Abstract
The early detection of osteoporosis risk enhances the lifespan and quality of life of an individual. A reasonable in-vivo assessment of trabecular bone strength at the proximal femur helps to evaluate the fracture risk and henceforth, to understand the associated structural dynamics on occurrence of osteoporosis. The main aim of our study was to develop a framework to automatically determine the trabecular bone strength from clinical femur CT images and thereby to estimate its correlation with BMD. All the 50 studied south Indian female subjects aged 30 to 80 years underwent CT and DXA measurements at right femur region. Initially, the original CT slices were intensified and active contour model was utilised for the extraction of the neck region. After processing through a novel process called trabecular enrichment approach (TEA), the three dimensional (3D) trabecular features were extracted. The extracted 3D trabecular features, such as volume fraction (VF), solidity of delta points (SDP) and boundness, demonstrated a significant correlation with femoral neck bone mineral density (r = 0.551, r = 0.432, r = 0.552 respectively) at p < 0.001. The higher area under the curve values of the extracted features (VF: 85.3 %; 95CI: 68.2–100 %, SDP: 82.1 %; 95CI: 65.1–98.9 % and boundness: 90.4 %; 95CI: 78.7–100 %) were observed. The findings suggest that the proposed framework with TEA method would be useful for spotting women vulnerable to osteoporotic risk.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
‘Osteoporosis’ is an abnormality characterized by reduction in quality and density of bone, which eventually leads to a frail skeleton and frequent fracture risks. The diagnosis of osteoporosis using dual energy x-ray absorptiometry (DXA) has been a standard clinical method. The utilisation of DXA for bone mineral density (BMD) assessment has been confined only to urban areas in the most developing countries of the world like India [1]. Though the trabecular BMD is a broadly utilised method for assessing the osteoporotic fracture risk and therapeutic efficiency, it does not consistently predict individual fracture risk and describe the physiopathology of osteoporotic alterations [2, 3]. Thus, the quantitative analysis of trabecular bone architecture and the clarification of connections between morphological parameters and bone strength have been an important research focus in the field of osteoporosis. The few studies highlighted the relationship between the structural measures of trabecular bone and BMD in the recent past [4–12]. These studies have utilised the images acquired by in-vivo or in-vitro [8, 10] using various diagnostic imaging modalities at different skeletal sites. In earlier attempts, the peripheral sites were analysed in-vivo, since they involved low dosages of radiation, and simple protocol; central body site, such as proximal femur was more often preferred for in-vitro analysis [8, 10, 13].
The trabecular structure extracted from high resolution CT and MR images of femur specimen described the bone strength of an individual and aided in osteoporosis diagnosis [10–13]. The in-vivo analysis on the spine trabecular structure extracted from CT images lead to vertebral fracture risk assessment; the extracted trabecular measures were compared with QCT measured BMD [14, 15]. The 3D trabecular parameters were extracted from high resolution clinical multi detector row CT image for fracture risk assessment at spine and compared against DXA measured BMD [16]. The efficacies of therapy on trabecular micro-structure at spine were monitored in patients with severe osteoporosis [17]. However, the structural assessment from clinical CT image at proximal femur neck has not been studied in-vivo.
The recent observations relating to the perception of bone quality highlighted the bone architecture as a key determinant of trabecular bone strength at the proximal femur [14–17]. The bone strength, determination techniques has incorporated the image processing algorithms; the global thresholding (GT) algorithm was widely used for background intensity removal because of its simple computation [6–12, 14–18]. This technique is not capable of utilising spatial or other image characteristics which leads to uniform nature of implementation irrespective of morphological or intensity related information. The major hindrance in this technique is that when the effect of noise is extraneous: pixels which are not the part of interested region of analysis are included [6, 8, 18]. Hence, there is a need to develop an alternative trabecular enhancement technique for better depiction of trabecular measures from clinical CT image. This simplifies the diagnosis of osteoporosis, where osteoporotic fracture of the proximal femur is legibly prone and substantially increase mortality risk [19]. The sensible in-vivo estimate of trabecular bone strength at proximal femur neck aids, to quantify the fracture risk and to understand the accompanied structural dynamics when osteoporosis progresses [20–24]. Hence, the present study is aimed to develop a framework to automatically determine the trabecular bone strength from clinical femur CT images in-vivo.
Materials and methods
Data collection
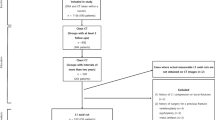
The present study is an attempt to initiate a novel approach to osteoporosis related disorders on the public health grounds. The SRM Hospital and Medical Research Center, SRM University, Chennai, India, gave its consent to arrange an osteoporotic screening camp at the end of the year 2010. The prescribed document was signed by every participant who attended the camp as a means of written voluntary consent. The institutional ethical clearance committee certified the protocol adopted for the study. Participants with disorders such as Paget’s diseases, hypo and hyperthyroidism, rheumatoid arthritis, diabetes, pregnancy, chronic liver and severe trauma-induced fractures as well as osteoporotic fractures were not considered for the study. Finally, 50 south Indian women (Mean ± SD age = 49.3 ± 13.4 years) aged 30 to 80 years were considered for whom examination and analysis was performed.
The BMD quantifications of the right femur for neck (N−BMD) and total femur (T−BMD) were performed with a standard narrow fan beam DXA scanner with multi-view image reconstruction (Lunar DPX Prodigy, GE Lunar Corporation, Madison, WI, USA) using standard measurement procedures. A control phantom was scanned every day prior to the measurements for quality assurance determinations. Then BMD T-score values were calculated using the following WHO standard formula (Eq.1):
The study population was divided into following two groups based on calculated T-score values of measured femur neck BMD: (1) normal subjects (n = 18, age = 41.3 ± 10.6 years), whose T-score ≥ −1 and (2) at-risk of osteoporosis subjects (n = 32, age = 53.8 ± 12.8 years), whose T-score < −1.
For each study subject, a standard CT image of the right proximal femur was obtained using a CT machine (Siemens Somatom, AS, Germany) utilising tube factors of 80 to 120 kV and 20 mAs by a single experienced imaging technologist. The image was acquired at reconstruction related aspects of slice thickness (0.5 mm), rotation time (single, 0.3 s) and rate of acquisition (128 slice/second). The study subjects were imaged in supine position with the pelvis and both legs stretched frontward and the big toes touching each other, causing in slight internal rotation (12° to 15°) of the femur. The images were acquired within the stipulated period of 3 days after acquiring DXA measurements.
CT image processing
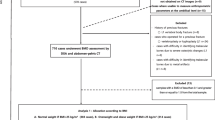
The detailed flow of the proposed CT image quantification system is outlined in the Fig. 1. The slices of coronal direction were selected from each CT image set (mean 33 ± 3 coronal slices per CT image set).
Femur region extraction
The following is the proposed partially automated femur region extraction aided by a trained radiological assistant and a model based segmentation technique. Though the shape of the femoral neck region may vary in the subsequent slices, its location remains unchanged. Hence, the center slice of each CT image set is selected and the contours are manually initialised. Then the active contour model (ACM) is applied only on the center slice of each CT image set [25] with contour area criterion for terminating the iterations [26]. The final fitted contour regions in each CT image set are then cropped and utilised for further neck extraction procedures. Figure 2a illustrates the initialised and the final fit contours of the femur region in a (processed) sample CT slice.
Automatic neck region extraction
Neck boundary detection
The next phase involved the application of a circular Hough transform (CHT) to the segmented femur bone in order to identify the femur head region. When the circle, obtained by CHT fits along with the periphery of the femur head region, which intersects the femur neck regions at points R 1 and R 2 as shown in Fig. 2b. These two points are combined to form the reference line (R) and further with the arc of the circle to form a circular segment. A tangent line which is parallel to the reference line is drawn and which passes through the point p 1 . Then a perpendicular line is drawn from the point p 1 and to intersect the reference line and the intersecting point is denoted as p 2 . The distance between p 1 and p 2 is denoted as y. The line between p 1 and p 2 is further extended on both the sides for the distances x and z, where y = x + z and x = z; the points are noted as p 3 and p 4 respectively . Then two lines (B 1 and B 2 ) parallel to the reference line are drawn through the points p 3 and p 4 respectively. These two lines are considered as the boundaries of the neck region.
Neck region cropping
The spatial location of the identified boundary lines and the initialized contour points (Fig. 2c) are projected across the slices; the ACM was applied on each slice to identify the boundary between the muscle region and cortical bone based on the fitted contour point sets C 1 and C 2 . The iteration was terminated when the number of points moved is lesser than a threshold or when the number of iterations reaches a maximum threshold [25]. The final fitted contours are cropped from each slice and stored in a new stack.
Cortical bone removal
The extracted neck region contains cortical bone. The minimal distance boundary method was used to remove the cortical bone region as depicted in the Fig. 2d. The technique employed to remove the cortical bone region is as follows: first, from the top right corner pixel, the control propagates in parallel with the B 1 line to identify the first black pixel; the number of pixels propagated is called propagation distance d 1 . Similarly, the propagation distances for subsequent lines towards B 2 are called d 2 , d 3 … d n . The subsequent task was to find the minimum propagation distance, which was fixed to be the appropriate thickness of the cortical bone and it has been removed on the C 2 side. The same procedure was applied on the C 1 side also to remove the cortical bone. As a result, the processed slices contained only trabecular bone.
Trabecular bone segmentation
Trabecular bone enhancement
The novel trabecular enrichment approach (TEA) was applied to the trabecular bone region on the femur neck of each slice. The TEA method is as follows: The gray level of the radiographic image, acquired from several subjects, normally varied due to fatty tissue projection and radiological artefacts which corresponded to the low frequency noise of the image. Hence, the method proposed by Geraets et al. was used to eliminate the low frequency noise [27]. Then intensity normalisation using the histogram specification technique detailed by Debashis et al. was used to increase the information (trabecular structure enhancement) and reduce the obscurity [28]. This procedure was applied independently to each individual 3 × 3 sub-block in order to obtain a normalised image. Then, the local contrast enhancement transformation, incorporating point transformation was applied; the goal was to localise the distribution around the mean of the intensity which provides considerable enhancement within each sub-block of 3 × 3 [29].
Binarisation
The enhanced image was binarised using a locally adaptive binarisation method (thresholding). The binarisation was performed as follows: pixel value was assigned one for those belonging to the trabecular bone, when its value was greater than the mean intensity value of the present block (16 × 16); else zero for those pixels belonging or representing to the hollow region.
Trabecular 3D reconstruction
The next phase was to visualise the trabecular bone architecture by reconstructing the segmented trabecular bone using the gradient based volume rendering reconstruction technique [30]. Initially, the segmented femur neck slices were arranged in a stack and surface shading calculation were performed at every voxel with local gradient vectors; opacity of every voxel was computed by applying surface classification operators. The trabecular structure was reconstructed by incorporating the calculated shading, opacity and projecting it onto the picture plane with bilinear interpolation. The performance was faster as it involved simple computation.
Trabecular feature extraction
Trabecular volume fraction2D
The neck region extracted from the center slice in the stack of each binarised CT image set was utilised for calculating the trabecular volume fraction (2D). It is the ratio between the apparent bone volume (BV) to the total volume (TV), which represents the number of segmented white pixels (belongs to trabecular bone) over the total number of pixels in the neck region and is designated as app.BV/TV [6].
3D volume fraction (VF 3D )
The segmented binary slices (contains only neck trabecular bone information) are arranged in a stack; this signifies a 3D numerical representation of the trabecular structure. The total number of white pixels and black pixels in the stack are independently calculated and designated as trabecular bone volume (TB 3D ) and volume of the hollow region (H 3D ) respectively. The 3D volume fraction (VF 3D ) is computed by finding the ratio between TB 3D and, summations of TB 3D and H 3D (Eq.2).
Skeletonisation: The next step involved the implementation of the iterative thinning algorithm for all the binarised CT slices in order to extract the additional trabecular bone features. The parallel thinning with two sub-iteration algorithm was used for skeletisation of every binarised CT slice [31].
3D delta point set (DP3DS)
The bifurcation points and the connecting segments are different topological connectivity elements in the trabecular structure; disrupted trabecular structure results in either crossing point or isolation point [13]. The properties of crossing number (CN) as detailed in the Fig. 3 and Eq.3 is used to classify the present pixel as bifurcation point or others based on Eq.4 [32].
The bifurcation points (represented as delta points) in each skeletonised CT slice in the stack were identified by applying the above procedure and were marked as true pixels (white color) in a new stack; this signifies a 3D numerical representation of delta points. Herein, the total number of 3D connected delta points within each 26 neighbour (sub-volume of 3 × 3 × 3) was computed. In our dataset, the six connected components were the lengthiest connected set of delta points and the 1, 2 connected 3D delta point set did not contribute significantly in the classification. Hence, every 3 to 6 connected delta point sets (Fig. 4) were considered for further analysis.
Solidity of 3D delta points (SDP3D)
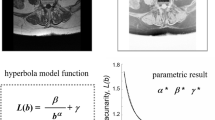
The cumulative, 3 to 6 connected 3D delta points are designated as CDP 3d . The difference in volume of trabecular bone over the volume of hallow region is compared with arbitrary reference of VF 3D . This was well placed in the trabecular bone structure of healthy subjects. This means, the greater the amount of trabecular bone closer to the reference, relatively lesser the risk of bone loss (RBL TB ). The ratio between CDP 3d and RBL TB is designated as solidity of 3D delta points. Increased value of RBL TB represents a low degree of trabecular thickness or higher rate of trabecular bone loss (Eq.5).
Confidence level (CL)
Dominants of 3 to 6 connected delta points over the 1 and 2 connected delta points are considered with high confidence level and vice versa are considered with a low confidence level (Eq.6).
Where
- DP1 − 2 :
-
Count of 1, 2 connected delta points
- DP3 − 6 :
-
Count of 3 to 6 connected delta points
Boundness
A measure of trabecular bone strength represents the volumetric distribution of trabecular structure throughout the volume of interest and is defined by the architectural connectivity of delta points in the 3D structure. Boundness is interpreted in Eq. 7.
Statistical analysis
Data were analysed using SPSS software package version 17.0 (SPSS Inc., Chicago, USA). All data were expressed as mean ± SD. The Pearson correlation analysis was used to find the correlation of extracted trabecular features against measured BMD values. The measured values in each subgroup were compared using a Student’s t-test.
Results
The CT image dataset of each woman was processed in order to automatically segment the trabecular bone of the femur neck region and its feature extraction (eqs. (2)–(7)) as described in the methodology sector. Further, the extracted image features are statistically analysed with DXA measured BMD values.
The resulting images of automatically segmented femur neck region from a sample CT slice, segmented trabecular bone and its corresponding skeletons are displayed in Fig. 5. The reconstructed 3D trabecular bone of the segmented neck region of a normal and osteoporosis samples are displayed in Fig. 6; the close approximation of trabecular bone in 3D space visually confirms the neck regions are properly extracted. Table 1 details the Pearson’s correlation results of extracted image features (TEA) against age and DXA measured BMD values. The trabecular volume fraction (2D) exhibited a significant correlation with respect to age and BMD at p < 0.05; the rest, image features (VF 3D , SDP 3D and Boundness) exhibited a significant correlation with BMD at p < 0.001 and with age at p < 0.05 respectively. Table 2 enumerates the study outcomes based on t-test for the extracted trabecular image features (3D) by a standard method (p < 0.05) and the proposed TEA method (p < 0.001) among normal and at-risk groups respectively. The results of ROC analysis are detailed in Table 3 and Fig. 7. The AUC values were predominant in the 3D trabecular features (Boundness: 90.4 %, SDP 3D : 82.6 % and VF 3D : 87.5 %), whereas, the value was considerably decremented in trabecular volume fraction 2D (69.7 %).
The proposed technique was implemented with MATLAB 8 software in a Windows background (WINDOWS 7); it took 120 s on an Intel E5620 (2.4GHz) and 3 GB of RAM to extract features for each dataset on an average.
Discussion
The trabecular volume fraction (app.BV/TV) was extensively analysed for describing the structural measure of trabecular bone and correlated with BMD [6, 8, 10, 16, 33, 34]. The studies on peripheral skeletal sites were immensely analysed using every possible existing imaging techniques [6, 16, 33]; on the contrary, the studies on the proximal femur site are limited. Previous structural measuring methods were mainly based on either the 2D (center slice) approach using high resolution radiological images or by the 3D approaches based on in-vitro analysis of the proximal femur [10, 13, 34]. Irregular and more anisotropic nature of trabecular structure leads to inaccurate illustration of morphological representation when a single slice (center slice) information is considered [10]. On the other hand, in-vitro analyses of the femur bone in living subjects are nearly impossible. Hence, the present study was designed to automatically extract the femur neck region from clinical CT images, segment the trabecular bone and, analyse the relationship between the extracted trabecular measures (3D) and DXA measured BMD; further, the results obtained by the standard trabecular bone segmentation algorithm (GT) and the proposed TEA method were compared. Our results reflected the high degree of reproducibility and consistency by means of the active contour model implemented for femur bone segmentation and neck region extraction.
Significance of trabecular architecture
Trabecular volume fraction (app.BV/TV) was an alternate density measure which was developed by considering only the center slice on the stack. It neither depicted the density distribution of the trabecular bone content present in the entire stack, nor the location specific density in the volume of interest [6]. Alternatively, we proposed a measure of the 3D volume fraction by considering the entire femur neck volume of interest for better definition of trabecular bone density. The degree of trabecular thickness at the 3D delta point defines the strength of the trabecular structure which gives the solidity of 3D delta points. The higher degree of trabecular content represents the dominance of bone content and well connected multi joint trabecular structure. The dominance of trabecular connectivity helps to improve the reliability of the solidity in the evaluation of boundness. The solidity considers only multiple delta point connectivity and neglects the closeness between the delta point sets. A high solidity with lower volume resulted in few strong and solid trabecular bone junctions (delta points) with a higher degree of inter trabecular space. Hence, the solidity was scaled by the 3D volume fraction to ensure high solid nature of trabecular bone with closeness.
The proposed TEA method vs. global thresholding
The detection of binarisation threshold becomes a difficult task in low resolution images with monomodal intensity histogram. The causes of this monomodal are owing to the partial volume effects and noise. The subjective determination of the threshold is more challenging and affects the estimation of app.BV/TV [35]. Link et al. described about the major challenges in deriving morphological parameters from high resolution MRI and multi-slice spiral CT images using a gray level thresholding method and obtained better significant results in high resolution MR images [36]. Similarly, Majumdar et al. reported that the utilisation of GT technique produced a loss of thickness and overestimation of trabeculae [6]; similar results were evident on CT images by utilising the global and local threshold techniques demonstrating insignificant outcomes [8, 37]. Our study found a significant improvement in recognising the trabecular morphology using the TEA method than the GT method (Table 2). Similarly, Akgundogdu et al. utilised hybrid skeleton graph analysis method to extract morphological parameters from femur trabecular bone and successfully classified osteoporotic as well as osteoarthritis samples [13]. The Pearson correlation results revealed a higher level of significance (p < 0.001) when the extracted (TEA) trabecular features (3D volume fraction: r = 0.551; boundness: r = 0.552) were correlated with femur neck BMD value (Table 1); whereas, the same features depicted lower degree of significance (p < 0.05) when GT was utilised. The parameter such as 3D volume fraction and boundness extracted using TEA method displayed 23 and 13 % superiority respectively, than GT method in distinguishing the low BMD subjects from normal subjects (Table 3).
Clinical application
Our results indicated that the extracted trabecular measures demonstrated a significant correlation with BMD. Hence, the combinational morphological features of the trabecular bone extracted from the clinical CT images could eventually be helpful in osteoporosis diagnosis. They are very useful in situations, where the mobility of the patient is a concern for DXA scans or additional information required for fracture treatment. Though the low radiation protocol (clinical CT image) is suited to depict the better morphology of a trabecular bone; it is not inferred as a substitution for high resolution radiological imaging systems. However, our results obtained on spatial resolutions attainable make the latent and feasibleness of using clinical CT in connection with digital skeletonisation.
The present study had limitations in spatial resolution and locating the region of interest, which is to locate the gap between the femur head region and the acetabulum to initialise the contour as it is sensitive to manual control. That is, however, initially the application of the active contour method was to identify proximal femur; subsequently Hough transform eliminates inaccuracies introduced in the gap close to the acetabulum. Since the objective was not to measure the size of the trabeculae or microstructure the approximated measures were considered; however, this hurdle can be minimised with high resolution imaging.
Conclusion
This is the first study attempted to segment and visualise the trabecular bone architecture from clinical femur CT images; in-vivo estimation of trabecular bone strength by utilising partially automated framework (semi-automated femur region segmentation, automated neck region extraction, trabecular bone enhancement and segmentation, feature extraction). The extracted novel 3D trabecular features (3D volume fraction, solidity of 3D delta points and boundness) demonstrated a significant (p < 0.001) correlation with the BMD values. The features extracted using the novel TEA method demonstrated superior results than the features extracted using the standard GT method. The findings suggest that the proposed framework with TEA method would be useful in a clinical environment where the mobility of patients is a concern for DXA scan or addition information required during fracture treatment.
References
International Osteoporosis Foundation. “The Asian Audit: Epidemiology, Costs and Burden of Osteoporosis in Asia2009”. (http://www.iofbonehealth.org/facts-and-statistics.html), 2009.
Arnold, J. S., Amount and quality of trabecular bone in osteoporotic vertebral fractures. Clin. Endocrinol. Metab. 2:221, 1973.
Kleerekoper, M., Villanueva, A. R., Stanciu, J., Rao, D. S., and Parfitt, A. M., The role of three-dimensional trabecular microstructure in the pathogenesis of vertebral compression fractures. Calcif. Tissue Int. 37:594, 1985.
Lang, T. F., Guglielmi, G., vanKuijk, C., DeSerio, A., Cammisa, M., and Genant, H. K., Measurement of bone mineral density at the spine and proximal femur by volumetric quantitative computed tomography and dual-energy x-ray absorptiometry in elderly women with and without vertebral fractures. Bone 30:247–250, 2002.
Bousson, V., LeBras, A., Roqueplan, F., Kang, Y., Mitton, D., Kolta, S., Bergot, C., Skalli, W., Vicaut, E., Kalender, W., Engelke, K., and Laredo, J. D., Volumetric quantitative computed tomography of the proximal femur: Relationships linking geometric and densitometric variables to bone strength-role for compact bone. Osteoporos. Int. 17:855–864, 2006.
Majumdar, S., Genant, H., Grampp, S., Newitt, D., Truong, V., Lin, J., and Mathur, A., Correlation of trabecular bone structure with age, bone mineral density and osteoporotic status: In vivo studies in the distal radius using high resolution magnetic resonance imaging. J. Bone Miner. Res. 12:111–118, 1997.
Mueller, D., Link, T. M., Monetti, R., Bauer, J., Boehm, H., Seifert-Klauss, V., Rummeny, E. J., Morfill, G. E., and Raeth, C., The 3D based scaling index algorithm: A new structure measure to analyze trabecular bone architecture in high-resolution MR images in vivo. Osteoporos. Int. 17:1483–1493, 2006.
Bauer, J. S., Kohlmann, S., Eckstein, F., Mueller, D., Lochmuller, E. M., and Link, T. M., Structural analysis of trabecular bone of the proximal femur using multislice computed tomography: A comparison with dual X-ray absorptiometry for predicting biomechanical strength in vitro. Calcif. Tissue Int. 78(2):78–89, 2006.
Showalter, C., Clymer, B. D., Richmond, B., and Powell, K., Three dimensional texture analysis of cancellous bone cores evaluated at clinical CT resolutions. Osteoporos. Int. 17:259–266, 2006.
Link, T. M., Vieth, V., Langenberg, R., Meier, N., Lotter, A., Newitt, D., and Majumdar, S., Structure analysis of high resolution magnetic resonance imaging of the proximal femur: In vitro correlation with biomechanical strength and BMD. Calcif. Tissue Int. 72(2):156–165, 2003.
Link, T. M., Majumdar, S., Lin, J. C., Newitt, D., Augat, P., Ouyang, X., Mathur, A., and Genant, H. K., A comparative study of trabecular bone properties in the spine and femur using high resolution MRI and CT. J. Bone Miner. Res. 13(1):122–132, 1998.
Issever, A. S., Vieth, V., Lotter, A., Meier, N., Laib, A., Newitt, D., Majumdar, S., and Link, T. M., Local differences in the trabecular bone structure of the proximal femur depicted with high-spatial-resolution MR imaging and multisection CT. Acad. Radiol. 9(12):1395–1406, 2002.
Akgundogdu, A., Jennane, R., Aufort, G., Benhamou, C. L., and Ucan, O. N., 3D image analysis and artificial intelligence for bone disease classification. J. Med. Syst. 34(5):815–828, 2010.
Chevalier, F., Laval-Jeantet, A. M., Laval-Jeantet, M., and Bergot, C., CT image analysis of the vertebral trabecular network in vivo. Calcif. Tissue Int. 51(1):8–13, 1992.
Gordon, C. L., Lang, T. F., Augat, P., and Genant, H. K., Image-based assessment of spinal trabecular bone structure from high-resolution CT images. Osteoporos. Int. 8(4):317–325, 1998.
Ito, M., Ikeda, K., Nishiguchi, M., Shindo, H., Uetani, M., Hosoi, T., and Orimo, H., Multi-detector row CT imaging of vertebral microstructure for evaluation of fracture risk. J. Bone Miner. Res. 20(10):1828–1836, 2005.
Graeff, C., Timm, W., Nickelsen, T. N., Farrerons, J., Marín, F., Barker, C., and Glüer, C. C., Monitoring teriparatide-associated changes in vertebral microstructure by high-resolution CT in vivo: Results from the EUROFORS study. J. Bone Miner. Res. 22(9):1426–1433, 2007.
Baum, T., Carballido-Gamio, J., Huber, M. B., Müller, D., Monetti, R., Räth, C., Eckstein, F., Lochmüller, E. M., Majumdar, S., Rummeny, E. J., Link, T. M., and Bauer, J. S., Automated 3D trabecular bone structure analysis of the proximal femur--prediction of biomechanical strength by CT and DXA. Osteoporos. Int. 21(9):1553–1564, 2010.
Tromp, A. M., Ooms, M. E., Popp-Snijders, C., Roos, J. C., and Lips, P., Predictors of fractures in elderly women. Osteoporos. Int. 11:134–140, 2000.
Johnell, O., Kanis, J. A., Oden, A., Johansson, H., DeLaet, C., Delmas, P., Eisman, J. A., Fujiwara, S., Kroger, H., Mellstrom, D., Meunier, P. J., Melton, L. J., III, O’Neill, T., Pols, H., Reeve, J., Silman, A., and Tenenhouse, A., Predictive value of BMD for hip and other fractures. J. Bone Miner. Res. 20:1185–1194, 2005.
Abrahamsen, B., Vestergaard, P., Rud, B., Bärenholdt, O., Jensen, J. E., Nielsen, S. P., Mosekilde, L., and Brixen, K., Ten-year absolute risk of osteoporotic fractures according to BMD T score at menopause: The Danish Osteoporosis Prevention Study. J. Bone Miner. Res. 21:796–800, 2006.
Black, D. M., Greenspan, S. L., Ensrud, K. E., Palermo, L., McGowan, J. A., Lang, T. F., Garnero, P., Bouxsein, M. L., Bilezikian, J. P., and Rosen, C. J., The effects of parathyroid hormone and alendronate alone or in combination in postmenopausal osteoporosis. N. Engl. J. Med. 349:1207–1215, 2003.
Black, D. M., Steinbach, M., Palermo, L., Dargent-Molina, P., Lindsay, R., Hoseyni, M. S., and Johnell, O., An assessment tool for predicting fracture risk in postmenopausal women. Osteoporos. Int. 12:519–528, 2001.
Kanis, J. A., Borgstrom, F., DeLaet, C., Johansson, H., Johnell, O., Jonsson, B., Oden, A., Zethraeus, N., Pfleger, B., and Khaltaev, N., Assessment of fracture risk. Osteoporos. Int. 16:581–589, 2005.
Kass, M., Witkin, A., and Terzopoulos, D., Snakes: Active contour models. Int. J. Comput. Vis. 1(4):321–331, 1988.
Wai-Pak, C., Kin-Man, L., and Wan-Chi, S., An adaptive active contour model for highly irregular boundaries. Pattern Recogn. 34(2):323–331, 2001.
Geraets, W. G. M., VanderStelt, P. F., Netelenbos, C. J., and Elders, P. J., A new method for automatic recognition of the trabecular pattern. J. Bone Miner. Res. 5:227–233, 1990.
Debashis, S., and Sankar, K. P., Automatic exact histogram specification for contrast enhancement and visual system based quantitative evaluation. IEEE Trans. Image Process. 20(5):1211–1220, 2011.
Sapthagirivasan, V., and Anburajan, M., Diagnosis of osteoporosis by extraction of trabecular features from hip radiographs using support vector machine: An investigation panorama with DXA. Comput. Biol. Med. 43(11):1910–1919, 2013.
Levoy, M., Display of surfaces from volume data. IEEE Comput. Graph. Appl. 8(3):29–37, 1988.
Guo, Z., and Hall, R. W., Parallel thinning with two sub-iteration algorithms. Commun. ACM 32(3):359–373, 1989.
Rutovitz, D., Pattern recognition. J. R. Stat. Soc. Ser. A 129(4):504–530, 1966.
Majumdar, S., Link, T. M., Augat, P., Lin, J. C., Newitt, D., Lane, N. E., and Genant, H. K., Trabecular bone architecture in the distal radius using magnetic resonance imaging in subjects with fractures of the proximal femur. Osteoporos. Int. 10(3):231–239, 1999.
Saha, P. K., Xu, Y., Duan, H., Heiner, A., and Liang, G., Volumetric topological analysis: A novel approach for trabecular bone classification on the continuum between plates and rods. IEEE Trans. Med. Imaging 29(11):1821–1838, 2010.
Newitt, D. C., vanRietbergen, B., and Majumdar, S., Processing and analysis of in vivo high-resolution MR images of trabecular bone for longitudinal studies: Reproducibility of structural measures and micro-finite element analysis derived mechanical properties. Osteoporos. Int. 13(4):278–287, 2002.
Link, T. M., Vieth, V., Stehling, C., Lotter, A., Beer, A., Newitt, D., and Majumdar, S., High-resolution MRI vs. multislice spiral CT: Which technique depicts the trabecular bone structure best? Eur. Radiol. 13(4):663–671, 2003.
Link, T. M., Majumdar, S., Lin, J. C., Augat, P., Gould, R. G., Newitt, D., Ouyang, X., Lang, T. F., Mathur, A., and Genant, H. K., Assessment of trabecular structure using high resolution CT images and texture analysis. J. Comput. Assist. Tomogr. 22:15–24, 1998.
Acknowledgments
The authors wish to thank the NIELIT Chennai Centre and the management of SRM Hospital and Research Centre, Kattankalathur, Chennai, India for providing the required facilitating infrastructure. They also wish to thank Mr. D. Kathirvelu, Assistant Professor, Biomedical Engineering department, SRM University for his kind assistance.
Conflict of interest
Sapthagirivasan, Anburajan and Janarthanam declare that there is no conflict of interest related to this article. All costs and related expenditures for this study have been managed by the first author only.
Author information
Authors and Affiliations
Corresponding authors
Additional information
This article is part of the Topical Collection on Systems-Level Quality Improvement
Rights and permissions
About this article
Cite this article
Sapthagirivasan, V., Anburajan, M. & Janarthanam, S. Extraction of 3D Femur Neck Trabecular Bone Architecture from Clinical CT Images in Osteoporotic Evaluation: a Novel Framework. J Med Syst 39, 81 (2015). https://doi.org/10.1007/s10916-015-0266-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-015-0266-7