Abstract
One of the major issues in time-critical medical applications using wireless technology is the size of the payload packet, which is generally designed to be very small to improve the transmission process. Using small packets to transmit continuous ECG data is still costly. Thus, data compression is commonly used to reduce the huge amount of ECG data transmitted through telecardiology devices. In this paper, a new ECG compression scheme is introduced to ensure that the compressed ECG segments fit into the available limited payload packets, while maintaining a fixed CR to preserve the diagnostic information. The scheme automatically divides the ECG block into segments, while maintaining other compression parameters fixed. This scheme adopts discrete wavelet transform (DWT) method to decompose the ECG data, bit-field preserving (BFP) method to preserve the quality of the DWT coefficients, and a modified running-length encoding (RLE) scheme to encode the coefficients. The proposed dynamic compression scheme showed promising results with a percentage packet reduction (PR) of about 85.39% at low percentage root-mean square difference (PRD) values, less than 1%. ECG records from MIT-BIH Arrhythmia Database were used to test the proposed method. The simulation results showed promising performance that satisfies the needs of portable telecardiology systems, like the limited payload size and low power consumption.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transmission of biomedical signals through wireless body area sensor (WBAN) technology promises to offer efficient real-time health monitoring services and automated diagnostic procedures. The collected biomedical signals over a long period of time require a large amount of storage capabilities. Thus, compression is introduced to reduce the storage space and transmission cost. At the same time, the critical diagnostic information and signal quality need to be preserved while compression. The Electrocardiogram (ECG) signal is a commonly monitored biomedical signal that assesses the heart condition. From the ECG signal, cardiologists can detect arrhythmias that may reveal the existence of some cardiac diseases. Since cardiologists need huge amount of ECG data for diagnosis, this ECG data have to be compressed while storage [1].
In case of remote monitoring through wireless transmission using portable devices, compression is necessary to reduce the payload packets, transmission rate and power consumed by the RF antenna [2, 3]. Some of the current hardware and software technologies used for WBAN nodes might introduce some limitations in the implementation of complex compression algorithms, which contain multiplication and complex math functions like logarithm (log) and square-root (sqrt). Therefore, the algorithm should be designed with low complexity to reduce the computational power since the sensors are powered by low-power batteries.
Discrete Wavelet Transform (DWT) method is widely employed for biomedical signal analysis due its powerful capabilities in analyzing signals in both time and frequency domains [4,5,6,7,8,9,10]. The DWT decomposes the original signal into subband coefficients that represent the measure of similarity in frequency content between the signal and the chosen wavelet function [11]. Decomposing the ECG signals into subband coefficients allows high separation between the noise and the dominant ECG morphology like the QRS waveform. In addition, in subbands the ECG’s iso-electric pauses and redundant data are clearly visible. Thus, their statistical redundancy can be highly encoded without losing significant diagnostic information. The compression procedure is conducted on these subband coefficients using different coding techniques such as: the set partitioning in hierarchical tree (SPIHT) coding [6, 12], vector quantization (VQ) [13], construction of the classified signature and envelope vector sets (CSEVS) [14] and energy package efficiency (EPE) [15]. These DWT-based compression techniques achieved high compression performance compared to other methods that code the redundancy of the original time signal.
The main challenges associated with real-time ECG compression methods are: the quality of the reconstructed signal, the compression ratio (CR), the execution time and computational power. The CR is an important factor to transmit the data as fast as possible [16]. Some of the encoding methods can be complex to implement on FPGA’s or basic microcontrollers since they require high computational costs and power. Thus, Chan et al. proposed a simple and forward encoding scheme to compress the DWT coefficients using bit-field preserving (BFP) and running length encoding (RLE) algorithms [7], and it was tested and validated on an FPGA system [17]. The DWT-BFP-RLE method is simple to implement since it allows forward data processing compared to other methods that require sorting and heavy computations.
For critical-time medical applications, the payload packets of the telecardiology systems are typically designed with small sizes of 40 to 60 bytes to increase the delivery rate [18]. In practice, to fit the ECG segment into a payload packet, it is compressed at the expense of losing some diagnostic information. In fact, the error and CR are in trade-off relationship i.e., increasing the CR results in higher error. Thus, the designer of the WBAN system must account for the duration (or number of samples) taken during transmission, because huge number of ECG samples requires higher CR to fit into the maximum payload available. In some work in literature [4, 19, 20], the error is controlled by a closed loop to ensure that the reconstructed signal can be used for clinical evaluation. The closed loop involves manipulation of some compression parameters to attain the payload constrain. Generally, these methods simply attain the size of the compressed segment by varying the compression parameters (which compromise on the quality of the reconstructed signal) to push it into the maximum payload available.
By reviewing literature, effective solutions were not found yet for the issue of controlling the size of compressed ECG data while retaining the clinical quality. Thus, we were motivated to design a new ECG compression scheme, based on DWT-BFP-RLE compression algorithm [7], which ensures that the compressed segments fit into the available payload packet while maintaining the CR to preserve the diagnostic information. By considering the size of the payload packet this method overcomes the limitations of the other methods. The proposed method automatically divides the ECG block of size N s into segments while fixing the other compression parameters. For validation, ECG records from MIT-BIH Arrhythmia Database are used.
This paper is organized as follows: first, “Methods” describes the discrete wavelet transform based compression method, and “Proposed dynamic compression scheme” introduces the proposed dynamic compression scheme for filling the payload packets. “Evaluation scheme” describes the performance matrices and datasets used to validate the method and then “Experimental results and discussion” presents the results and discusses the outcomes. Finally, “Conclusions” draws the conclusions and discusses the future work.
Methods
Discrete wavelet transform
Wavelet transform (WT) is a signal processing tool that represents the signal in both time and frequency domains. WT provides a good localization resolution and is suitable for the analysis of non-stationary signals like ECGs [21]. The discrete wavelet transform (DWT) is a special form of the continuous wavelet transform (CWT) which uses discrete wavelet functions [22].
The CWT is the correlation between a wavelet function ψ a,b (t) and a time domain signal f(t) given by:
where a is the scaling coefficient and b is the translation coefficient, and both are real continuous numbers with a > 0 and b ≥ 0. The wavelet function is defined as:
The DWT removes the redundancy of CWT by discretizing the wavelet coefficients as
where a 0 > 1, b 0 > 0, (m,n) ∈ Z, where n represents the discrete time index. Usually, a 0 is set to 2 and b 0 to 1, to have a dyadic grid function which produces dyadic (hyperbolic) grids instead of linear grids [11]. Dyadic grids are the simplest and most efficient way of discretization. Consequently, Eq. 2 can be written as
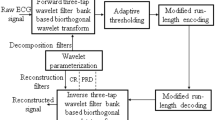
This DWT equation can be implemented efficiently using Mallat implementation algorithm [22], which is based on orthogonal and biorthogonal filter banks. It decomposes the signal into subbands using two types of finite impulse response (FIR) filter arrangements; high-pass filters with coefficients h(n) and low-pass filters with coefficients g(n). The high-pass filter produces the wavelet detail coefficients d n and the low-pass filter produces the wavelet approximation coefficients a n , where n = 1,2,..J, and J is the highest decomposition level as shown in Fig. 1.
Using the low-pass filter, the approximation coefficient a 1 can be further decomposed into two subbands in the next decomposition level and so on up to d J and a J [23]. A down-sampling operation by a factor of 2 (↓2) is applied after each filter.
The subband coefficients resulted from the filter bank structure are further encoded. The lower subband coefficients of the ECG signal contain most of the energy spectrum, hence more bits have to be preserved in these subbands. On the other hand, since the higher subband coefficients have more noise-like signals which are not important, less bits are preserved and encoded. As a result, this encoding technique can achieve high compression performance. The original signal can be reconstructed from the encoded subband coefficients using reconstruction filters, which are mirrors of the decomposition filters. This type of filters is called quadrature mirror filters (QMF) [24,25,26]. In this work, the commonly used filter banks for ECG analysis such as Daubechies (Db) [27], Biorthogonal (bior), Coiflet (coif), Symmlet (sym), Morlet (morl) are used.
Compression based on DWT and thresholding
ECG decomposition using wavelet filter banks
Decomposition levels 4 to 6 are commonly used in DWT-based ECG compression methods since most of the ECG energy is confined to these bands [8]. Figure 2 shows a decomposed ECG signal using J = 5. It is clear that the first three subbands (d 1, d 2 and d 3) contain noise and high-frequency ECG morphologies, like QRS complex peaks. The two remaining subbands contain low-frequency ECG components such as P, T and U waveforms. Thus, using wavelet functions that have similar morphology to the QRS complex, such as Daubechies (Db) wavelet, minute details of ECG signals can be preserved while only keeping a small number of coefficients, leading to an efficient coding [28, 29]. Generally, the compression performance is highly dependent on the morphology of the mother wavelet and the ECG information of interest.
In this work, the mean of the approximation coefficient a 4 is subtracted before threshodling and encoding, and then it is added later on at the reconstruction stage. This step is similar to removing the mean of the original signal before decomposition, but it is much faster in case of using FPGAs or microcontrollers due to the less number of samples in a 4 compared to the original signal.
Thresholding and bit-preserving
After decomposing the signal into subband coefficients using DWT, the signal part which has low power will produce very low coefficient values. These insignificant coefficients can be set to zeros without losing significant information [30]. A threshold value (λ) is calculated to define the level of the insignificant coefficients. This threshold is obtained either globally for all of the subbands or locally for each subband. The local, or adaptive, threshold showed better results compared to the global threshold [31].
One of the commonly used adaptive thresholds is provided in Eq. 5, which is proposed by [32].
where, σ is the standard deviation of the signal’s noise and \(N_{d_{n}}\) is the number of samples in the n th subband (d n ). However, this standard deviation computation method is generally used for offline methods since it requires post-processing of the whole signal [33]. Thus, it is not adapted for real-time algorithms.
In [7], Chan et al. proposed a new thresholding technique based on the bit-field preserving scheme. This scheme depends on the number of bits of interest (BOI) in the significant coefficients. BOI can be any stream of bits of length I S b between bit 0 to M-1. Preserving few bits from the coefficients will introduce a truncation error while decompression/reconstruction at the expense of increasing the CR. Thus, the bit-field preserving (BFP) scheme is used to reduce the truncation error by preserving most of the bits. BFP involves adding a rounding coefficient C r o u n d to each subband coefficient. In this work, thresholding and BFP are performed as follows:
-
i.
Find the bit-depth value of each subband (B S b ), which is the most signicant bit of the maximum coefficient in the subband, B S b = M S B(|m a x(d n )|).
-
ii.
Select the desired preserved bit-length (I S b ) for each subband coefficient.
-
iii.
Calculate the rounding coefficient as \(C_{round} = 2^{B_{Sb}-I_{Sb}}\).
-
iv.
Apply a round-off mechanism by adding C r o u n d to each coefficient in the subband.
-
v.
Calculate the new bit-depth (B n ). If the new B n is greater than current B S b , the B S b is updated as B n .
-
vi.
Calculate the new subbands encoding thresholds as \(\lambda _{Sb} = 2^{B_{n}-I_{Sb}+1}\).
-
vi.
Apply the encoding thresholds λ S b to each subband coefficient by setting the insignificant coefficients, less than the absolute value of threshold, to zeros (Fig. 2).
-
viii.
For each significant coefficient, greater than λ S b , extract the BOI, BOI length and B n .
More detailed description of the BFP technique and the round-off mechanism is found in [7, 17]. In this work, the BFP technique accounts for the sign of the coefficients as follows
The compression performance is controlled by the preserved-lengths I S b , where S b stands for the subband coefficients d 1, d 2,.., d J and a J . As shown from Fig. 3a, BOI are the bits from B S b + 1 to B S b − I S b + 1 and I S b is the length of these bits. In this works, each BOI is stored into one byte and the same for BOI range. Thus, I S b is no more than 7 bits (i.e. bits 0 to 6 hold the extracted bits and bit 7 for the sign bit). The negative coefficient is treated as the positive coefficient by taking the absolute value, but the sign bit will be set to 1.
Encoding and data mapping
The encoding step is conducted in three steps. First, for each significant coefficient, greater or equal to λ S b , a one (1) is sent to the significant map (SM) and the BOI including the sign bit at the (B S b + 1)th are sent to the BOI packet. On the other hand, for each insignicant coefficient a zero (0) is sent to the SM but no BOI are extracted. Thus, the generated SM packet will hold 1’s and 0’s that indicate the order of significant and insignificant bytes in each subband, respectively. Second, each 8 bits from the SM are bundled as one byte. Thus, each subband with N samples will have N/8 SM packets.
To illustrate the thresholding, BFP and SM bits generation step, Fig. 4 shows an example of these steps on one of the decomposed ECG subbands. The C r o u n d is calculated from the maximum absolute coefficient to conduct the BFP step and then λ S b is calculated after the round-off procedure. The reconstructed subband shows the amount of change in each coefficient and the restored significant coefficients. In addition, Fig. 4 shows the SM values corresponding to each coefficient along with the final SM byte representation. Since SM is holding a lot of redundant 0’s, it can be further compressed using running-length encoding (RLE) scheme and this is the third step in the encoding process. RLE replaces the consecutive zero bytes with only two bytes; a byte with a zero value and a byte representing the number of consecutive zeros, i.e. SM bytes= [1 0 0 0 0 0 7 0 0 3 0 0 0 2 0], can be encoded as SMe= [1 0 5 7 0 2 3 0 3 2 0 1]. Usually, the last two subbands (a J and d J ) have fewer samples and less consecutive zeros, and thus RLE method is not applied to their SM bytes.
Packetization
Before transmission, the encoded and extracted data have to be packetized in a clear format which allows fast reconstruction at the receiver side. The final packetized compressed segment is illustrated Fig. 3b. The final compressed packet holds the headers, BOI, SM and the mean of a J . Table 1 shows the sizes of the packets in the compressed segment which holds the following information in sequence:
-
i.
The Number of Samples (N s ): Indicates the total number of ECG samples taken for compression. To point out for 32, 64, 128, 256, 512 and 1024 ECG samples, the N s value is set to 0, 1, 2, 3, 4, or 5, respectively.
-
ii.
The Range of BOI (BOI Range): indicates the range of I S b of the subband. The 4 MSB holds the low bit range (B S b + 1) and the 4 LSB holds the high bit range (B S b − I S b + 1).
-
iii.
The Number of BOI (BOI Size): indicates the number BOI of the significant coefficients in bytes.
-
iv.
Bits-of-Interests (BOI): holds the BOI of the subband.
-
v.
The size of SMe or SM (SMe/SM Size): the number of bytes that holds the significant map or the encoded significant map.
-
vi.
Significant Map Stream in Bytes (SMe/SM): holds the SM and SMe bytes of the subband.
-
vii.
The Mean of a J (Mean): the subtracted mean of the approximation subband a J .
We assume that the packets arrive in sequence at the receiver side and ready for decompression and decoding. The packets’ error detection task is conducted by the communication link, which adds the cyclic redundancy check (CRC) protocol to the packets before transmission [34]. To decompress the packets, only the type of the wavelet filter and level of decomposition have to be provided, while the other information are retrieved from the compressed packets.
Wavelet selection to avoid signal distortion
The present study addresses the issue of the limited payload size in the telecardiology systems and how to overcome this issue by dividing the ECG data into smaller segments. Hence, the selected wavelet function has to count for the case of small segments. The length of the wavelet filter (L) has to be no more than twice the length of the data segment (N s ). Otherwise, the signal will be distorted at the edges [35, 36]. In the case of the decomposed subbands, it is enough to check the length of the last decomposed subband, \(N_{d_{J}}\) or \(N_{a_{J}}\). This criterion can be simply expressed as
where, \(N_{d_{J}}\) is equal to N s /2J and it corresponds to the length of the last decomposed subband coefficient using J decomposition levels.
For example, if the data length is N s = 64 and the decomposition level J = 4. Then, the signal will be decomposed into 5 subband coefficients C = [d 1,d 2,d 3,d 4,a 4] of lengths \(N_{d_{n}} =\left [ 32, 16, 8, 4, 4 \right ]\). Since \(N_{d_{4}}\) is equal to 4, then the selected wavelet filter has to be of length L ≤ 2 × 4 = 8. Db4 with L = 8 is suitable in this case but daubechies with higher order will cause distortion to the signal, although increasing the order of the FIR filter increases the efficiency of the filtering scheme.
We partially solve the issue of distortion by initializing the length of the subband coefficient as \(N_{d_{j}}+L\) but mainly we try not to use the wavelet filters with \(L > 2 N_{d_{J}}\).
Proposed dynamic compression scheme
In this study, the DWT-BFP-RLE compression algorithm described in “Compression based on DWT and thresholding” has been adopted and modified for the portable mobile telecardiology systems. Since, the new scheme tackles the case of the limited payload size, it is endowed with automatic mechanism that controls the size of the compressed segments. The modified scheme is called “DWT Dynamic Compression Scheme”. It dynamically checks whether the size of the compressed segment fits into the available payload before transmission. The main objective of this scheme is to maintain the compression level instead of increasing it and losing the diagnostic information. Figure 5 shows the general structure of the proposed dynamic compression scheme. The scheme is summarized in the following steps:
-
Step 1.
Store N s = 2n ECG samples into a buffer block. Compress the samples using DWT-BFP-RLE compression algorithm and packetize them as described in “Packetization”.
-
Step 2.
Check the size of the compressed segment. If the size of the compressed segment is less than or equal to the maximum allowable number of bytes (M), transmit the segment. Note that in case the compressed segment is < M, zeros are padded to the segment to fill the payload.
-
Step 3.
Otherwise, if the compressed segment > M, divide the uncompressed ECG samples stored in the buffer into two new segments each with size N 2 = N s /2.
-
Step 4.
Check the size of the new segments N 2 before compression; if the size of samples (2 × N 2 Bytes) is less than M bytes, packetize and send them without compression by adding one header which is N s = 0. The zero value indicates that the data was not compressed and it does not require decompression at the receiver side.
-
Step 5.
Otherwise, apply the compression algorithm onto each segment separately, but in the correct sequence, where the first half of the data is to be compressed and transmitted first.
-
Step 6.
Go back to step 2 and repeat the process onto the new divided segments until all of the data are transmitted.
The total number of generated compressed packets depends on the I S b values and the selected wavelet. Using these two parameters we try to compromise between the compression performance and the total number of packets generated.
According to wavelet selection criteria in “Wavelet selection to avoid signal distortion”, this scheme is modified to check the competence of the wavelet with the desired decomposition level J and the size of the ECG block N s .
Evaluation scheme
Dataset
To validate the proposed scheme, ECG records from MIT-BIH Arrhythmia Database (MITDB) are used [37]. This database contains 48 half an hour two-channel ambulatory ECG recordings, obtained from 47 subjects studied by the BIH Arrhythmia Laboratory. The records are digitally sampled at 360 Hz using 11-bit resolution over a ± 5m V range, which means that the bit-length is I E C G = 11. To prepare the ECG signal for compression, the data of range ± 5m V was mapped to its raw ADC format of -1024 to 1023 to imitate the format of the packets in the real hardware implementation (microcontrollers or FPGAs).
Performance metrices
To test the performance of the proposed compression algorithm, three standard measures are adopted; the compression ratio, the error or distortion amount between the original and the reconstructed signal, and the quality of the algorithm. The compression ratio (CR) is defined as the ratio between the number of bits representing the original signal and the number of bits of the compressed packet. CR is calculated as follows:
where N s is the block size or the number of ECG samples taken for compression and I E C G is the number of bits in each ECG sample and it is equal to 11. N C o m p represents the number of bytes in the compressed segment and they are multiplied by 8 to get the total number of bits in them.
The percentage root-mean square difference (PRD) is the most commonly used error measure in literature [1]. PRD represents the distortion amount between the original ECG signal x o r g and the reconstructed/decompressed signal x r e c and it is defined by:
where N s denotes the block size. The mean of x o r g and x r e c is removed since it can mask the real performance of the algorithm [38]. To show the correlation between the compression performance and the amount of distortion, a quality score (QS) measure was introduced by [39]. Practically, QS is the ratio between CR and PRD as given by Eq. 9 and it represents the quality of the method.
The higher the quality score, the better is the compression performance.
In order to evaluate the dynamic compression scheme, we introduced a new measure, called the packet reduction (PR) determined by Eq. 10, which measures the percentage amount of reduction in the number of generated packets while compression.
where, N R a w is the number of raw transmitted packets in case of no compression and N C o m p r e s s i o n is the number of compressed packets generated by the dynamic compression scheme for the same number of the raw samples. Since each sample has a resolution of 11-bits, which requires 2 bytes to hold it, then N R a w = 2 × N s .
Experimental results and discussion
Before running the proposed dynamic compression scheme, the modified DWT-BFP-RLE compression algorithm is validated using four different decomposition levels (J = 3 to 6) and five different commonly used mother wavelet functions (Db4, Db5, sym4, sym6 and bior4.4). Then, the compression parameters are selected to validate the dynamic compression scheme. The preserved lengths I S b are selected to be short for the higher subbands to reduce the noise level and to pick up the important coefficients only. While for the lower subbands I S b lengths are set to be large up to 6 for a J to preserve the bits of the important coefficients as much as possible. Thus, \(I_{d_{1}}\), \(I_{d_{2}}\), \(I_{d_{3}},...I_{d_{J}}\) and \(I_{a_{J}}\)are set to: [1, 2, 3, 6], [1, 2, 3, 4, 6], [1, 2, 3, 4, 5, 6] and [1, 2, 3, 4, 5, 5, 6] for the decomposition levels of 3, 4, 5 and 6, respectively.
Before conducting the compression algorithm, the whole records of MITDB are divided into blocks of size 1024. This means that the size of the final subband coefficient \(N_{d_{6}}\)= 16 samples. According to “Wavelet selection to avoid signal distortion”, the wavelet filters Db5, sym6 and bior4.4 with L = 10, 12 and 10, will distort the signal. To avoid this distortion, we have studied their behavior until J = 5.
To obtain the average compression results, the compression algorithm is performed on the sub-blocks first. Second, the average performance matrices (CR and PRD) of each record are obtained as the blocks averages. Finally, The total average of the database is calculated from the records’ averages. Figure 6a illustrates the average CRs corresponding to different wavelets and J values. It is clear that sym4 and bior4.4 wavelets give the highest CR results followed by sym6 and Db4, while Db5 shows the lowest CR. Sym4 and bior4.4’s CRs range between 6.86:1 and 8.81:1. In general, the highest CRs are achieved by the 5th decomposition level (J = 5). The trend of the CR curves indicates that between J = 4 to 6, the values are close to each other, while for J = 3 the CR values are low because only two subbands are encoded which are d 1 and d 2.
From Fig. 6b it is quite clear that the PRD error increases steadily with the decomposition level. This means that the more subbands are encoded, the more detail coefficients are discarded in the higher levels. Bior4.4 achieved the lowest PRD values of 1.14%, 1.25% and 1.29% at J = 3-5. In general, the wavelets maintained close PRD values at each decomposition level.
To select the best wavelet and decomposition level, the QS measure is obtained as illustrated in Fig. 7. Based on the QS results, bior4.4 shows the highest QS values and the peak values are 7.60 and 7.59 at J = 4 and 5, respectively. It is also noted that sym4 gives very close QS performance to bior4.4 and the peak values are 7.54 and 7.49 at J = 4 and 5 as well. Therefore, we decided to adopt both bior4.4 and sym4 wavelets with J = 4 to study our proposed dynamic compression scheme described in “Proposed dynamic compression scheme”.
Different preserved lengths were also studied in this work and it was concluded that increasing the length of the preserved lengths increases the quality of the reconstructed signal but decreases the CR. On the other hand, setting short preserved lengths increases the CR but decreases the quality of the signal. The effect of changing the preserved lengths on the first 1024 samples of the compressed signal is shown in Fig. 8, where the signal is mostly preserved in the first and second case with smoothed low-level noise. In the third case, the signal lost some details and some peaks were attenuated due to the very low preserved lengths.
Comparison to other methods
Previous works in literature took the first 10 minutes of the MITDB record 117 for evaluation [6]. Here we adopted the same record and the block size was set to N s = 1024. A comparison between the proposed method and other developed methods is given in Table 2. The comparison results show that modified DWT-BFP-RLE scheme outperforms the other developed methods by achieving small PRD errors with relatively the same CRs. The QS results validate the good compression performance and low reconstruction error compared to the other methods.
Performance of dynamic compression scheme
To validate the dynamic compression scheme, it is assumed that the maximum available payload size (M) is 70 bytes, which imitates the experimental setup proposed in reference [42]. Thus, according to Eq. 7, the CR should not be less than 1.26:1, 2.51:1, 5.03:1, 10.06:1 and 20.11:1 for N s = 64, 128, 256, 512 and 1024, respectively. Moreover, our main objective is to compromise between the compression performance and the number of generated compressed packets N C o m p r e s s i o n . Practically, we are looking for the proper wavelet function, I S b combination and ECG block length N s which are going to produce the highest QS values with less number of compressed segments N C o m p r e s s i o n .
According to the literature and our experimental studies, different values of N s result in different values of QS and N C o m p r e s s i o n using the same I S b as shown in Fig. 9. This implies that the optimal N s value can not be selected based on the values of QS and N C o m p r e s s i o n . Accordingly, the size of N s is selected based on the desired CR. In this study we assumed that the CR should be with an average value of 5.00:1 and thus N s = 256 samples (C R = (N s × 11)/(8 × 70) = 5→N s = 254.5 ≈ 256). Using J= 4, the bior4.4 filter might introduce some distortion in segments with sizes less than 256. Thus, sym4 is preferred in this case but the results of bior4.4 will be presented to give a clear view about their effect.
Using MITDB, 8 ECG signals, records 100, 102, 104, 109, 111, 117, 119 and 207, were selected for performance evaluation. To mimic the operation of real-time monitoring in telecardiology systems, it is assumed that continuous 5 minutes of ECG signal is transmitted and this signal has to be divided, compressed and packetized before transmission. The number of packets required to send 5 minutes of raw ECG data (un-compressed) using a payload of of size 70 bytes is N R a w ≈ 3,086 packets (5 minutes × 60 seconds × 360 Hz × 2 Bytes / 70 Bytes), since each ECG sample is represented by 2 bytes.
The dynamic compression scheme, described in “Proposed dynamic compression scheme”, was applied on the ECG records using both sym4 and bior4.4 mother wavelets with J = 4 levels and using six different I S b combinations: [1, 2, 4, 5, 6], [2, 3, 3, 4, 6], [2, 3, 3, 5, 6], [3, 3, 3, 5, 6], [2, 4, 4, 5, 6] and [3, 4, 4, 5, 6]. Table 3 shows the average evaluation results of the 8 ECG records. From the results we can conclude that N C o m p r e s s i o n is inversely related to CR. Thus, sym4 with lower preserved lengths at I S b = [1, 2, 4, 5, 6] achieves the highest CR value of 4.89:1 and the lowest N C o m p r e s s i o n value of 524 packets, while it achieves 4.88:1 CR and 528 N C o m p r e s s i o n for bior4.4. In addition, since PR is a function of N C o m p r e s s i o n , it is highest for the lowest N C o m p r e s s i o n . Figures 10, 11, and 12 show the evaluation results of each ECG record.
From Fig. 10, it is clear that QS at I S b = [1, 2, 4, 5, 6] gives better quality score compared to the other I S b combinations, especially for bior4.4. However, sym4 achieves better overall QS performance. At I S b = [2, 3, 3, 5, 6], sym4 achieves high QS compared to I S b = [2, 3, 3, 4, 6] and [3, 3, 3, 5, 6]. For low I S b combination, CR go up to 6.00:1 for both sym4 and bior4.4 as shown in Fig. 11a. From Fig. 11b, sym4 shows lower PRD results compared to bior4.4 and it has a linear decreasing trend as I S b values increases. The maximum PRD is less than 1.00% and it reaches a minimum value of 0.28%.
From Fig. 12a, we can see that N C o m p r e s s i o n is inversely related to CR for all of the ECG records studied. PR in Fig. 12b ranges from 85.39% to 59.85% at 5.91:1 and 2.16:1 CR, respectively. These PR values are sufficient enough to reduce the power consumed in the real hardware systems. Also, the dynamic compression scheme generated less number of packets ranging between 461 and 1239 packets compared to the raw data which require 3,086 packets. This reduction boosts the battery’s life of the portable telecardiology devices by reducing the power consumed using transmission. However, the user of the scheme have to seek a good trade-off between the number of compressed packets and the compression performance.
To reveal the visual quality of the dynamic compression scheme, the original ECG signal and the reconstructed signal using different I S b combination obtained with sym4 and bior4.4 for MITDB records 100, 117 and 207 are shown in Figs. 13, 14, 15, 16 and 17. Figure 13 demonstrates the results of the first 1,536 samples (4.26 seconds) of MITDB record 117 using I S b = [2, 3, 3, 5, 6]. The lines in the plots indicates how the signal was divided into packets, where it is clear that the scheme generated 9 and 8 compressed packets using bior4.4 and sym4, respectively. In fact, we need 44 packets each with 70 bytes payload to transmit raw 1,536 samples, which means that we reduced the amount of packets by 20.45% and 18.18%, and the PR achieved 79.45% and 81.81% using bior4.4 and sym4, respectively. Furthermore, the average and standard deviation of CR and the total PRD (at the right side of the plots) show that sym4 gives better compression performance.
The results of using different I S b combination on the same ECG record is shown in Fig. 14. I S b = [3, 3, 3, 5, 6] resulted in N C o m p r e s s i o n = 7 packets for both bior4.4 and sym4. Although bior4.4 produces higher average CR than sym4, sym4 produces lower PRD value. The same procedure is conducted on MITDB record 100 and the results are clear in Figs. 15 and 16, but some of the generated packets are distorted at the edges in case of bior4.4 wavelet, while sym4 maintained the quality of the signal. This distortion is also clear in Fig. 17 on MITDB record 207 using I S b = [2, 4, 4, 5, 6].
The introduced distortion in the reconstructed packets, indicated by the red dashed circles, is because of the small size of the ECG segment, N = 64, compared to the size of the wavelet filter, which is L = 10 for bior4.4. In fact, the I S b combination selected in Figs. 16 and 17 generated a lot of packets, up to 18 N C o m p r e s s i o n , with few number of samples and from it the distortion was generated. The distortion can lead to losing some diagnostic information and introducing extra artifacts. Therefore, from the demonstration results it is recommended to adopt wavelet filters with suitable lengths when implementing the proposed DWT dynamic compression scheme to preserve the diagnostic information.
Conclusions
In this work, we introduced a novel ECG compression scheme that considers the limited size of the payload packets in low power portable telecardiology systems. The scheme controls the length of the compressed block dynamically while maintaining the CR and the diagnostic information. It is based on the DWT method, which decomposes the signal into subband coefficients, RLE scheme to encode the data and BFP method to preserve the coefficients before encoding. For efficient transmission, a packetization scheme for the compressed data was designed to minimize the headers and increase the space for compressed data.
The proposed scheme achieved expectation by its ability of controlling the size of the compressed packets dynamically, by checking if the compressed packet is exceeding the maximum available payload. The scheme automatically divides the ECG block of samples into smaller segments while preserving the CR instead of increasing it, where increasing it might increase the residual error in the reconstructed signal. Experimental results obtained from using the MIT-BIH Arrhythmia Database showed that the efficiency of the system can be highly increased by reducing the number of packets generated. In addition, the obtained results showed the superiority of the proposed scheme by achieving a packet reduction up to 85.39% at PRD values less than 1%. Moreover, it was shown that the length of the wavelet filter is crucial in case of compressing small segments to avoid distortion in the reconstructed signal. Nevertheless, further improvement can be done on this method to achieve higher CR and QS. Our future direction is to implement the method on ultra-low power hardware since the initial indication shows promising results.
References
Jalaleddine, S., Hutchens, C.G., Strattan, R.D., and Coberly, W.A., ECG Data compression techniques-a unified approach. IEEE Trans. Biomed. Eng. 37(4):329–343, 1990.
Yazicioglu, R.F., Torfs, T. , Penders, J., Romero, I., Kim, H., Merken, P., Gyselinckx, B., Yoo, H.-J., and Van Hoof, C. : In: Engineering in Medicine and Biology Society, 2009. EMBC 2009 Annual International Conference of the IEEE, IEEE, Piscataway, pp 3205–3208, 2009
Marcelloni, F., and Vecchio, M., An efficient lossless compression algorithm for tiny nodes of monitoring wireless sensor networks. Comput. J. 52(8):969–987, 2009.
Djohan, A., Nguyen, T.Q., and Tompkins, W.J.: In: Engineering in Medicine and Biology Society, 1995., IEEE 17th Annual Conference, vol 1, IEEE, Piscataway, pp 167–168 , 1995
Liu, J.-H., Hung, K.-C., and Wu, T.-C., Ecg compression using non-recursive wavelet transform with quality control. Int. J. Electron. 103(9):1550–1565, 2016.
Lu, Z., Kim, D.Y., and Pearlman, W.A., Wavelet compression of ECG signals by the set partitioning in hierarchical trees algorithm. IEEE Trans. Biomed. Eng. 47(7):849–856, 2000.
Chan, H.-L., Siao, Y.-C., Chen, S.-W., and Yu, S.-F., Wavelet-based ECG compression by bit-field preserving and running length encoding. Comput. Methods Prog. Biomed. 90(1):1–8, 2008.
Manikandan, M.S., and Dandapat, S., Wavelet-based electrocardiogram signal compression methods and their performances: a prospective review. Biomed. Signal Process. Control 14:73–107, 2014.
Jayachandran, E. S., P. J. K., and R. A. U., Analysis of myocardial infarction using discrete wavelet transform. J. Med. Syst. 34(6):985–992, 2010. https://doi.org/10.1007/s10916-009-9314-5.
Bessmeltcev, V., and Katasonov, D., Application of discrete wavelet transform with changing presentation of coefficients in data compression in mobile ecg monitoring systems. Biomed. Eng. 50(1):35–39, 2016.
Addison, P.S., The illustrated wavelet transform handbook: introductory theory and applications in science, engineering, medicine and finance. Boca Raton: CRC press, 2002.
Rajankar, S., and Talbar, S., A quality-on-demand electrocardiogram signal compression using modified set partitioning in hierarchical tree. SIViP 10(8):1559–1566, 2016.
Sahraeian, S., and Fatemizadeh, E.: Wavelet-based 2-D ECG data compression method using SPIHT and VQ coding. In: EUROCON The International Conference on Computer as a Tool, IEEE Piscataway, pp 133–137, 2007
Gurkan, H., Compression of ECG signals using variable-length classifıed vector sets and wavelet transforms. EURASIP Journal on Advances in Signal Processing 2012(1):1–17, 2012.
Allen, V.A., and Belina, J.: ECG Data compression using the discrete cosine transform (DCT). In: Proceedings of Computers in Cardiology 1992, IEEE, Piscataway, pp 687–690, 1992
Cho, G.-Y., Lee, S.-J., and Lee, T.-R., An optimized compression algorithm for real-time ecg data transmission in wireless network of medical information systems. J. Med. Syst. 39(1):1–8, 2015.
Lee, H.-W., Hung, K.-C., Wu, T.-C., and Ku, C.-T., A modified run-length coding towards the realization of a RRO-NRDPWT-based ecg data compression system. EURASIP Journal on Advances in Signal Processing 2011 (1):1–8, 2011.
Liang, X., and Balasingham, I.: Performance analysis of the ieee 802.15. 4 based ECG monitoring network. In: Proceedings of the 7th IASTED International Conferences on Wireless and Optical Communications, pp. 99–104, 2007
Benzid, R., Messaoudi, A., and Boussaad, A., Constrained ecg compression algorithm using the block-based discrete cosine transform. Digital Signal Process. 18(1):56–64, 2008.
Blanchett, T., Kember, G., and Fenton, G., KLT-Based quality controlled compression of single-lead ecg. IEEE Trans. Biomed. Eng. 45(7):942–945, 1998.
Ahmed, S.M., Al-Shrouf, A., and Abo-Zahhad, M., ECG Data compression using optimal non-orthogonal wavelet transform. Med. Eng. Phys. 22(1):39–46, 2000.
Mallat, S.G., A theory for multiresolution signal decomposition: the wavelet representation. IEEE Trans. Pattern Anal. Mach. Intell. 11(7):674–693, 1989.
Abbate, A., DeCusatis, C., and Das, P.K., Wavelets and subbands: fundamentals and applications. 1st edn. Berlin: Springer Science & Business Media, 2002.
Croisier, A., Esteban, D., and Galand, C.: Perfect channel splitting by use of interpolation/decimation/tree decomposition techniques. In: International Conference on Information Sciences and Systems, vol 2, Patras, Greece, pp 443–446, 1976
Vaidyanathan, P.P., Multirate digital filters, filter banks, polyphase networks, and applications: a tutorial. Proc. IEEE 78(1):56–93, 1990.
Vaidyanathan, P.P., Multirate systems and filter banks. India: Pearson Education, 1993.
Daubechies, I., Orthonormal bases of compactly supported wavelets. Commun. Pure Appl. Math. 41:909–996, 1988.
Merah, M., Abdelmalik, T., and Larbi, B., R-peaks detection based on stationary wavelet transform. Comput. Methods Prog. Biomed. 121(3):149–160, 2015.
Boutaa, M., Bereksi-Reguig, F., and Debbal, S., ECG Signal processing using multiresolution analysis. J. Med. Eng. Technol. 32(6):466–478, 2008.
Abo-Zahhad, M., Al-Ajlouni, A.F., Ahmed, S.M., and Schilling, R.J., A new algorithm for the compression of ECG signals based on mother wavelet parameterization and best-threshold levels selection. Digital Signal Process. 23(3):1002–1011, 2013.
Poornachandra, S., Wavelet-based denoising using subband dependent threshold for ECG signals. Digital signal process. 18(1):49–55, 2008.
Donoho, D.L. , and Johnstone, I.M. : Threshold selection for wavelet shrinkage of noisy data. In: Engineering in Medicine and Biology Society, 1994. Engineering Advances: New Opportunities for Biomedical Engineers. Proceedings of the 16th Annual International Conference of the IEEE Piscataway, pp. A24–A25, 1994
Quotb, A., Bornat, Y., and Renaud, S., Wavelet transform for real-time detection of action potentials in neural signals. Frontiers in Neuroengineering 4(7):10, 2011.
Alesanco, A., Istepanian, R., and Garcia, J.: The effects of transmission errors in ECG real-timewavelet compression codecs, In: Computers in Cardiology, 2005, IEEE, Piscataway, pp 45–48 , 2005
Déprez, F., Rioul, O., and Duhamel, P.: Border recovery for subband processing of finite-length signals. application to time-varying filter banks. In: 1994 IEEE International Conference on Acoustics, Speech, and Signal Processing, 1994. ICASSP-94, vol 3, IEEE, Piscataway, pp III133, 1994
Jensen, A., and la Cour-Harbo, A., Ripples in mathematics: the discrete wavelet transform. Berlin: Springer Science & Business Media, 2001.
Goldberger, A.L., Amaral, L.A., Glass, L., Hausdorff, J.M., Ivanov, P.C., Mark, R.G., Mietus, J.E., Moody, G.B., Peng, C.-K., and Stanley, H.E., Physiobank, physiotoolkit, and physionet components of a new research resource for complex physiologic signals. Circulation 101(23):e215–e220, 2000.
Blanco-Velasco, M., Cruz-Roldán, F., Godino-Llorente, J.I., Blanco-Velasco, J., Armiens-Aparicio, C., and López-Ferreras, F., On the use of prd and cr parameters for ecg compression. Med. Eng. Phys. 27(9): 798–802, 2005.
Fira, C.M., and Goras, L., An ECG signals compression method and its validation using NNs. IEEE Trans. Biomed. Eng. 55(4):1319–1326, 2008.
Hilton, M.L., Wavelet and wavelet packet compression of electrocardiograms. IEEE Trans. Biomed. Eng. 44 (5):394–402, 1997.
Al-Shrouf, A., Abo-Zahhad, M., and Ahmed, S.M., A novel compression algorithm for electrocardiogram signals based on the linear prediction of the wavelet coefficients. Digital Signal Process. 13(4):604–622, 2003.
Touati, F., Erdene-Ochir, O., Mehmood, W., Hassan, A., Mnaouer, A.B., Gaabab, B., Rasid, M.F.A., and Khriji, L.: An experimental performance evaluation and compatibility study of the bluetooth low energy based platform for ECG monitoring in wbans. Int. J. Distrib. Sens. Netw. 2015:9, 2015
Acknowledgements
Authors would like to express sincere appreciation to Qatar National Research Fund ”NPRP Grant #4-1207-2-474“. The in-kind support of Sultan Qaboos University is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors certify that they have NO conflict of interest in the subject matter or materials discussed in this manuscript.
Additional information
This article is part of the Topical Collection on Mobile & Wireless Health
Rights and permissions
About this article
Cite this article
Al-Busaidi, A.M., Khriji, L., Touati, F. et al. Wavelet-based Encoding Scheme for Controlling Size of Compressed ECG Segments in Telecardiology Systems. J Med Syst 41, 166 (2017). https://doi.org/10.1007/s10916-017-0817-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-017-0817-1