with the semantic relevance metric. The second step is to weight each retrieved ROI based on its visual similarity to the queried ROI. The probability is computed to predict the likelihood of the queried ROI depicting a malignant lesion. In order to verify the feasibility of the proposed algorithm, a lung nodule dataset including 366 nodule regions of interest (ROIs) is assembled from LIDC-IDRI lung images on CT scans. Three groups of texture features are implemented to represent a nodule ROI. Our experimental results on the assembled lung nodule dataset show good performance improvement over existing popular classifiers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Computer-aided diagnosis (CAD) of lung cancer is an extremely important task, as lung cancer is the leading cause of cancer mortality around the world [1]. Early detection and diagnosis are critical as the chance of recovering in the early phase of the cancer illness. Low-radiation dose computer tomography (CT) scans is one of the most common screenings for lung cancer [2, 3]. Thus, a critical issue is the diagnosis of a pulmonary nodule as benign or malignant with CT.
In general, CAD systems extract the features of a lung cancer image and apply a classifier to test the malignancy. Usually the diagnosis performance of CAD systems is assessed through the receiver operating characteristic (ROC) curve or the area under the ROC curve (AUC). Lu et al. [4] developed an intelligent system for lung cancer diagnosis with 32 samples. They obtained an AUC of 0.99. Orozco et al. [5] used 11 features and SVM as classifier. They obtained a value of 0.805 when evaluating 23 malignant nodules and 22 non-nodules. Shen et al. [6] explored multi-crop convolutional neural networks to handle the classification of lung nodule malignancy suspiciousness. Its AUC value reached 0.93.
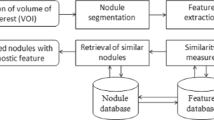
Content-based image retrieval (CBIR) is one of the CAD methods, which can help doctors diagnose a given case by retrieving a selection of similar annotated cases from large medical image repositories [7]. Gundreddy et al. [8] proposed a two-step CBIR scheme for classification of breast lesions, in which two features were used to represent the breast lesions. Jiang et al. [9] developed a scalable image retrieval CAD system to assist radiologists in evaluating the likelihood of malignancy of mammographic masses. Tsochatzidis et al. [10] proposed a new texture descriptor to capture mass properties, and applied CBIR to diagnose mammographic masses. Dubey et al. [11] used CBIR method to assist lung diagnosis, however, they just focused on the feature characteristic of lung CT. Ma et al. [12] explored a context-sensitive similarity measure method to retrieve CT imaging signs. Few of them concentrated on lung nodule classification.
There are two important processes in CBIR, feature extracting and similarity metric. The feature extracting is an important task which can be sufficient to describe the image accurately. The image similarity includes semantic relevant and visual similarity [13, 14]. The semantic relevant depends on the malignant or benign label of masses, which means that if the two masses are both malignant masses, they are similar on the semantic. Visual similarity is the feature similarity, means that the retrieved image must be similar to the query image from human’s perspective. However, at present, many researchers only use semantic similarity metric, ignoring the visual similarity metric [13]. This leads to that, when they are applied to image retrieval problems, images ranked at the top of a retrieval list may not be visually similar to the query image. Therefore, the conclusion will make doctors be less likely to trust the system. As a result, the growing vast repositories of clinical imaging data cannot be searched effectively for similar images on the basis of descriptions of similarity measurement. Our group has studied similarity metric of lung nodules in Ref. [14]. This paper synthetically considers semantic relevance and visual similarity, without discussing the importance of each other. However, according to [13], semantic relevance plays an important role, and visual similarity is an important complement. Our study is consistent with this idea. Semantic relevance is used for discarding the semantic irrelevant lung nodule ROIs. And then visual similarity is used to retrieve the similar nodule. Moreover, our study takes into account multivariate texture features (local binary pattern feature, Gabor feature, and Haralick feature), but Ref. [14] does not.
The goal of our study is to develop a new two-step content-based image retrieval (TSCBIR) scheme for computer- aided diagnosis of lung nodules and to propose a new similarity metric method to evaluate the similarity between the query lung nodule and reference lung nodule dataset. First, a lung nodule dataset was assembled from the LIDC-IDRI lung CT database. Second, three groups of features were implemented to represent a nodule ROI. Third, a two-step CBIR (TSCBIR) approach was proposed to classify lung nodules. At last, AUC value and classification accuracy were used as the performance assessment index.
Materials
Lung nodule dataset
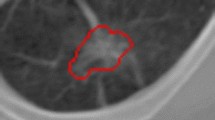
For developing and testing a new TSCBIR scheme in this study, a lung nodule dataset was assembled from an existing completed reference database of LIDC-IDRI lung images on CT scans [15]. The nodules which defined as any lesion were “nodule ≥ 3 mm”. The red arrow in Fig. 1 points to the nodule of the lung CT image.
The lung nodule dataset was assembled as follows:
-
1)
Xml file processing. Based on the annotation of xml, the slice with the largest nodule marked by more than three thoracic radiologists was selected.
-
2)
Outline drawing. For a nodule, each radiologist gives an outline annotation. According to the nodular boundary coordinate, the boundary on the CT slice was drawn.
-
3)
Outline fusion. Three or four nodule boundaries were fused to a reference ground truth based on the Simultaneous Truth and Performance Level Estimation (STAPLE) algorithm [16].
-
4)
Nodule extracting. The nodule was extracted from the lung CT on the basis of the reference ground truth boundary. Figure 1 shows the workflow of lung nodule dataset assembling.
The malignancy of a nodule can be divided into five ratings, with 1 representing highly unlikely for cancer and 5 representing highly suspicious for cancer. For each nodule, the average rating of four thoracic radiologists was computed. In this study, if the rating > 3.5, we labeled this nodule as malignant; if the rating < 2.5, we labeled this nodule as benign. With the process described above, at last, 366 lung nodule dataset was obtained, in which 191 radiologists identified malignant nodules and 175 radiologists identified benign nodules.
Computational features
In this study, for each lung nodule ROI, three different types of widely used 2D texture features were extracted: local binary pattern (LBP) feature [17], Gabor feature [18], gray level co-occurrence matrix (GLCM) feature (Haralick feature) [19]. The number of features for each feature group is summarized in Table 1.
The first group of feature was LBP feature. It was first proposed as a texture descriptor for images [20]. The LBP calculated the relationships between the central pixel and each neighbor pixel, and it returned binary code for each central pixel. The LBP features were computed with the local binary pattern feature histogram calculated from the coded image.
The second feature group was computed based on Gabor filters. A Gabor filter was obtained by modulating a sinusoid with a Gaussian functional and further being discretized over orientation and frequency. We convolved the image with 12 Gabor filters: three frequencies (0.3, 0.4, and 0.5) and four orientations (0°, 45°, 90°, and 135°). We then computed means and standard deviations from the 12 response images, and 24 Gabor features for each image were obtained.
The third group of feature was Haralick feature, which can be calculated from the gray level co-occurrence matrix. A relative displacements (d = 1 pixel) and four different angles (θ = 0°, 45°, 90°, and 135°) were considered for computational feature extraction. Thus, we calculated a set of four values for each of the preceding 13 measures, except the maximal correlation coefficient feature. For each image, the mean and standard deviation of each of these 13 measures were computed, so 26 features were generated in this group.
Methods
Distance metric learning for similarity metrics
As mentioned above, the image similarity includes semantic relevance and visual similarity. The semantic relevance depends on the malignant or benign label of nodules, which means that if two nodules are both malignant nodules, they are similar on the semantic. Visual similarity is the feature similarity, which means that the retrieved images should look like the query image. Most distance metric learning algorithms essentially preserve only the semantic similarity among data points by learning a distance metric with the given pairwise constraints. However, visual similarity is of equal importance.
Semantic relevance
Let dataset C = {x 1, x 2, …, x n } be a collection of image data points, where n is the number of samples in the collection. Each x i ∈ R m is a data vector where m is the number of features.
In this study, a Mahalanobis distance is learned to measure the image semantic relevance. The Mahalanobis distance can be computed by the formula as follows [14]:
Thus, a matrix A for computing the Mahalanobis distance is required to learn. According to the pairwise constraints [21], the dataset can be divided into two parts. The set of equivalence constraints denoted by
and the set of inequivalence constraints denoted by
The data points connected by equivalence constraints should be close in the new feature space, and the data points connected by inequivalence constraints should be kept far away in the new feature space. Therefore, the semantic similarity metric is obtained by optimizing the formula as:
Where β is the tradeoff parameter. y i = A T x i ∈ R k (k > m) is the new feature representation from the x i , this formula means that the feature representations of the data points in the same class should be closer, and the data points in different class should be far away.
Imposing A T A = I to the Eq. (2), the transformation matrix A can be obtained through eigendecomposition in Eq. (2). In this case, the solution of A is constructed by the d eigenvectors associated with the d smallest eigenvalues. Then the Mahalanobis distance can be computed by the formula as follows:
This Mahalanobis distance preserves the semantic relevance.
Visual similarity
The visual similarity is the feature similarity, which means that the retrieved image must be similarity compared to the queried image from human’s perspective. In this study, European distance is used to measure visual similarity. A smaller distance means the higher degree of similarity.
Content-based retrieval scheme for lung nodules classification
With the two distance metrics, a new CBIR scheme is explored to classify lung nodules. A two-step similarity metric approach is applied to retrieve similar reference nodule ROIs. In this approach, Mahalanobis distance is used to discard the semantic irrelevant reference ROI in the first step and European distance can more focus on the visual similarity in the second step. Therefore, our scheme first retrieves for K most similar ROIs by computing the Mahalanobis distance between the queried ROI and each of the reference nodule ROI in our dataset. The K most similar ROIs correspond to the reference ROIs with smallest Mahalanobis distance to the queried ROI. In the second step, the scheme weight each retrieved ROI based on its European distance to the queried ROI. The weighting factor is calculated as:
F(i) represents the features of ith reference ROI. F(q) represents the features of the queried nodule.
Finally, the probability that the queried ROI is malignant is computed. The formula is as follows (M is the number of malignant nodules and B is the number of benign nodules):
With this scheme, when giving a threshold of S q (such as S T = 0.5), if S q is above S T , we can figure this queried nodule is malignant, otherwise, it is benign.
Experimental setup
To avoid the bias caused by unblalanced texture feature values, all the extracted features are normalized with the mean and the standard deviation computed from the 366 lung nodule ROIs in the dataset.
For fair comparison of the algorithms, 200 randomly selected lung nodules are chosen from the reference library to serve as the training dataset for the training experiment. The remaining nodules are used as testing dataset.
The ability of the proposed scheme for classification between benign and malignant nodules is evaluated with AUC, classification accuracy and p value from t − test.
ROC curve can be obtained by varying the threshold of the probability for predicting malignancy. And AUC is used to evaluate the classification performance. Classification accuracy is able to be calculated as:
The correctly classified samples are according to the threshold S T = 0.5.
In the subsequential figures, each experiment is repeated 10 times with randomly selecting training images. Thus, the AUC value and classification accuracy showed in the figures are mean value over these 10 runs.
Results and discussion
Parameter configuration
There are several parameters of our proposed scheme needed to be set beforehand for experiments. We investigated the sensitivity of β in Eq. (2) for the TSCBIR scheme with the combined features. We varied β with [10−8, 10−6, 10−4, 10−2, 100, 102, 104, 106, 108]. The mean AUC of our dataset are shown in Fig. 2. It can be seen that TSCBIR scheme prefers a large value of β. This experiment demonstrates that the performance of TSCBIR scheme will be very stable when this parameter value greater than or equal to 1.
We then investigated the effect of parameter K in Eq. (4) for TSCBIR scheme in image classifying. The number of retrieved reference ROIs (K) is set within the range [5, 10, 15, 20]. Figure 3 reports the mean AUC with different K. According to the figure, the performance curve in Fig. 3 has small fluctuations. When K = 10, the mean AUC value reaches the maximum.
We evaluated the performance of the proposed scheme at different dimensions for the transformation matrix A. Figure 4 shows the mean AUC with different feature dimensions. When dimension varies in a wide range, the curve has a slight–variability. This indicates that the classification performance is not sensitive to feature dimension.
Feature analysis and classification
The texture feature analysis plays a very important role in computer-aided classification. In section 2.2, three types of texture features have been obtained. The classification performance of TSCBIR scheme is analyzed with different types of features (Haralick features, Gabor features, LBP features and the combined features). Table 2 shows the AUC value and accuracy comparison of differentiating lung nodules using different feature groups. The combined features have the largest AUC value, followed by Haralick, LBP, Gabor features. However, the accuracy of the combined features is worse than the accuracy of Haralick at S T = 0.5. Table 3 displays statistically significant difference between different feature groups at the 5% significance level about the AUC value. An unpaired t-test is used to compute the p value. The data analysis illustrates that the classification performance between the combined features and Haralick, Gabor, LBP features has a statistically significant difference. However, the classification performance between the LBP features and Haralick, Gabor features had no statistically significant difference (p=0.0733 and 0.0637, respectively).
Classification performance
To illustrate and verify the feasible of our scheme on lung nodule diagnosis comprehensively, the performance of TSCBIR scheme was compared with the existing classifiers reported for classification of benign and malignant tumor lesions: (i) support vector machine (SVM) [5], which was used to differentiate malignant nodules and non-nodules; (ii) extreme learning machine (ELM) [22], which was applied to classify breast masses; (iii) orthogonal projection learning for KDPDM (KDPDMorth) [14], which was used to classification of benign and malignant nodules. Semantic similarity metric alone was also included as a comparative reference (denoted as “Semantic”), the probability of a queried ROI is malignant was computed as the ratio between the number of the malignant nodules and the number of the retrieved nodules. Specially, the nodules were represented by the combined features.
Before the classification experiments were carried out, some parameters needed to be optimized which would further improve the classification performance. The number of hidden nodes of ELM cannot be infinite in real implementation. We tested the number within the range [200,400,600,800 1000]. When the number was set as 1000, the training and testing performances of ELM kept almost fixed. Fivefold cross validation (5-CV) was used to optimize the SVM parameters. We extracted 365 nodular samples for the experiments. The data were divided into five groups on average. The classifiers were trained with 4 folds and tested with the remaining fold, so looped five times. The best value of penalty parameter c = 4 and kernel function parameter g = 0.03125 would be used in the following experiments. The experimental results are listed in Tables 4 and 5.
From Table 4, it can be observed that the proposed scheme TSCBIR is effective on the assembled lung nodule dataset. More importantly, TSCBIR outperforms the classical classifiers (e.g., SVM and ELM). Moreover, the comparison of KDPDMorth and TSCBIR illustrates that semantic relevance plays an important role, and visual similarity is an important complement.
Table 5 shows statistically significant difference between our scheme and existing classifiers at the 5% significance level. An unpaired t-test is used to compute the p value. The data analysis illustrates that the classification performance between TSCBIR and Semantic, SVM, ELM, KDPDMorth has a statistically significant difference.
Overall performance
We evaluated the total performance of TSCBIR scheme using a leave-one-nodule-out method with a classification threshold of S T = 0.5. The leave-one-nodule-out validation method selected one nodule as the queried nodule and the remaining 365 nodules in the dataset were used as reference nodules. This process was repeated 366 times so that each nodule was used as the queried nodule once in the whole process. The results are showed in Fig. 5. The AUC value is 0.986 and the classification accuracy is 0.918. Table 6 shows the confusion matrix of lung nodule classification. Both of the classes obtained higher than 80% classification rates. 14.1% of malignant nodules were misclassified as benign nodules while few benign nodules were misclassified as the malignant class. Table 7 displays the recall, precision and F-score of lung nodules classification. Overall, the results show relatively good classification performance.
Conclusions
We have developed a two-step CBIR approach for classification of lung nodules, and showed it to be capable of yielding excellent retrieval results. The assembled dataset is based on the LIDC-IDRI CT images. With the lung nodule dataset, we can retrieve the nodule malignancy, query the nodule characteristics noted by the radiologists based on the retrieval results, such as calcification, sphericity, internal structure.
In this study, three groups of features are implemented to represent a nodule ROI. Experiment results have demonstrated that the combined features have a better description of the lung nodule for classification. The Mahalanobis distance which preserving the semantic relevance and European distance which describing visual similarity are first proposed to assess the malignancy of lung nodules using content-based image retrieval. The experiment results also demonstrate that our proposed scheme had a better classification performance than the state-of-the-art classifiers.
References
Siegel, R.L., Miller, K.D., and Jemal, A., Cancer statistics, 2015. CA Cancer J Clin. 65:5–29, 2015. https://doi.org/10.3322/caac.21254.
Armato, R.S., Mclennan, G., Bidaut, L., et al., The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys. 38(2):915–931, 2011. https://doi.org/10.1118/1.3528204.
Huang, P., Park, S., Yan, R., et al., Added value of computer-aided CT image features for early lung cancer diagnosis with small pulmonary nodules: a matched case-control study. Radiology. 5:162725, 2017. https://doi.org/10.1148/radiol.2017162725.
Lu, C., Zhu, Z., and Gu, X., An intelligent system for lung cancer diagnosis using a new genetic algorithm based feature selection method. J Med Syst. 38(9):97–105, 2014. https://doi.org/10.1007/s10916-014-0097-y.
Orozco, H., Villegas, O., Sánchez, V., Domínguez, H., and Alfaro, M., Automated system for lung nodules classification based on wavelet feature descriptor and support vector machine. BioMed Eng. 14(1):9, 2015. https://doi.org/10.1186/s12938-015-0003-y.
Shen, W., Zhou, M., Yang, F., Yu, D., Dong, D., Yang, C., Zang, Y., and Tian, J., Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recogn. 61:663–673, 2017. https://doi.org/10.1016/j.patcog.2016.05.029.
Kumar, A., Kim, J., Cai, W., Fulham, M., and Feng, D., Content-based medical image retrieval: a survey of applications to multidimensional and multimodality data. J Digit Imaging. 26:1025–1039, 2013. https://doi.org/10.1007/s10278-013-9619-2.
Gundreddy, R.R., Tan, M., Qiu, Y., Cheng, Liu, H., and Zheng, B., Assessment of performance and reproducibility of applying a content-based image retrieval scheme for classification of breast lesions. Med Phys. 42:4241–4249, 2015. https://doi.org/10.1118/1.4922681.
Jiang, M., Zhang, S., Li, H., and Metaxas, D.N., Computer-aided diagnosis of mammographic masses using scalable image retrieval. IEEE T Biomed Eng. 62(2):783–791, 2015. https://doi.org/10.1109/TBME.2014.2365494.
Tsochatzidis, L., Zagoris, K., Arikidis, N., et al., Computer-aided diagnosis of mammographic masses based on a supervised content-based image retrieval approach. Pattern Recogn. 71:106–117, 2017. https://doi.org/10.1016/j.patcog.2017.05.023.
Dubey, S.R., Singh, S.K., and Singh, R.K., Local wavelet pattern: a new feature descriptor for image retrieval in medical CT databases. IEEE T Image Process. 24(12):5892–5903, 2015. https://doi.org/10.1109/TIP.2015.2493446.
Ma, L., Liu, X., Gao, Y., et al., A new method of content based medical image retrieval and its applications to CT imaging sign retrieval. J Biomed Inform. 66:148–158, 2017. https://doi.org/10.1016/j.jbi.2017.01.002.
Liu, Y., Jin, R., Lily, M., Sukthankar, R., Goode, A., Zheng, B., Hoi, S.C.H., and Satyanarayanan, M., A boosting framework for visuality-preserving distance metric learning and its application to medical image retrieval. IEEE T Pattern Anal. 32:30–44, 2010. https://doi.org/10.1109/tpami.2008.273.
Wei, G., Ma, H., Qian, W., and Qiu, M., Similarity measurement of lung masses for medical image retrieval using kernel based semisupervised distance metric. Med Phys. 43(12):6259–6269, 2016. https://doi.org/10.1118/1.4966030.
Armato, S.G., et al., The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys. 38(2):915–931, 2011. https://doi.org/10.1118/1.3528204.
Warfield, S.K., Zou, K.H., and Wells, W.M., Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE T Med Imaging. 23(7):903–921, 2004. https://doi.org/10.1109/TMI.2004.828354.
Ojala, T., Pietikäinen, M., and Mäenpää, T., Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell. 24(7):971–987, 2002. https://doi.org/10.1109/tpami.2002.1017623.
Wei, C. H., Li, Y., and Li, C. T., Effective extraction of Gabor features for adaptive mammogram retrieval. in 2007 I.E. International Conference on Multimedia and Expo (IEEE, New York, NY, 2007), pp. 1503–1506.
Haralick, R.M., Shanmugam, K., and Dinstein, I., Textural features for image classification. IEEE Trans Syst Man Cybern. 6:610–621, 1973.
Sluimer, I.C., van Waes, P.F., Viergever, M.A., and Ginneken, B.v., Computer-aided diagnosis in high resolution CT of the lungs. Med Phys. 30(12):3081–3090, 2003. https://doi.org/10.1118/1.1624771.
Liu, Y., and Jin, R., Distance metric learning: a comprehensive survey, technical report. Report No. UCB/CSD-02-1206, 2006.
Xiong, Y., Luo, Y., Huang, W., Zhang, W., Yang, Y., and Gao, J., A novel classification method based on ICA and ELM: a case study in lie detection. Bio-Med Mater Eng. 24(1):357–363, 2014. https://doi.org/10.3233/BME-130818.
Acknowledgements
The research is supported by the Recruitment Program of Global Experts (Grant no. 01270021814101/022), the National Natural Science Foundation of China (No. 61702087; No. 81671773; No. 61672146; No. 81473708), the Fundamental Research Funds for the Central Universities (Grant no. N150408001).
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Wei, G., Cao, H., Ma, H. et al. Content-based image retrieval for Lung Nodule Classification Using Texture Features and Learned Distance Metric. J Med Syst 42, 13 (2018). https://doi.org/10.1007/s10916-017-0874-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-017-0874-5