Abstract
A novel Schiff base ligand, chromone-3-carbaldehyde-aminophenazone (L) and its Ln(III) (Ln = La, Yb) complexes were synthesized and characterized by physicochemical methods. The interaction between the ligand, Ln(III) complexes and calf thymus DNA in physiological buffer (pH = 7.10) was investigated by using UV–vis spectroscopy, fluorescence spectra, ethidium bromide experiments and viscosity measurements, indicating that the studied compounds can all bind to DNA via an intercalation binding mode and the complexes have stronger binding affinity than the free ligand alone. Furthermore, antioxidant activity of the ligand and its complexes was determined by superoxide and hydroxyl radical scavenging methods in vitro, suggesting that Ln(III) complexes inhibit stronger antioxidant activity than the ligand alone and some standard antioxidants, such as mannitol and vitamin C.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, a great number of experiments indicate that the binding between small molecules and DNA can greatly help understand drug–DNA interactions and design new and promising anticancer drugs for clinical use [1, 2], and then the interaction of drug molecules with DNA has become more and more active during the past several years [3]. DNA binding metal complexes are being investigated in many laboratories because of their utility as DNA structural probes, DNA dependent electron transfer probes, and DNA foot printing, sequence-specific cleaving agents; especially they can be used as potential anticancer drug [4–6]. Generally, metal complexes bind to DNA in a non-covalent way, such as intercalation, groove-binding and external electrostatic binding for cations [7]. Considerable useful applications of the metal complexes require that they can bind to DNA via an intercalation binding mode inducing cellular degradation [8–10]. The intercalation ability not only correlates to the coordination geometry and ligand donor atom type [11, 12], but also relates with the metal ion type and its valence [13]. Among all the metal complexes, lanthanide complexes are widely investigated since the lanthanide complexes with tetracycline, phenanthroline [14], adriamycin and pyridine [15–17] have already been synthesized as a probe to study nucleic acids. Additionally, the importance of reactive oxygen species (ROS) which are involved in the pathogeneses of various diseases, such as lifestyle-related diseases including hypertension and photoaging due to exposure to ultraviolet radiation has attracted considerable attention over the past decades [18, 19]. In the human body, superoxide and hydroxyl radicals are produced by the reduction of oxygen and over production of free radicals are considered to be the main contributor to oxidative stress [20]. Recently, studies suggest that many metal complexes also exhibited interesting antioxidant activities [21, 22]. Therefore, it is significant to search for new metal complexes as potential antioxidants.
Flavonoids, occurring widely throughout the plant kingdom, are one of the most representative families of plant secondary metabolites and display a remarkable spectrum of biological activities, such as developing the purpose of drug delivery or target identification [23, 24]. It should be especially noted that developing flavonoids as anticancer agents has interested medicinal chemists for many years. Chromones usually are indentified as a kind of flavonoids that are ubiquitous in nature especially in plants. They are oxygen-containing heterocyclic compounds with a benzoannelated γ-pyrone ring, with the parent compound being chromone (4H-chromone-4-one, 4H-1-benzopyran-4-one). Molecules containing the chromone structure have a wide range of biological activities including tyrosine and protein kinase C inhibitors, antifungal, antiallergenic, antiviral, antitublin, antihypertensive, anticancer agents, as well being active at benzoazepine receptors, lipoxygenase, cyclooxygenase and modulating P-glycoprotein-mediated multidrug resistance [25–27].
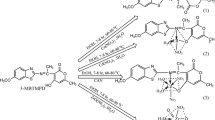
In order to give a deep research to chromone ramifications and their lanthanide complexes, in this paper, we synthesized and characterized a novel ligand, chromone-3-carbaldehyde-aminophenazone (L, Fig. 1) and its Ln (III) complexes. We described a comparative study of the interaction of novel Ln(III) complexes and the ligand with calf thymus DNA (CT DNA) using electronic absorption spectroscopy, fluorescence spectra, ethidium bromide experiments and viscosity measurements for the first time. The antioxidant activity of the ligand and its Ln(III) complexes was also investigated. Information obtained from our study would be helpful to understand the mechanism of interactions of chromone derivatives and their complexes with nucleic acid and should be useful in the development of potential probes of DNA structure and conformation, and new therapeutic reagents for some certain diseases.
Experimental Section
Materials and Instrumentation
Nitroblue tetrazolium (NBT), methionine (MET), vitamin B2 (VitB2), EB and CT DNA were purchased from Sigma Chemical Co. Ethylenediaminetetraaceticacid (EDTA), safranin, 4-aminophenazone and Ln(NO3)3•6H2O [Ln(III) = La and Yb] were produced in China. All chemicals used were of analytical grade. EDTA-Fe(II) and KH2PO4-Na2HPO4 buffers were prepared with doubly distilled water.
All the experiments involved with the interaction of the ligand and its lanthanide complexes with CT DNA were carried out in doubly distilled water buffer containing 5 mM Tris [Tris(hydroxymethyl)-aminomethane] and 50 mM NaCl and adjusted to pH 7.10 with hydrochloric acid. Solution of CT DNA in Tris–HCl buffer gave ratios of UV absorbance of about 1.8–1.9:1 at 260 and 280 nm, indicating that the CT DNA was sufficiently free of protein [28]. The CT DNA concentration per nucleotide was determined spectrophotometrically by employing an extinction coefficient of 6,600 M−1 cm−1 at 260 nm [29]. The ligand and its complexes were dissolved in a solvent mixture of 1% DMF and 99% Tris–HCl buffer (pH 7.10) at the concentration 1.0 × 10−5 M.
Carbon, hydrogen and nitrogen were determined using an Elemental Vario EL analyzer. The metal contents of the complexes were determined by titration with EDTA (xylenol orange tetrasodium salt used as an indicator and hexamethylidynetetraimine as buffer). The melting point of the ligand was determined on a Beijing XT4-100X microscopic melting point apparatus (the thermometer was not corrected). The IR spectra were obtained in KBr discs on a Therrno Mattson FTIR spectrophotometer in the 4,000–400 cm−1 region. 1 H NMR spectra were recorded on a Varian VR 200 MHz spectrometer in CDCl3-d with TMS (tetramethylsilane) as internal standard. Mass spectra were performed on an APEX II FT-ICR MS instrument using methanol as mobile phase. All conductivity measurements were performed in DMF solution with a DDS-11C conductometer at 25.0 °C. The UV–vis spectra were recorded on a Shimadzu UV-240 spectrophotometer. Fluorescence measurements were recorded on a Hitachi RF-4500 spectrofluorophotometer at ambient temperature. The antioxidant activity was performed in DMF solution with a 721-E spectrophotometer.
Preparation of the Ligand (L) and its Ln(III) Complexes
Preparation of the Ligand (L)
Scheme of the synthesis of the ligand is shown in Fig. 1. 3-carbaldehyde chromone was prepared according to the literature methods [30]. To a solution of 3-carbaldehyde chromone (0.870 g, 5 mmol) in ethanol (50 mL) at warming was added 4-aminophenazone (1.117 g, 5.5 mmol) in ethanol (20 mL). The reaction mixture was boiled on an oil bath for 8 h with stirring. After that, the yellow solution was concentrated and cooled, a yellow solid was separated out and filtered, washed with massive water and ethanol, and then recrystallized from DMF and water to give the ligand. Yield: 75.63%. Mp: 269−271 °C. Anal Calcd for C21H17N3O3 requires C, 70.18; H, 4.77; N, 11.69. Found: C, 70.23; H, 4.82; N, 11.66. IR νmax (cm−1): ν(carbonyl)C=O: 1656, ν(hydrazonic)C=O: 1631, νC=N: 1591 cm−1. λmax(nm): 350, 277 nm. L (CDCl3-d 200 MHz): δ 9.85 (1 H, s, 9-H), 8.67 (1 H, s, 2-H), 8.30 (1 H, d, 5-H), 7.72 (1 H, d, 8-H), 7.25–7.45 (7 H, m, 6, 7, 13, 13′, 14, 14′, 15-H), 3.16 (3 H, s, 11-H), 2.41 (3 H, s, 12-H).
Preparation of the Ln(III) Complexes
The ligand (144 mg, 0.40 mmol) was dissolved in the mixture solution of ethanol and chloform (20 mL). After 5 min, La(III) nitrate (87 mg, 0.20 mmol) was added quickly and the solution was refluxed on a water bath with stirring, and the reaction temperature was kept at 90 °C for 10 h. A yellow precipitate, the La(III) complex, was separated from the solution by suction filtration, purified by washing several times with ethanol and dried for 24 h in a vacuum. The Yb(III) complex was prepared by the same way. Anal Calcd for La(III) complex C42H34N9O15La requires: C, 48.33; H, 3.28; N, 12.08; La, 13.31; Found: C, 48.36; H, 3.23; N, 12.03; La, 13.24. Yield: 82%. IR νmax (cm−1): ν(carbonyl)C=O: 1634, ν(hydrazonic)C=O: 1611, νC=N: 1,569 cm−1, \( {\nu^{{_{{{\rm{N}}{{\rm{O}}_{{3}}}}}}}} \): 1479, 1385, 1320, 1184, 698, νM-O: 631, νM-N: 433. λmax (nm):327, 271 cm−1. Λ m (S cm2 mol−1): 87.5. Anal Calcd for Yb(III) complex C42H34N9O15Yb requires: C, 46.80; H, 3.18; N, 11.70; Yb, 16.05; Found: C, 46.78; H, 3.25; N, 11.69; Yb, 16.08. Yield: 76%. IR ν(carbonyl)C=O: 1632, ν(hydrazonic)C=O: 1615, νC=N: 1,577 cm−1, \( {\nu^{{_{{{\rm{N}}{{\rm{O}}_{{3}}}}}}}} \): 1479, 1383, 1321, 1188, 702, νM-O: 636, νM-N: 423. λmax (nm): 328, 270 cm−1. Λ m (S cm2 mol−1): 99.8.
DNA Binding Studies
Electronic Absorption Titration
Electronic absorption titration experiments were performed with fixed concentration drugs (10 μM), while gradually increasing the concentration of CT DNA. When measuring the absorption spectra, an equal amount of CT DNA was added to both the complex solutions and the reference solution to eliminate the absorbance of CT DNA itself. Each sample solution was scanned in the range of 190–500 nm.
Fluorescence Spectra
To compare quantitatively the affinity of the compounds bound to DNA, K b values of the compounds to DNA were obtained by the luminescence titration method. Fixed amounts of compound were titrated with increasing amounts of DNA, over a range of DNA concentrations from 2.5 to 35 μM. Excitation wave lengths of 357 nm for La(III) complex, 359 nm for Yb(III) complex and 335 nm for L were used, and fluorescence emission intensities were monitored at 455 nm, 458 nm and 452 nm, respectively. The concentration of the bound compound was calculated using Eq [31]:
where C t is the total compound concentration, F is the observed fluorescence emission intensity at given DNA concentration, F 0 is the intensity in the absence of DNA, and F max is the fluorescence of the totally bound compound. Binding data were cast into the form of a Scatchard plot of r/C f versus r, where r is the binding ratio C b/[DNA]t and C f is the free compound concentration. All experiments were conducted at room temperature in a buffer containing 5 mM Tris–HCl (pH 7.10) and 50 mM NaCl concentrations.
Further support for the ligand and complexes binding to DNA via intercalation is given through the emission quenching experiment. EB is a common fluorescent probe for DNA structure and has been employed in examinations of the mode and process of metal complexes binding to DNA [32]. A 2.0 mL solution of 4 μM DNA and 0.32 μM EB (at saturating binding levels) was titrated by 2.5–50 μM the lanthanide complexes and ligand. Quenching data were analyzed according to the Stern−Volmer equation which could be used to determine the fluorescent quenching mechanism:
Where F 0 and F are the fluorescence intensity in the absence and presence of drug at [Q] concentration respectively; K q is the quenching constant and [Q] is the quencher concentration. Plots of F 0/F versus [Q] appear to be linear and K q depends on temperature [33].
Viscosity Measurements
Viscosity experiments were conducted on an Ubbelodhe viscometer, immersed in a thermostated water-bath maintained to 25.0 ± 0.1 °C. Titrations were performed for the complexes (0.5–4 μM), and each compound was introduced into a DNA solution (5 μM) present in the viscometer. Data were presented as (η/η 0)1/3 versus the ratio of the concentration of the compound and DNA, where η is the viscosity of DNA in the presence of the compound and η 0 is the viscosity of DNA alone [34, 35].
Antioxidant Activity
In antioxidant activity experiments the superoxide radicals \( \left( {{{\hbox{O}}_{{2}}}^{{ \bullet - }}} \right) \) were produced by the system MET/VitB2/NBT [36]. The amount of \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \) and suppression ratio for \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \) can be calculated by measuring the absorbance at 560 nm, because NBT can be reduced quantitatively to blue formazan by \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \). The solution of MET, VitB2 and NBT were prepared with 0.067 M phosphate buffer (pH = 7.8) at the condition of avoiding light. The tested compounds were dissolved in DMF. The reaction mixture contained MET (0.01 mol L−1), NBT (4.6 × 10−5 mol L−1), VitB2 (3.3 × 10−6 mol L−1), phosphate buffer solution (0.067 mol L−1) and the tested compound (the final concentration: \( {{\hbox{C}}_{\rm{i}}}_{{({\rm{i}} = {1} - {5})}} = 0.{4},\;{1}.0,\;{2}.0,\;{4}.0,\;{6}.0\;\mu {\hbox{M}} \)). After incubating at 30 °C for 10 min and illuminating with a fluorescent lamp for 3 min, the absorbance (Ai) of the samples was measured at 560 nm. The sample without the tested compound and avoiding light was used as the control. The suppression ratio for \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \) was calculated from the following expression:
where Ai = the absorbance in the presence of the ligand or its complexes, A0 = the absorbance in the absence of the ligand or its complexes.
In antioxidant activity experiments the hydroxyl radical (HO•) in aqueous media was generated through the Fenton reaction [37]. The solution of the tested compounds was prepared with DMF. The reaction mixture contained 2.5 mL 0.15 M phosphate buffer (pH = 7.4), 0.5 mL 114 μM safranin, 1 mL 945 μM EDTA–Fe(II), 1 mL 3% H2O2 and 30 μL the tested compound solution (the final concentration: \( {{\hbox{C}}_{{{\text{i}}\;({\rm{i}} = {1}-{6})}}} = {1}.0,\;{2}.0,\;{3}.0,\;{4}.0,\;{5}.0,\;{6}.0\;\mu {\hbox{M}} \)). The sample without the tested compound was used as the control. The reaction mixtures were incubated at 37 °C for 60 min in a water-bath. Absorbances (Ai, A0, Ac) at 520 nm were measured. The suppression ratio for HO• was calculated from the following expression:
Where Ai = the absorbance in the presence of the tested compound; A0 = the absorbance in the absence of the tested compound; Ac = the absorbance in the absence of the tested compound, EDTA–Fe(II) and H2O2.
The antioxidant activity was expressed as the 50% inhibitory concentration (IC50). IC50 values were calculated from regression lines where: x was the tested compound concentration in μM and y was percent inhibition of the tested compounds.
Results and Discussion
Structure of the Ln(III) Complexes
The complexes were prepared by direct reaction of ligand with the appropriate mole ratios of Ln(III) nitrate in the mixture solution of chloroform and ethanol. The yields were good to moderate. The desired Ln(III) complexes were separated from the solution by suction filtration, purified by washing several times with ethanol. The complexes are air-stable for extended periods and soluble in DMSO and DMF; slightly soluble in ethanol and methanol; insoluble in benzene, water and diethyl ether. The molar conductivities of the complexes are around 87.5–99.8 S cm2 mol−1 in DMF solution, showing that the two complexes are 1:1 electrolytes [38].
The IR spectra of the complexes are similar and IR spectra usually provide a lot of valuable information on coordination reactions. All the spectra are characterized by vibrational bands mainly due to the C = O, C = N, M-O, M-N and NO3 groups. The ν(carbonyl)C=O and ν(hydrazonic)C=O vibrations of the free ligand appear at 1,656 and 1,631 cm−1, respectively; whereas for the complexes these peaks shift to 1,634, 1,611 and 1,632, 1,615 cm−1, Δν(ligand-complexes) is equal to 22, 20 and 24, 16 cm−1, respectively. In the complexes, the band at 631 cm−1 or so is assigned to νM–O, demonstrating that the oxygen of carbonyl has formed a coordinative bond with the rare earth ions [39]. The different shifts of the wavenumbers indicate that the Ln-O (carbonyl) bond is a little stronger than the Ln-O (hydrazonic) bond. The band at 1,591 cm−1 for the free ligand is assigned to νC=N stretch, which shifts to about 1,569 cm−1 for the complexes, and Δν(ligand-complexes) is equal to 22 cm−1. Weak bands at 433 cm−1 or so are assigned to νM–N in the complexes. These shifts and new bands further confirm that the nitrogen of the imino-group bonds to the rare earth ions [40]. The absorption bands of the coordinated nitrates were observed at about 1479 (νas) and 817 (νs) cm−1. The ν3 (E′) free nitrates appear at 1,385 cm−1 in the spectra of the rare earth complexes. In addition, the separation of the two highest frequency bands |ν4–ν1| is approximately 195 cm−1, and accordingly the coordinated \( {\hbox{N}}{{\hbox{O}}_{{3}}}^{ - } \) anion in the complexes is a bidentate ligand [41].
The study of the electronic spectra in the ultraviolet and visible (UV–vis) ranges for the rare earth complexes and ligand was carried out in a buffer solution containing 5 mM Tris–HCl (pH 7.10) and 50 mM NaCl concentrations. The electronic spectra of the free ligand have a strong band at λmax = 277 nm and a medium band at λmax = 350 nm. The complexes also yield two bands and the two bands are shifted to 270 and 328 nm or so. These indicate that the rare earth complexes are formed.
The electrospray ionization (ESI) mass spectra of La(III) and Yb (III) complex were made. The mass spectra of La(III) complex show peaks at 1043, 919 and 718, which can be assigned to the fragments [La(III) complex + H]+, [La(III) complex - 2NO3]2+ and [La(III) complex - 3NO3 - La], respectively. And the mass spectra of Yb(III) complex show peaks at 1077 and 718 which can be assigned to the fragments [Yb(III) complex + H]+ and [La(III) complex - 3NO3 - La], respectively.
In conclusion, the elemental analyses, molar conductivities and other structural analyses indicate that the novel ligand and lanthanide ions can form mononuclear ten-coordination [LnL2·(NO3)2]NO3 [Ln(III) = La and Yb, L is chromone-3-carbaldehyde-aminophenazone] complexes with 1:2 metal-to-ligand stoichiometry at the Ln(III) centers.
DNA Binding Studies
Electronic Absorption Spectroscopy
Before reacting the ligand and its Ln(III) complexes interacting with CT DNA, their solution behavior in buffer solution at room temperature was monitored by UV–vis spectroscopy for 24 h. Liberation of the ligand and its complexes was not observed under these conditions. These suggest that the ligand and its complexes are stable under the conditions studied.
The binding abilities of the ligand and Ln(III) complexes with CT DNA can be characterized by measuring their effects on the UV–vis spectroscopy. Intercalative mode of binding usually results in hypochromism and red shift because of the strong stacking interaction between an aromatic chromophore and DNA base pairs. The extent of the spectral change is related to the strength of binding [32]. The electronic absorption spectra of the free ligand and its complexes in the absence and presence of CT DNA are given in Fig. 2. As shown, in the presence of DNA, the absorption bands of the ligand, La(III) and Yb(III) complexes at 350. 327 and 328 nm exhibited hypochromism of 43.89%, 53.95% and 45.80%, respectively. The hypochromism observed for the lanthanide complexes are all accompanied by a red shift of about 7 nm. The binding of complexes to DNA led to decrease in the absorption intensities with a small amount of red shifts in the UV–vis absorption spectra. After intercalating the base pairs of DNA, the π* orbital of the intercalated ligand can couple with the π orbital of the DNA base pairs, thus, decreasing the π→π* transition energy and resulting in the bathochromism. On the other hand, the coupling π orbial is partially filled by electrons, thus, decreasing the transition probalities and concomitantly resuting in the hypochromism [42]. All these results suggest that the complexes can interact with CT DNA quite probably by intercalating the compounds into DNA base pairs, so we can conclude that the free ligand and its metal complexes can interact with CT DNA via a same mode (intercalation), and the complexes bind to DNA more strongly than the free ligand alone.
Electronic absorption spectra of the ligand (a), La(III) (b) and Yb(III) (c) complexes (10 μM) in the absence (top spectrum) and presence of increasing amounts of CT DNA (2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0 and 22.5 μM; subsequent spectra). Arrows show the absorbance changes upon increasing DNA concentration
EB (a typical indicator of intercalation) quenching assay was performed to further test the binding modes of DNA to these investigated compounds [43]. Figure 3 shows the electronic absorption spectra when EB was employed. The absorbance of Tris–HCl buffer solution is highest when there is only EB alone. After adding CT DNA into the buffer solution, the absorbance decreases. However, it increases when the ligand and metal complexes were added dropwise into the buffer solution. In addition, the maximal absorption of EB at 479 nm decreases and shifts to 494 nm in the presence of CT DNA, and that is characteristic of intercalation mode. Curve (B) in Fig. 3 shows the absorption of a mixture solution of EB, DNA and the prepared compounds. It is also found that the absorption at 494 nm has increased comparing with curve (C). These may result from two reasons: (1) EB bound to the free ligand and metal complexes strongly, resulting in the decrease of the amount of EB intercalated into DNA; (2) there exists competitive intercalation between the free ligand and metal complexes and EB with DNA, so releasing some free EB from DNA–EB system. However, the former reason could be precluded because there were no new absorption peaks appearing. Thus, we can come to a conclusion that the prepared compounds can displace EB from its DNA-EB system. Quite similar results are also observed for other Ln(III) complexes.
Fluorescence Spectra
Figure 4 exemplifies the steady-state emission spectra of 10 μM solutions of lanthanide complexes in Tris–HCl buffer and the spectra show an increase in the emission intensity with successive addition of CT DNA at room temperature. According to the Scatchard equation, a plot of r/C f versus r gave K b values of (7.80 ± 1.06) × 105, (2.92 ± 0.40) × 106, (1.91 ± 0.23) × 106 M−1 from the fluorescence data for the free ligand, La(III) and Yb(III) complexes, respectively. Results obtained from fluorescence spectra suggest that all the compounds are protected from solvent water molecules by the hydrophobic environment inside the DNA helix and that the metal complexes can be protected more efficiently than the free ligand alone. Since the hydrophobic environment inside the DNA helix reduces the accessibility of solvent water molecules to the compound and the compound mobility is restricted at the certain binding site, a decrease of the vibrational modes of relaxation results. The binding of the compounds to DNA leading to a significantly increase in emission intensity also agrees with those observed for other intercalators [44]. The fluorescence spectra show that the lanthanide complexes can bind to DNA more strongly and deeply than the free ligand alone, The higher binding affinities of the complexes are probably attributed to the extension of the π system of the intercalated ligand due to the coordination of metal ions, which also leads to a planar area greater than that of the free ligand and the coordinated ligand penetrating more deeply into and stacking more strongly with the base pairs of DNA.
The emission enhancement spectra of the free ligand (a), La(III) (b) and Yb(III) (c) complexes (10 μM) in the absence (bottom spectrum) and presence of increasing amounts of CT DNA (2.5, 5.0, 7.5, 10.0, 12.5 and 15.0 μM; subsequent spectra). Arrows show the emission intensity changes upon increasing DNA concentration. Inset: Scatchard plot of the fluorescence titration data of ligand and rare earth complexes, \( {K_{\rm{b}}}\left( {\hbox{a}} \right) = \left( {{7}.{8}0\pm {1}.0{6}} \right) \times {1}{0^{{5}}} \); \( {K_{\rm{b}}}\left( {\hbox{b}} \right) = \left( {{2}.{92}\pm 0.{4}0} \right) \times {1}{0^{{6}}}{{\hbox{M}}^{{ - {1}}}} \); \( {K_{\rm{b}}}\left( {\hbox{c}} \right) = \left( {{1}.{91}\pm 0.{23}} \right) \times {1}{0^{{6}}}{{\hbox{M}}^{{ - {1}}}} \)
The DNA binding mode of all the compounds was further monitored by a fluorescent EB displacement assay. EB is one of the most sensitive fluorescence probes that can bind to DNA and the fluorescence of EB increases after intercalating into DNA base pairs [45]. Competitive binding of other drugs to DNA and EB system will result in the decrease in the fluorescence intensity of the DNA-EB system. The reduction of the fluorescence emission intensity gives a criterion to investigate the DNA binding propensity of the complexes and the stacking interaction (intercalation) between the adjacent DNA base pairs [46]. Figure 5 shows the emission spectra of the DNA–EB system with increasing amounts of the Ln(III) complexes and free ligand. The addition of the compounds to the DNA-bound EB solutions caused obvious reduction in emission intensities and the quenching of EB bound to DNA by the rare earth complexes is in good agreement with the linear Stern-Volmer equation. In the plots of F 0/F versus [Q], K q values are given by the ratio of the slope to the intercept and K q values for the ligand, La(III) and Yb(III) complexes are (1.75 ± 0.03) × 104, (5.54 ± 0.12) × 104 and (2.26 ± 0.05) × 104 M−1. The results show that all the compounds can bind to DNA and the complexes bind to DNA more strongly than the free ligand, which also validate the electronic absorption spectral results. Since these changes indicate only one kind of quenching process, it may be concluded that the complexes and free ligand can bind to DNA via the same mode (intercalation).
The emission spectra of DNA-EB system λex = 525 nm, \( {\lambda_{\rm{em}}} = {55}0 - {65}0\;{\hbox{nm}} \), in the presence of 0, 2.5, 5, 7.5, 10, 12.5, 15, 17.5, 20.0, 22.5, 25, 27.5, 30.0, 32.5, 35.0, 37.5, 40.0, 42.5, 45.0, 47.5 and 50.0 μM free ligand (a) and La(III) (b) complex, and 0, 2.5, 5, 7.5, 10, 12.5, 15, 17.5, 20.0, 22.5, 25, 27.5, 30.0, 32.5 and 35.0 μM Yb(III) (c) complex. Arrows show the emission intensity changes upon increasing ligand and complexes. Inset: Stern-Volmer plot of the fluorescence titration data of ligand and complexes, \( {K_{\rm{q}}}\left( {\hbox{a}} \right) = \left( {{1}.{75} \pm 0.0{3}} \right) \times {1}{0^{{4}}}\;{{\hbox{M}}^{{ - {1}}}} \); \( {K_{\rm{q}}}\left( {\hbox{b}} \right) = \left( {{5}.{54}\pm 0.{12}} \right) \times {1}{0^{{4}}}\;{{\hbox{M}}^{{ - {1}}}} \); \( {K_{\rm{q}}}\left( {\hbox{c}} \right) = \left( {{2}.{26} \pm 0.0{5}} \right) \times {1}{0^{{4}}}\;{{\hbox{M}}^{{ - {1}}}} \)
Viscosity Measurements
Viscosity titration measurements were carried out to clarify the interaction modes between the investigated compounds and CT DNA. Viscosity measurements are very sensitive to changes in the length of DNA, as viscosity is proportional to L 3 for rod-like DNA of length L, and then viscosity measurements are regarded as the least ambiguous and the most critical tests of a binding model in solution in the absence of crystallographic structural data [47]. A classical intercalation model demands that the DNA helix must lengthen as base pairs are separated to accommodate the binding complexes, leading to the increase of DNA viscosity, as for the behaviors of the known DNA intercalators [48]. In contrast, a partial and/or non-classical intercalation of the complex could bend (or kink) the DNA helix, reducing its viscosity concomitantly [47]. In addition, some compounds such as [Ru(bpy)3]2+, which interacts with DNA by an electrostatic binding mode, have no influence on DNA viscosity [49]. To further elucidate the binding mode of the present complexes, viscosity measurements were carried out by keeping [DNA] = 5 μM and varying the concentration of the compounds.
The changes in relative viscosity of CT DNA in the presence of the ligand and complexes are shown in Fig. 6. As seen from Fig. 6, upon increasing amounts of the ligand and complexes, the relative viscosities of DNA increase steadily with increasing concentrations of ligand and complexes. The increased degree of viscosity, which may depend on the binding affinity of compounds to DNA, following the order of La(III) complex > Yb(III) complex > L. These results suggest that all the title complexes and ligand intercalate between the DNA base pairs and the binding affinities of the complexes are higher than that of the free ligand, which is consistent with the above experimental results.
On the basis of all the spectroscopic studies together with the viscosity measurements, we find that the lanthanide complexes and free ligand can bind to CT DNA via an intercalative binding mode (Fig. 7) and the complexes bind to CT DNA more strongly and deeply than the free ligand alone.
Antioxidant Activity
Due to the ligand and its Ln(III) complexes exhibit good DNA binding affinity, it is considered worthwhile to investigate their antioxidant activity. Reactive oxygen species (ROS), such as superoxide anion and hydroxyl radical, are usually generated by all aerobic cells during normal oxygen metabolism, and cumulative information obtained has proved that the oxidation induced by ROS is involved in the pathogenesis of various diseases through direct effects on DNA directly and by acting as a tumor promoter [50]. Then in this paper, antioxidant activity of the prepared ligand and Ln(III) complexes was investigated by comparing their scavenging effects on superoxide anion and hydroxyl radical.
Figure 8 and Table 1 depict the inhibitory effect of the compounds on \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \). The average suppression ratio for \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \) increases with the increasing amount of the compound concentration in the range of tested concentration. The average suppression ratio of the ligand (IC50 = 23.214 μM) is the least in all the compounds and La(III) complex (IC50 = 0.492 μM) is the more effective inhibitor than Yb(III) complex (IC50 = 1.438 μM). It is clear that the scavenger effect on \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \) can be enhanced by the formation of metal-ligand complexes and the metal ions also affect the ability. The metal ions may have different and selective nature for scavenging \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \). Due to the lower IC50 values, they are potential drugs to eliminate the radicals.
The data of suppression ratio for HO• are shown in Fig. 9 and Table 2. The inhibitory effect of all the compounds is marked and the average suppression ratio for HO• increases with the increasing compounds concentration. The average suppression ratio of the ligand is the poorest in all the compounds, La(III) complex (IC50 = 0.861 μM) and Yb(III) complex (IC50 = 1.167 μM) are the more effective inhibitors than the ligand (IC50 = 2.605 μM), which illuminates that because of the formation of coordination compounds, the scavenger effect can be enhanced and the rare earth metal ions, La(III) and Yb(III) also affect the scavenger ability. From the above depictions, we can easily find that Ln(III) complexes exhibit considerable scavenging activity due to the chelation of organic molecules to the lanthanide ions which exert different and selective effects on scavenging radicals of the biological system. We also find that complexes are better inhibitor than mannitol which is usually used as special scavenger for HO•. It is reported that IC50 value of ascorbic acid (Vc, a standard agent for non-enzymatic reaction) for HO• is 1.537 mg mL−1 (8.727 mmol), and the scavenging effect for \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \) is only 25% at 1.75 mg mL−1 (9.94 mmol) [51]. Notably, the investigated complexes have much stronger scavenging abilities for HO• and \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \) radicals. It was believed that the information obtained form present work would be useful to develop new potential antioxidants and therapeutic agents for some diseases.
Conclusion
In summary, a new ligand, chromone-3-carbaldehyde-aminophenazone Schiff base, and its Ln(III) complexes have been prepared and characterized. Their DNA binding properties and antioxidant activities were studied systematically. Experimental results indicate that the ligand and its complexes can bind to DNA via an intercalation binding mode and the complexes have better DNA binding affinity than the free ligand alone. Furthermore, the complexes have more active scavenging effects on \( {{\hbox{O}}_{{2}}}^{{ \bullet - }} \) and HO• than the standard antioxidants, such as mannitol and vitamin C. Our work clearly indicates that rare earth-based complexes have many potential practical applications, such as understanding the mechanisms of interaction between compounds and DNA, the development of potential anticancer drugs and new antioxidants.
References
Blasius R, Moucheron C, Mesmasker AK (2004) Eur J Inorg Chem 20:3971
Zhang GW, Guo JB, Pan JH, Chen XX, Wang JJ (2009) J Mol Struct 923:114
Riahi S, Ganjali MR, Bagheri M (2009) Mater Sci Eng, C 29:1808
Erkkila KE, Odem DT, Barton JK (1999) Chem Rev 99:2777
Metcalfe C, Thomas JA (2003) Chem Soc Rev 32:215
Xiong Y, Ji LN (1999) Coord Chem Rev 185:711
Li Y, Yang ZY, Wang MF (2009) Eur J Med Chem 44:4585
Li Y, Yang ZY (2010) J Fluoresc 20:329
Wang J, Yang ZY, Yi XY, Wang BD (2009) J Photochem Photobiol, A Chem 201:183
Long EC (1993) J Inorg Organomet Polym 3:3
Mahadvan S, Palaniandavar M (1997) Inorg Chim Acta 254:291
Xu H, Zheng KC, Deng H, Lin LJ, Zhang QL, Ji LN (2003) New J Chem 27:1255
Asadi M, Safaei E, Ranjbar B, Hasani L (2004) New J Chem 28:1227
Bisi CC, Carugo O (1989) Inorg Chim Acta 159:157
Ci YX, Li YZ, Chang WB (1991) Anal Chim Acta 248:589
Horne DA, Dervan PB (1990) J Am Chem Soc 112:2435
Moser HE, Dervan PB (1987) Science 238:645
Yan MX, Li YQ, Meng M, Ren HB, Kou Y (2006) Biochem Biophys Res Commun 347:192
Cai L (2006) Free Radic Biol Méd 41:851
Alexandrova ML, Bochev G (2005) Free Radic Biol Méd 39:297
Li Y, Yang ZY, Wang MF (2010) J Fluoresc 20:891
Li Y, Yang ZY (2009) Inorg Chim Acta 362:4823
Havsteen BH (2002) Pharmacol Ther 96:67
Huang W, Liu MZ, Li Y, Tan Y, Yang GF (2007) Bioorg Med Chem 15:5191
Sosnovskikh VY (2003) Russ Chem Rev 72:489
Li Y, Yang ZY, Wu JC (2010) Eur J Med Chem 45:5692
Walenzyk T, Carola C, Buchholz H, König B (2005) Tetrahedron 61:7366
Cai YZ, Luo Q, Sun M, Corke H (2004) Life Sci 74:2157
Heim KE, Tagliaferro AR, Bobilya DJ (2002) J Nutr Biochem 13:572
Nohara A, Umetani T, Sanno Y (1973) Tetrahedron Lett 14:1995
Howe GM, Wu KC, Bauer WR, Lippard SJ (1976) Biochem 15:4339
Pyle AM, Rehmann JP, Meshoyrer R, Kumar CV, Turro NJ, Barton JK (1989) J Am Chem Soc 111:3051
Eftink MR, Ghiron CA (1981) Anal Biochem 114:199
Eriksson M, Leijon M, Hiort C, Norden B, Graeslund A (1994) Biochem 33:5031
Xiong Y, He XF, Zou XH, Wu JZ, Chen XM, Ji LN, Li RH, Zhou JY, Yu KB (1999) J Chem Soc, Dalton Trans: Inorg Chem 1:19
Winterbourn CC (1979) Biochem J 182:625
Winterbourn CC (1981) Biochem J 198:125
Geary WJ (1971) Coord Chem Rev 7:81
Marchetti F, Pettinari C, Pettinari R, Cingolani A, Leonesi D, Lorenzotti A (1999) Polyhedron 18:3041
Narang KK, Singh VP (1993) Transit Met Chem 18:287
Wang BD, Yang ZY, Zhang DW, Wang Y (2006) Spectrochim Acta Part A 63:213
Bai GY, Dong B, Lü YY, Wang KZ, Jin LP, Gao LH (2004) J Inorg Biochem 98:2011
Wilson WD, Ratmeyer L, Zhao M, Strekowski L, Boykin D (1993) Biochem 32:4098
Xu H, Zheng KC, Lin LJ, Li H, Gao Y, Ji LN (2004) J Inorg Biochem 98:87
Le-Pecq JB, Paoletti C (1967) J Mol Biol 27:87
Lee M, Rhodes AL, Wyatt MD, Forrow S, Hartley JA (1993) Biochem 32:4237
Satyanarayana S, Dabrowiak JC, Chaires JB (1993) Biochem 32:2573
Liu JG, Zhang QL, Shi XF, Ji LN (2001) Inorg Chem 40:5045
Liu JG, Ye BH, Li H, Zhen QX, Ji LN, Fu YH (1999) J Inorg Biochem 76:265
Towyz RM (2004) Hypertension 44:248
Xing RG, Yu HH, Liu S, Zhang WW, Zhang QB, Li ZE, Li PC (2005) Bioorg Med Chem 13:1387
Acknowledgements
This work is supported by the National Natural Science Foundation of China (20975046) and Gansu NSF (0710RJZA012).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, Y., Yang, Zy., Li, Tr. et al. Synthesis, Characterization, DNA Binding Properties and Antioxidant Activity of Ln(III) Complexes with Schiff Base Ligand Derived from 3-Carbaldehyde Chromone and Aminophenazone. J Fluoresc 21, 1091–1102 (2011). https://doi.org/10.1007/s10895-010-0782-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-010-0782-2