Abstract
A novel Schiff-base ligand (H5L), hesperetin-2-hydroxy benzoyl hydrazone, and its copper (II), zinc (II) and nickel (II) complexes (M·H3L) [M(II) = Cu, Zn, Ni], have been synthesized and characterized. The ligand and Zn (II) complex exhibit green and blue fluorescence under UV light and the fluorescent properties of the ligand and Zn (II) complex in solid state and different solutions were investigated. In addition, DNA binding properties of the ligand and its metal complexes have been investigated by electronic absorption spectroscopy, fluorescence spectra, ethidium bromide displacement experiments, iodide quenching experiments, salt effect and viscosity measurements. Results suggest that all the compounds bind to DNA via an intercalation binding mode. Furthermore, the antioxidant activity of the ligand and its metal complexes was determined by superoxide and hydroxyl radical scavenging methods in vitro. The metal complexes were found to possess potent antioxidant activity and be better than the free ligand alone and some standard antioxidants like vitamin C and mannitol.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA binding metal complexes have been extensively investigated during the past several decades because they can be used as potential anticancer drugs, DNA structural probes, DNA-dependent electron transfer probes, DNA footprinting, sequence-specific cleaving agents and so on [1–3]. In order to develop new potential DNA targeting antitumor drugs, the binding mechanisms of metal complexes to DNA should be further studied. Basically, metal complexes interact with double helix DNA in either non-covalent or covalent way and the former way includes three binding modes, i.e., intercalation, groove binding and external static electronic effects. Among these interaction modes, intercalation is one of the most important DNA binding modes because numerous biological experiments have demonstrated that DNA is the primary intracellular target of anticancer drugs due to the intercalation between small molecules and DNA, which can cause DNA damages in cancer cells, blocking the division of cancer cells and resulting in cell death [4–6]. It was reported that DNA binding mode and affinity are affected by a number of factors, such as planarity of ligands [7, 8], the coordination geometry, the ligand donor atom type [9, 10], the metal ion type and its flexible valence [11, 12].
Flavonoids are a broadly distributed class of naturally occurring pigments present in vascular plants and are responsible for most of the coloring in nature [13]. Up to now, more than 6,000 flavonoids have already been identified, although only a smaller number is important from a dietary point of view [14]. Recently, flavonoids receive considerable attention in the literature, mainly due to their biological and physiological importance [15]. They are potentially antioxidant, antibacterial, anticancer, anti-inflammatory and antiallergenic agents since they can stimulate or inhibit a wide variety of enzyme systems as pharmacological agents [16–19]. The best-known and studied flavonoids are quercetin, morin and rutin, whereas hesperetin is one kind of the lesser-known flavonoids. Hesperetin (5,7,3′-trihydroxyl-4′-methoxyl-flavanone) usually occurs ubiquitously in plants, fruits, flowers and foods of plant origin [20]. As an important bioactive Chinese traditional medicine, hesperetin has multiple biological and pharmacological activities, including antioxidant properties [21–23], inhibition of cancer development [24, 25], effects on the blood–brain barrier [26, 27] and signal transduction pathways [28], etc.
In order to give a deep research to hesperetin ramifications and their metal complexes, in this paper, a novel ligand, hesperetin-2-hydroxy benzoyl hydrazone (H5L, Fig. 1) and its transition metal complexes were synthesized and characterized. We described a comparative study of the interaction of the complexes and ligand with calf thymus DNA (CT DNA) using absorption spectroscopy, fluorescence spectra, iodide quenching experiments, salt effect and viscosity measurements for the first time. In addition, the antioxidant activity of the ligand and its metal complexes was determined by superoxide and hydroxyl radical scavenging method in vitro. Furthermore, the fluorescent properties of the ligand and Zn (II) complex in solid state and different solutions were investigated. Information obtained from our work would be helpful to understand the interaction mechanism of hesperetin hydrazones and their complexes with DNA and useful in the development of potential probes of DNA structure and conformation, and new therapeutic reagents for some certain diseases.
Experimental section
Materials and physical measurements
Ethidium bromide (EB) and CT DNA were purchased from Sigma Chemical Co. All chemicals and solvents used were of analytical reagent grade and used without further purification unless otherwise noted.
All the experiments involved with the interaction of the ligand and its transition metal complexes with CT DNA were carried out in doubly distilled water buffer containing 5 mM Tris [Tris(hydroxymethyl)-aminomethane] and 50 mM NaCl and adjusted to pH 7.1 with hydrochloric acid. Solution of CT DNA in the buffer gave ratios of UV absorbance of about 1.8–1.9:1 at 260 and 280 nm, indicating that the CT DNA was sufficiently free of protein [21]. The CT DNA concentration per nucleotide was determined spectrophotometrically by employing an extinction coefficient of 6,600 M−1 cm−1 at 260 nm [22]. The ligand and its complexes were dissolved in a solvent mixture of 1% DMF and 99% Tris–HCl buffer (pH 7.1) at the concentration 1.0 × 10−5 M.
Carbon, hydrogen and nitrogen were determined using an Elemental Vario EL analyzer. The metal contents of the metal complexes were determined by titration with EDTA (xylenol orange tetrasodium salt used as an indicator and hexamethylidynetetraimine as buffer). The melting points of the compounds were determined on a Beijing XT4-100X microscopic melting point apparatus (the thermometer was not corrected). The IR spectra were obtained in KBr discs on a Therrno Mattson FTIR spectrophotometer in the 4,000–400 cm−1 region. 1H NMR spectra were recorded on a Varian VR 200 MHz spectrometer in DMSO-d 6 with TMS (tetramethylsilane) as an internal standard. Mass spectra were performed on a APEX II FT-ICR MS instrument using DMF as mobile phase. All conductivity measurements were performed in DMF solution with a DDS-11C conductometer at 25.0°C. The UV-vis spectra were recorded on a Shimadzu UV-240 spectrophotometer. Fluorescence measurements were recorded on a Hitachi RF-4500 spectrofluorophotometer at room temperature.
Preparation of the ligand and complexes
Synthesis of the ligand (H5L)
Scheme of the synthesis of the ligand is shown in Fig. 1. Hesperetin (3.02 g, 10 mmol) and 2-hydroxy benzoyl hydrazine (1.82 g, 12 mmol) were dissolved in ethanol (50 mL). Acetic acid (1.0 mL) was added into the solution. The solution was refluxed on an oil bath for 24 h with stirring and a yellow precipitate formed. The yellow precipitate was filtered, washed several times with ethanol and recrystallized from DMF and water to give the ligand. Yield: 83.6%. M. p.: 264–267°C. Anal Calcd for C23H20N2O7 requires C, 63.30; H, 4.62; N, 6.42. Found: C, 63.19; H, 4.73; N, 6.46. λ max (nm): 368, 353, 332 nm.
Synthesis of the complexes
The ligand (43.6 mg, 0.1 mmol) was dissolved in ethanol (20 ml) and Cu(NO3)2•3H2O (24.5 mg, 0.1 mmol) was then added. After 5 min, triethylamine (20.2 mg, 0.2 mmol) was added into the solution and the solution was stirred for one day at room temperature. Then a blue precipitate, Cu (II) complex was separated by suction filtration. It was purified by washing several times with ethanol, and then dried for 48 h in vacuum. The Zn (II) complex and Ni (II) complex were also synthesized as the above method. Anal Calcd for Cu (II) complex C23H18N2O7Cu requires C, 55.48; H, 3.64; N, 5.63; Cu, 12.76; Found: C, 55.38; H, 3.60; N, 5.68; Cu, 12.73. Yield: 73.2%. λ max (nm): 412, 387, 372 nm. Λ m (S cm2 mol−1): 6.6. Anal Calcd for Zn (II) complex C23H18N2O7Zn requires C, 55.27; H, 3.63; N, 5.61; Zn, 13.08; Found: C, 55.33; H, 3.52; N, 5.68; Zn, 13.13. Yield: 79.3%. λ max (nm): 408, 384, 372 nm. Λ m (S cm2 mol−1): 7.4. Anal Calcd for Ni (II) complex C23H18N2O7Ni requires C, 56.02; H, 3.68; N, 5.68; Ni, 11.90; Found: C, 56.07; H, 3.62; N, 5.63; Ni, 12.03. Yield: 75.4%. λ max (nm): 410, 369, 358 nm. Λ m (S cm2 mol−1): 6.1.
DNA binding studies
Electronic absorption spectroscopy
Electronic absorption titration experiments were performed with fixed concentration drugs (10 μM), while gradually increasing the concentration of CT DNA. When measuring the absorption spectra, an equal amount of CT DNA was added to both the compound solutions and the reference solution to eliminate the absorbance of CT DNA itself. Each sample solution was scanned in the range of 190–500 nm.
Fluorescence spectra
To compare quantitatively the affinity of the compounds bound to DNA, the intrinsic binding constants (K b) of the compounds to DNA were obtained by the luminescence titration method. Fixed amounts of compound were titrated with increasing amounts of DNA, over a range of DNA concentrations from 2.5 to 20 μM. Scan speed was 480 nm/min, slit width was 5 nm. All experiments were conducted at a constant room temperature in a buffer solution containing 5 mM Tris-HCl (pH 7.1) and 50 mM NaCl concentrations. The concentration of the bound compound was calculated using Eq. [29].
where C t is the total compound concentration, F is the observed fluorescence emission intensity at given DNA concentration, F 0 is the intensity in the absence of DNA, and F max is the fluorescence of the totally bound compound. Binding data were cast into the form of a Scatchard plot of r/C f versus r, where r is the binding ratio C b/[DNA]t, C f is the free compound concentration and n is the binding site number.
Further support for the ligand and complexes binding to DNA via intercalation is given through the emission quenching experiment. EB is a common fluorescent probe for DNA structure and has been employed in examinations of the mode and process of metal complexes binding to DNA [30]. A 2.0 mL solution of 4 μM DNA and 0.32 μM EB (at saturating binding levels) was titrated by 2.5–30 μM the complexes and ligand. Quenching data were analyzed according to the Stern–Volmer equation which could be used to determine the fluorescent quenching mechanism:
Where F 0 and F are the fluorescence intensities in the absence and presence of compound at [Q] concentration, respectively; K q is the quenching constant and [Q] is the quencher concentration. Plots of F 0/F versus [Q] appear to be linear and K q depends on temperature [31].
Iodide quenching experiments
The fluorescence quenching efficiency is evaluated by Stern–Volmer equation, which varies with the experimental conditions. Quenching plots were constructed according to the following Stern–Volmer equation.
Where F 0 and F are the fluorescence intensities in the absence and presence of iodide at [I−] concentration, respectively; K sv is the quenching constant and [I−] is the concentration of iodide. Plots of F o/F versus [I−] appear to be linear and K sv was evaluated by linear least-squares analysis of the data according to the equation [32].
Salt effect
Fluorescence intensities were recorded in the absence and presence of DNA in the mixture solution of each compound and NaCl.
Viscosity measurements
Viscosity measurements were conducted on an Ubbelodhe viscometer, immersed in a thermostated water-bath maintained to 25.0 ± 0.1°C. Titrations were performed for the compounds (0.5–4 μM), and each compound was introduced into a DNA solution (5 μM) present in the viscometer. Data were presented as (η/η 0)1/3 versus the ratio of the concentration of the compound and DNA, where η is the viscosity of DNA in the presence of the compound and η 0 is the viscosity of DNA alone [33, 34].
Antioxidant activity
Superoxide radical scavenging activity
The superoxide radicals (O •−2 ) were produced by the system MET/VitB2/NBT [35]. The amount of O •−2 and suppression ratio for O •−2 can be calculated by measuring the absorbance at 560 nm, because NBT can be reduced quantitatively to blue formazan by O •−2 . The solution of MET, VitB2 and NBT were prepared with 0.067 M phosphate buffer (pH = 7.8) at the condition of avoiding light. The tested compounds were dissolved in DMF. The reaction mixture contained MET (0.01 mol L−1), NBT (4.6 × 10−5 mol L−1), VitB2 (3.3 × 10−6 mol L−1), phosphate buffer solution (0.067 mol L−1) and the tested compound (the final concentration: Ci (i=1–6) = 0.4, 1.0, 2.0, 4.0, 6.0, 8.0 μM). After incubating at 30°C for 10 min and illuminating with a fluorescent lamp for 3 min, the absorbance (Ai) of the samples was measured at 560 nm. The sample without the tested compound and avoiding light was used as the control. The suppression ratio for O •−2 was calculated from the following expression:
where Ai = the absorbance in the presence of the ligand or its complexes, A0 = the absorbance in the absence of the ligand or its complexes.
Hydroxyl radical scavenging activity
The hydroxyl radical (HO•) in aqueous media was generated through the Fenton reaction [36]. The solution of the tested compounds was prepared with DMF. The reaction mixture contained 2.5 mL 0.15 M phosphate buffer (pH = 7.4), 0.5 mL 114 μM safranin, 1 mL 945 μM EDTA–Fe(II), 1 mL 3% H2O2 and 30 μL the tested compound solution (the final concentration: Ci (i=1−6) = 1.0, 2.0, 3.0, 4.0, 5.0, 6.0 μM). The sample without the tested compound was used as the control. The reaction mixtures were incubated at 37°C for 60 min in a water-bath. Absorbances (Ai, A0, Ac) at 520 nm were measured. The suppression ratio for HO• was calculated from the following expression:
Where Ai = the absorbance in the presence of the tested compound; A0 = the absorbance in the absence of the tested compound; Ac = the absorbance in the absence of the tested compound, EDTA–Fe(II) and H2O2.
The antioxidant activity was expressed as the 50% inhibitory concentration (IC50). IC50 values were calculated from regression lines where: x was the tested compound concentration in mM and y was percent inhibition of the tested compounds.
Results and discussion
Characterization of the complexes
The complexes were prepared by direct reaction of the ligand with appropriate mole ratios of M (II) nitrate in ethanol. The complexes are stable in atmospheric conditions for extended periods and easily soluble in DMF and DMSO; slightly soluble in ethanol, methanol and acetone; insoluble in benzene, water and diethyl ether. The molar conductivities of the complexes are around 6.1–7.4 in DMF solution and in accord with them being formulated as non-electrolytes [37]. The elemental analyses and molar conductivities show that formulas of the complexes conform to M·H3L [M (II) = Cu, Zn, Ni].
IR spectra
The IR spectra of the ligand and its complexes are presented in Table 1. On the basis of the very similar spectra of the three complexes, it may be assumed that they have the similar coordination structures. All the spectra are characterized by vibrational bands mainly due to the O–H, C=O, C=N, NNH, M–O and M–N groups. The ν (O–H) for the ligand appears at 3,443 cm−1, and this peak shifts at 3,424 cm−1 or so for the complexes. The ν (C=O) vibration of the free ligand is at 1,632 cm−1, for the complexes, this peak shifts at 1,606 cm−1 or so; Δν (ligand-complexes) is equal to 26 cm−1. For the complexes, the band at 570 cm−1 or so is assigned to ν (M–O). It demonstrates that the oxygen of the carbonyl group has formed a coordinative bond with the metal ions [38]. δ (NNH) vibration of the free ligand appears at 1,517 cm−1, and for the complexes the vibrations are still at 1,519–1,524 cm−1, which indicates that the active hydrogen of the −NNH = still exists in the complexes. The band at 1,611 cm−1 for the free ligand is assigned to the ν (C=N) stretch, which shifts to 1,570 cm−1 or so for the complexes; Δν (ligand-complexes) is equal to 41 cm−1. Weak bands at 475 cm−1 or so are assigned to ν (M–N) for the complexes. These further confirm that the nitrogen of the imino-goup bonds to the metal ions [39].
1H NMR spectra
The 1H NMR spectra of the ligand and its Zn (II) complex are assigned as follows: H5L (DMSO-d 6, 200 MHz), δ (ppm) 3.12 (2H, bs, 3-H), 3.77 (3H, s, −OCH3), 5.09 (1H, dd, J = 3.0, 12.3 Hz, 2-H), 5.88 (1H, d, J = 2.2 Hz, 6-H), 5.93 (1H, d, J = 2.2 Hz, 8-H), 6.91 (5H, bs, −Ar-H), 7.37 (1H, dd, J = 1.8, 7.6 Hz, 4″-H), 7.91 (1H, dd, J = 1.8, 7.6 Hz, 3″-H), 9.10 (1H, s, 3′-OH), 10.01 (1H, s, 6″-OH), 11.33 (1H, s, 5-OH), 11.65 (1H, s, −NH-C=O), 12.86 (1H, s, 7-OH). Zn (II) complex 1H NMR (DMSO-d 6, 200 MHz), δ (ppm) 2.89 (2H, bs, 3-H), 3.78 (3H, s, −OCH3), 5.00 (1H, dd, J = 2.3, 10.2 Hz, 2-H), 5.55 (1H, d, J = 2.0 Hz, 6-H), 5.69 (1H, d, J = 2.0 Hz, 8-H), 6.86 (5H, bs, −Ar-H), 7.23 (1H, dd, J = 1.5, 7.6 Hz, 4″-H), 7.84 (1H, dd, J = 1.5, 7.6 Hz, 3″-H), 9.07 (1H, s, 3′-OH), 9.30 (1H, s, −NH-C=O), 13.94 (1H, s, 7-OH). In the complex, we can find hydrogen atoms of 5-OH and 6″-OH were displaced by the metal ion and a new complex was formed by coordination of zinc ion and ligand, which is also supported by the IR spectra.
UV-vis spectra
The study of the electronic spectra in the ultraviolet and visible (UV-vis) (Fig. 4) ranges for the metal complexes and ligand was carried out in a Tris-HCl buffer solution. The electronic spectra of the free ligand exhibit three absorption bands at 332 (band I), 353 (band II) and 368 (band III) nm (DMF was used as the solvent). Band I is related to ring A, band II to ring C and band III to ring B. The complexes also yield three bands and the three bands are shifted to 360, 380 and 410 nm or so. This shows that there is a large conjugate system formed by the ligand in the complexes. Therefore, the complexes should be chelates containing both a five-membered and a six-membered ring and the metal complex formation leads to the red shifts of the absorption spectra. All these indicate that the metal complexes are formed.
Mass spectra
The electrospay ionization (ESI) mass spectra of Cu(II) complex was made. The mass spectra of Cu(II) complex show peak at 537 (data not shown) which can be assigned to the fragment [Cu(II) complex + K]+.
Fluorescence spectra
The emission spectra of the ligand and Zn (II) complex in solid state (the excitation and emission slit widths were 1.5 nm) were recorded at room temperature. The ligand exhibits green fluorescence and the emission is readily observed with naked eyes under UV light. Whereas the Zn (II) complex exhibits blue fluorescence and the emission is also readily observed with naked eyes under UV light. However, the fluorescence intensities of the Cu (II) and Ni (II) complexes are much too weak in the solid state, which are not discussed in our present work. Fluorescence spectra of the ligand and Zn (II) complex in solid state are shown in Fig. 2. The Zn (II) complex and ligand in the solid state at room temperature have different emissions at 485 and 522 nm at the same excitation (350 nm), respectively. Because Zn(NO3)2 has no fluorescence, the observed blue fluorescence is attributed to the coordinated ligand. However, it is noteworthy that the emission wavelength of Zn (II) complex is slightly shorter than that of the free ligand and can probably be assigned to the intra-ligand fluorescent emission, and can probably be the result of coordination of the free ligand to Zn (II) that increases the ligand conformational rigidity [40]. Thus, Zn (II) complex may be used as an advanced material for blue light emitting diode devices.
The influence of solvents on the fluorescence intensities of the ligand and Zn (II) complex was also investigated. As shown in Table 2, the fluorescence intensities of Zn (II) complex in organic solvents are weaker than that of the powder. This may be due to the quenching process of solvent molecules in the solution. The energy transfer from the triplet excited state of the ligand to the emitting level of the zinc ion could not be carried out perfectly because the oscillatory motions of the entering solvent molecules may consume more energy. All these indicate that the polarity of the solvents affects the fluorescence of Zn (II) complex. Furthermore, the fluorescence intensity of Zn (II) complex, relative to that of the free ligand, is dramatically enhanced, which is clearly caused by the formation of coordination bonds between the ligand and zinc ion. In addition, the solution fluorescent spectra of the ligand and Zn (II) complex are highly solvent-dependent.
Structure of the complexes
Since the crystal structures of the metal complexes have not been obtained yet, we characterized the complexes and determined its possible structure by elemental analyses, molar conductivities, IR spectra, 1H NMR spectra, mass spectra, fluorescence studies and UV-vis spectra. The likely structure of the complexes is shown in Fig. 3.
DNA binding studies
Electronic absorption spectroscopy
Before reacting the ligand and its metal complexes interacting with CT DNA, its solution behavior in Tris-HCl buffer solution at room temperature was monitored by UV-vis spectroscopy for 24 h. Liberation of the ligand and its complexes was not observed under these conditions. These suggest that the ligand and its complexes are stable under the conditions studied.
Titration with electronic absorption spectroscopy is an effective method to investigate the binding mode of DNA with metal complexes [41]. In the intercalative binding mode, the π* orbital of the intercalated ligand can couple with the π orbital of the DNA base pairs, thus, decreasing the π→π* transition energy and resulting in the bathochromism. On the other hand, the coupling π orbital is partially filled by electrons, thus, decreasing the transition possibilities and concomitantly resulting in the hypochromism [42]. Generally, the binding of an intercalative molecule to DNA is accompanied by hypochromism and/or significant red-shift (bathochromism) in the absorption spectra due to the strong stacking interaction between the aromatic chromophore of the ligand and DNA base pairs with the extent of hypochromism and red-shift commonly consistent with the strength of the intercalative interaction [43]. Therefore, in order to obtain evidence for the binding ability of each compound to CT DNA, spectroscopic titration of compounds solutions with CT DNA should be performed [44].
The electronic absorption spectra of the free ligand and its metal complexes in the absence and presence of CT DNA are given in Fig. 4. As shown, there exists three bands at 368, 353, 332 nm for the free ligand and there are also three bands for the metal complexes. With increasing concentrations of CT DNA, the absorption band at 353 nm for the free ligand exhibited hypochromism of 28.1% and was accompanied by a small red-shift of 3 nm in λ max, from 353 to 356 nm. Upon addition of CT DNA, for the Cu(II), Zn(II) and Ni(II) complexes, the absorption bands at 369, 387 and 384 nm also show hypochromism of 43.2%, 33.2 and 29.2%, respectively. The hypochromism observed for the metal complexes are all accompanied by a small red shift by about 3 nm. All these results suggest that the compounds can interact with CT DNA quite probably by intercalating the compounds into DNA base pairs, so we can conclude that the free ligand and its metal complexes can interact with CT DNA via the same mode (intercalation).
Electronic absorption spectra of the free ligand (a), Cu (II) (b), Zn (II) (c) and Ni (II) (d) complexes (10 μM) in the absence (top spectrum) and presence of increasing amounts of CT DNA (2.5, 5.0, 7.5, 10.0, 12.5, and 15.0 μM; subsequent spectra). Arrow shows the absorbance changes upon increasing DNA concentration
In order to further test if the complexes could bind to CT DNA via the intercalationmod, EB (a typical indicator of intercalation) was employed [45]. Figure 5 shows the electronic absorption spectra when EB was employed. The absorbance of Tris-HCl buffer solution is highest when there is EB alone. After adding CT DNA into the buffer solution, the absorbance decreases. However, it increases when the ligand and metal complexes were added dropwise into the buffer solution. Figure 5 shows that the maximal absorption of EB at 479 nm decreased and shifted to 494 nm in the presence of DNA, which was characteristic of intercalation mode. Curve (B) in Fig. 5 was the absorption of a mixture solution of EB, DNA and the free ligand and metal complexes. It was found that the absorption at 494 nm or so increased comparing with curve (C). It could result from two reasons: (1) EB bound to the free ligand and metal complexes strongly, resulting in the decrease of the amount of EB intercalated into DNA; (2) there exists competitive intercalation between the free ligand and metal complexes and EB with DNA, so releasing some free EB from DNA–EB system. However, the former reason could be precluded because there were no new absorption peaks appearing.
Fluorescence spectra
The spectral results of the emission titrations for the metal complexes and free ligand with CT DNA are illustrated in the titration curves (Fig. 6). Compared to the free ligand and its metal complexes alone, the emission intensity increases with increasing concentrations of CT DNA. Results obtained from fluorescence spectra suggest that all the compounds are protected from solvent water molecules by the hydrophobic environment inside the DNA helix and that the metal complexes can be protected more efficiently than the free ligand alone. This implies that all the metal complexes and free ligand can insert between DNA base pairs deeply and the metal complexes can bind to DNA more strongly than the free ligand alone. Since the hydrophobic environment inside the DNA helix reduces the accessibility of solvent water molecules to the compound and the compound mobility is restricted at certain binding sites, a decrease of the vibrational modes of relaxation results. The binding of the compounds to DNA leading to a significantly increase in emission intensity also agrees with those observed for other intercalators [46]. According to the Scatchard equation, a plot of r/C f versus r gave K b values of (3.02 ± 0.25) ×105, (1.35 ± 0.05) ×106, (3.58 ± 0.38) ×106 and (2.07 ± 0.09) ×106 M−1 from the fluorescence data for the ligand, Cu(II), Zn(II) and Ni(II) complexes, respectively. The higher binding affinities of the metal complexes are probably attributed to the extension of the π system of the intercalated ligand due to the coordination of transition metal ions, which also leads to a planar area greater than that of the free ligand and the coordinated ligand penetrating more deeply into and stacking more strongly with the base pairs of DNA.
The emission enhancement spectra of free ligand (a), Cu (II) (b), Zn (II) (c) and Ni (II) (d) complexes (10 μM) in the presence of 0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5 and 20.0 μM DNA. Arrows show the emission intensity changes upon increasing DNA concentration. Inset: Scatchard plot of the fluorescence titration data of ligand and metal complexes, K b (a) = (3.02 ± 0.25) ×105 M−1; K b (b) = (1.35 ± 0.05) ×106 M−1; K b (c) = (3.58 ± 0.38) ×106 M−1; K b (d) = (2.07 ± 0.09) ×106 M−1
Binding modes of the compounds with DNA were further monitored by a fluorescent EB displacement assay. EB is one of the most sensitive fluorescence probes that can bind to DNA. The fluorescence of EB increases after intercalating into DNA base pairs. If the compounds intercalates into DNA, it leads to a decrease in the binding sites of DNA available for EB and resulting in the decrease in the fluorescence intensity of DNA-EB system [47]. The reduction of the emission intensity gives a measure of DNA binding propensity of the compounds and stacking interaction (intercalation) between the adjacent DNA base pairs [48, 49]. Steady-state emission quenching experiments are usually used to observe the binding mode of the metal complexes and free ligand to CT DNA. Figure 7 shows the emission spectra of the DNA–EB system with increasing amounts of the metal complexes and free ligand. The addition of the compounds to the DNA-bound EB solutions caused obvious reduction in emission intensities. The quenching plots illustrated that the quenching of EB bound to DNA by the complexes and free ligand is in good agreement with the linear Stern–Volmer equation. In the plots of F 0/F versus [Q], K q is given by the ratio of the slope to the intercept and K q values for the ligand, Cu (II), Zn (II) and Ni (II) complexes are (1.57 ± 0.12) ×105, (1.79 ± 0.06) ×105, (2.26 ± 0.07) ×105 and (2.10 ± 0.08) ×105 M−1, respectively. The data show that the interaction of the metal complexes with DNA is stronger than that of the free ligand, which is consistent with the electronic absorption spectral results. Since these changes indicate only one kind of quenching process, it may be concluded that the metal complexes and free ligand bind with DNA via the same mode (intercalation mode).
The emission spectra of DNA-EB system, in the presence of 0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, 25.0, 27.5 and 30.0 μM free ligand (a), Cu (II) (b), Zn (II) (c) and Ni (II) (d) complexes. Arrows show the emission intensity changes upon increasing ligand and complexes. Inset: Stern–Volmer plot of the fluorescence titration data of ligand and complexes. K q (a) = (1.57 ± 0.12) ×105 M−1; K q (b) = (1.79 ± 0.06) ×105 M−1; K q (c) = (2.26 ± 0.07) ×105 M−1; K q (d) = (2.10 ± 0.08) ×105 M−1
Iodide quenching experiments
The results of the emission titrations for the Zn (II) complex and potassium iodide with absence and presence of CT DNA are illustrated in the titration curves (Fig. 8). Compared to the Zn (II) complex alone, the emission intensity decreases with increasing concentrations of CT DNA. The quenching plots illustrated that the quenching studies of the Zn (II) complex are in good agreement with the linear Stern–Volmer equation.
a Fluorescence spectra of Zn (II) complex (10 μM) with increasing concentration of KI (5, 10, 15, 20, 25, 30 and 35 mM). b Fluorescence spectra of Zn (II) complex (10 μM) and DNA (20 μM) system with increasing concentration of KI (5, 10, 15, 20, 25, 30 and 35 mM). cInset: Stern–Volmer plot of the fluorescence titration data of Zn (II) complex. Effect of KI concentration (1: Zn(II) complex + KI; 2: Zn (II) complex + KI + CT DNA). Ksv (1) = 10.5 ± 0.63 M−1, Ksv (2) = 4.59 ± 0.22 M−1
In the aqueous solution, iodide and ferrocyanide anions quench the fluorescence of Zn (II) complex very efficiently, so we used potassium iodide as the quencher to determine the relative accessibilities of the free and bound Zn (II) complex. From Fig. 8, we can find that the addition of Zn (II) complex and CT DNA results in extensive quenching of the fluorescence intensity. The quenching data were plotted according to the Stern–Volmer equation and the slopes were calculated by the linear least-squares method. The observed quenching constants were 4.59 ± 0.22 and 10.5 ± 0.63 M−1 with and without CT DNA, respectively. The quenching of the Zn (II) complex fluorescence was in fact enhanced by a factor of about 3 when the Zn(II) complex was bound to the DNA helix. K sv values were used to deduce the interaction mode of the fluorescence probe with DNA. Higher binding constants should correspond to the better protection by the DNA and a stronger inhibition of quenching by anionic species. Thus, we can conclude that the metal complexes are intercalated into the DNA helix and they should be protected from the anionic quencher, owning to the base pairs above and below the intercalators [50].
Salt effect
The effects of the ionic strength on the Zn (II) complex fluorescence intensity was tested by the addition of a strong electrolyte, such as NaCl, instead of potassium iodide. Cations of the salts can neutralize the negatively charged phosphate. If the compound binds to DNA through an electrostatic interaction mode, the surface of DNA will be surrounded by the sodium ions with the increasing ionic strength. Then the compound has difficulty to approach DNA molecules and the strength of interaction with DNA decreases, and then the degree of fluorescence quenching also decreases [29]. As seen from Fig. 9, addition of NaCl to the Zn (II) complex in the absence and presence of CT DNA has few or no effects on the fluorescence intensity, showing that the interaction of the Zn (II) complex with CT DNA is not electrostatic interaction. Thus, we can come to a conclusion that the compounds interact with DNA is not via an electrostatic mode.
Viscosity measurements
In order to further clarify the interaction nature between the compounds and DNA, viscosity measurements were carried out. Optical photophysical probes provide necessary but not sufficient evidence to support a binding model. Hydrodynamic measurements that are sensitive to length change (i.e. viscosity and sedimentation) are regarded as the least ambiguous and the most critical tests of a binding model in solution in the absence of crystallographic structural data [51]. A classical intercalation model demands that the DNA helix must lengthen as base pairs are separated to accommodate the binding complexes, leading to the increase of DNA viscosity, as for the behaviors of the known DNA intercalators [52]. In contrast, a partial and/or non-classical intercalation of the complex could bend (or kink) the DNA helix, reducing its viscosity concomitantly [51]. In addition, some complexes such as [Ru(bpy)3]2+, which interacts with DNA by an electrostatic binding mode, have no influence on DNA viscosity [53]. The effects of all the compounds on the viscosity of CT DNA are shown in Fig. 10. The viscosities of DNA increase steadily with increasing concentrations of ligand and complexes and the extent of the increase observed for the ligand is smaller than those for the complexes. Viscosity measurements clearly show that all the compounds can intercalate between adjacent DNA base pairs, causing an extension in the DNA helix and thus increasing the viscosity of DNA and that the complexes can intercalate more strongly and deeply than the free ligand, leading to the greater increase in viscosity of the DNA with an increasing concentration of the complexes. The results obtained from the viscosity experiments validate those obtained from the spectroscopic studies.
On the basis of all the spectroscopic studies together with the viscosity measurements, we find that the transition metal complexes and free ligand can bind to CT DNA via an intercalative mode (Fig. 11) and the transition metal complexes bind to CT DNA more strongly and deeply than the free ligand.
Antioxidant activity
Due to the ligand and its transition metal complexes exhibiting good DNA binding affinity, it is considered worthwhile to investigate their antioxidant activity. Reactive oxygen species (ROS), such as superoxide anion (O •−2 ) and hydroxyl radical (HO•), are generated by all aerobic cells during normal oxygen metabolism, and cumulative information obtained has proved that the oxidation induced by ROS is involved in the pathogenesis of various diseases through direct effects on DNA directly and by acting as a tumor promoter [54–56]. In this paper, the antioxidant activity of the ligand and its transition metal complexes was studied by comparing their scavenging effect on superoxide radical and hydroxyl radical.
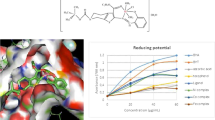
Figure 12 depicts the inhibitory effects of the ligand and transition metal complexes on O •−2 (a) and HO• (b). The antioxidant activity of the compounds is expressed as 50% inhibitory concentration (IC50 in μM). From Fig. 12(a), the inhibitory effect of the compounds studied on O •−2 is concentration dependent and the suppression ratio increase with increasing sample concentrations in the range tested. IC50 values of the ligand, Cu (II), Zn (II) and Ni (II) complexes are 118.06, 0.42, 1.93 and 4.46 μM, respectively. It is clear that the scavenger effect on O •−2 can be enhanced by the formation of metal-ligand coordination compounds and the nature of metal ions also affects the ability. As seen from Fig. 12(b), IC50 values of the ligand, Cu (II), Zn (II) and Ni (II) complexes are 85.63, 4.88, 8.36 and 39.46 μM, respectively. The metal complexes are better antioxidants than the ligand. Obviously, the metal complexes exhibit considerable scavenging activity due to the chelation of organic molecules to the metal ions and the metal ions also exert differential and selective effects on scavenging radicals of the biological system. Furthermore, we find that the metal complexes show scavenging HO• effects than mannitol which is usually used as special scavenger for HO• and it is IC50 value is 10.19 μM [57], indicating that the synthesized complexes are better potential antioxidants. In addition, it is reported that IC50 value of ascorbic acid (Vc, a standard agent for non-enzymatic reaction) for HO• is 1.537 mg mL−1 (8.727 mmol), and the scavenging effect for O •−2 is only 25% at 1.75 mg mL−1 (9.94 mmol) [58]. In addition, the magnetic properties influence on the reactivity of the metal complexes with superoxide anion and hydroxyl radicals, and we find complexes which are paramagnetic can scavenge the superoxide and hydroxyl radicals more greatly than complexes which are diamagnetic. Notably, the investigated ligand and complexes have much stronger scavenging abilities for HO• and O •−2 radicals. It was believed that the information obtained form present work would be useful to develop new potential antioxidants and therapeutic agents for some diseases.
Conclusion
In summary, a novel hesperetin Schiff-base ligand and three transition metal complexes have been synthesized and characterized. The ligand and Zn (II) complex can emit fluorescence in the solid state and their emission spectra are highly solvent-dependent. K b values obtained from the fluorescence titration data of the ligand, Cu (II), Zn (II) and Ni (II) complexes are (3.02 ± 0.25) ×105, (1.35 ± 0.05) ×106, (3.58 ± 0.38) ×106 and (2.07 ± 0.09) ×106 M−1, respectively, indicating that all the compounds can bind to DNA via the intercalation mode and metal complexes can bind to DNA more strongly than the free ligand. This is due to the chelating effect (metal ions to free ligand) which can enhance the planar functionality of the metal complexes and the metal complexes can insert and stack between the DNA base pairs more easily and deeply than the free ligand alone. It is noteworthy that Zn (II) complex can not only emit blue fluorescence but also possess better DNA binding ability, and it should be further studied in the future. Antioxidant activity was also studied by the scavenging superoxide and hydroxyl radical methods and experimental results indicate the metal complexes possess better antioxidant activity than the free ligand. Our present work would be helpful to understand the interaction mechanism of small molecule compounds binding to DNA and useful in the development of potential probes of DNA structure and conformation, new therapeutic reagents and antioxidants.
References
Erkkila KE, Odom DT, Barton JK (1999) Recognition and reaction of metallointercalators with DNA. Chem Rev 99(9):2777–2795
Haq I, Lincoln P, Suh D, Norden B, Choedhry BZ, Chaires JB (1995) Interaction of Δ- and Λ-[Ru(phen)2DPPZI2+ with DNA: a calorimetric and equilibrium binding study. J Am Chem Soc 117(17):4788–4796
Metcalfe C, Thomas JA (2003) Kinetically inert transition metal complexes that reversibly bind to DNA. Chem Soc Rev 32(4):215–224
Catherine H, Marguerite P, Michael RG, Stephanie Heinz S, Bernard M (2001) Preparation, characterization and crystal structures of manganese(II), iron(III) and copper(II) complexes of the bis[di-1, 1-(2-pyridyl)ethyl]amine (BDPEA) ligand; evaluation of their DNA cleavage activities. J Biol Inorg Chem 6(1):14–22
Li VS, Choi D, Wang Z, Jimenez LS, Tang MS, Kohn H (1996) Role of the C-10 substituent in mitomycin C-1-DNA bonding. J Am Chem Soc 118(10):2326–2331
Zuber G, Quada JC Jr, Hecht SM (1998) Sequence selective cleavage of a DNA octanucleotide by chlorinated bithiazoles and bleomycins. J Am Chem Soc 120(36):9368–9369
Kumar CV, Barton JK, Turro NJ (1985) Photophysics of ruthenium complexes bound to double helical DNA. J Am Chem Soc 107(19):5518–5523
Xu H, Zheng KC, Deng H, Lin LJ, Zhang QL, Ji LN (2003) Effects of ligand planarity on the interaction of polypyridyl Ru(II) complexes with DNA. Dalton Trans 3(11):2260–2268
Mahadevan S, Palaniandavar M (1997) Spectroscopic and voltammetric studies of copper(II) complexes of bis(pyrid-2-yl)-di/trithia ligands bound to calf thymus DNA. Inorg Chim Acta 254(2):291–302
Xu H, Zheng KC, Deng H, Lin LJ, Zhang QL, Ji LN (2003) Effects of the ancillary ligands of polypyridyl ruthenium(II) complexes on the DNA-binding behaviors. New J Chem 27(8):1255–1263
Mozaffar A, Elham S, Bijan R, Leila H (2004) Thermodynamic and spectroscopic study on the binding of cationic Zn(II) and Co(II) tetrapyridinoporphyrazines to calf thymus DNA: the role of the central metal in binding parameters. New J Chem 28(10):1227–1234
Chaires JB (1998) Energetics of drug-DNA interactions. Biopolymers 44(3):201–215
Knekt P, Jarvinen R, Seppanen R, Heliovaara M, Pukkala LT, Aromaa A (1997) Dietary flavonoids and the risk of lung cancer and other malignant neoplasms. Am J Epidemiol 146(3):223–230
Erlund I (2004) Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr Res 24(10):851–874
Sakaguchi Y, Maehara Y, Baba H, Kusumoto T, Sugimachi K, Newman RA (1992) Flavone acetic acid increases the antitumor effect of hyperthermia in mice. Cancer Res 52(12):3306–3309
Habtemariam S (1997) Flavonoids as inhibitors or enhancers of the cytotoxicity of tumor necrosis factor-alpha in L-929 tumor cells. J Nat Prod 60(8):775–778
Aljancic I, Vajs V, Menkovic N, Karadzic I, Juranic N, Milosavljevic S, Macura S (1999) Flavones and sesquiterpene lactones from Achillea atrata subsp. multifida: antimicrobial activity. J Nat Prod 62(6):909–911
Ko F, Chu C, Lin C, Chang C, Teng C (1998) Isoorientin-6″-O-glucoside, a water-soluble antioxidant isolated from Gentiana arisanensis. Biochim Biophys Acta 1389(2):81–90
Nkengfack A, Vouffo T, Fomun Z, Meyer M, Bergendorff SO (1994) Prenylated isoflavanone from the roots of Erythrina sigmoidea. Phytochemistry 36(4):1047–1051
Formica JV, Regelson W (1995) Review of the biology of quercetin and related bioflavonoids. Food Chem Toxicol 33(12):1061–1080
Cai YZ, Luo Q, Sun M, Corke H (2004) Antioxidant activity and phenolic compounds of 112 traditional Chinese medicinal plants associated with anticancer. Life Sci 74(17):2157–2184
Heim KE, Tagliaferro AR, Bobilya DJ (2002) Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. J Nutr Biochem 13(10):572–584
Li Y, Yang ZY, Wang MF (2009) Synthesis, characterization, DNA binding properties and antioxidant activity of Ln(III) complexes with hesperetin-4-one-(benzoyl) hydrazone. Eur J Med Chem 44(11):4585–4595
Cooray HC, Janvilisri T, Veen HW, Hladky SB, Barrand MA (2004) Interaction of the breast cancer resistance protein with plant polyphenols. Biochem Biophys Res Commun 317(1):269–275
So FV, Guthrie N, Chambers AF, Carroll KK (1997) Inhibition of estrogen receptor-positive MCF-7 human breast cancer cell proliferation by flavonoids in the presence and absence of excess estrogen. Cancer Lett 112(2):127–133
Mitsunaga Y, Takanaga H, Matsuo H, Naito M, Tsuruo T, Ohtani H, Sawada Y (2000) Effect of bioflavonoids on vincristine transport across blood-brain barrier. Eur J Pharmacol 395(3):193–201
Youdim KA, Dobbie MS, Kuhnle G, Proteggente AR, Abbott NJ, Rice-Evans C (2003) Interaction between flavonoids and the blood-brain barrier: in vitro studies. J Neurochem 85(1):180–192
Jim JO, Brown J, Fleming J, Harrison PR (2003) Effects of dietary flavonoids on major signal transduction pathways in human epithelial cells. Biochem Pharmacol 66(11):2075–2088
Howe GM, Wu KC, Bauer WR (1976) Binding of platinum and palladium metallointercalation reagents and antitumor drugs to closed and open DNAs. Biochemistry 15(19):4339–4346
Zeng YB, Yang N, Liu WS, Tang N (2003) Synthesis, characterization and DNA-binding properties of La(III) complex of chrysin. J Inorg Biochem 97(3):258–264
Efink MR, Ghiron CA (1981) Fluorescence quenching studies with proteins. Anal Biochem 114(2):199–227
Kumar CV, Turner RS, Asuncion EH (1993) Groove binding of a styrylcyanine dye to the DNA double helix: the salt effect. J Photochem Photobiol A Chem 74(2–3):231–238
Eriksson M, Leijon M, Hiort C, Norden B, Gradsland A (1994) Binding of Δ- and Λ-[Ru(phen)3]2+ to [d(CGCGATCGCG)]2 Studied by NMR. Biochemistry 33(17):5031–5040
Xiong Y, He XF, Zou XH, Wu JZ, Chen XM, Ji LN, Li RH, Zhou JY, Yu RB (1999) Interaction of polypyridyl ruthenium(II) complexes containing non-planar ligands with DNA. J Chem Soc Dalton Trans 1(1):19–24
Winterbourn CC (1979) Comparison of superoxide with other reducing agents in the biological production of hydroxyl radicals. Biochem J 182(2):625–628
Winterbourn CC (1981) Hydroxyl radical production in body fluids. Roles of metal ions, ascorbate and superoxide. Biochem J 198(1):125–131
Geary WJ (1971) Use of conductivity measurements in organic solvents for the characterization of coordination compounds. Coord Chem Rev 7(1):81–122
Marchetti F, Pettinari C, Leonesi D, Lorenzotti A (1999) Group 12 metal complexes of tetradentate N2O2-Schiff-base ligands incorporating pyrazole: synthesis, characterisation and reactivity toward S-donors, N-donors, copper and tin acceptors. Polyhedron 18(23):3041–3050
Narang KK, Singh VP (1993) Synthesis and characterization of cobalt(II), nickel(II), copper(II) and zinc(II) complexes with acetylacetone bis-benzoylhydrazone and acetylacetone bis-isonicotinoylhydrazone. Transition Met Chem 18(3):287–290
Dollberg CL, Turro C (2001) New quinone diimine complex of zinc with pH-dependent emission in the visible region. Inorg Chem 40(11):2484–2485
Kelly JM, Tossi AB, McConnell DJ, Uigin CO (1985) A study of the interactions of some polypyridylruthenium(II) complexes with DNA using fluorescence spectroscopy, topoisomerisation and thermal denaturation. Nucleic Acids Res 13(17):6017–6034
Pyle AM, Rehmann JP, Meshoyrer R, Kumar CV, Turro NJ, Barton JK (1989) Mixed-ligand complexes of ruthenium(II): factors governing binding to DNA. J Am Soc Chem 111(8):3051–3058
Chao H, Mei WJ, Huang QW, Ji LN (2002) DNA binding studies of ruthenium(II) complexes containing asymmetric tridentate ligands. J Inorg Biochem 92(3–4):165–170
Song YM, Wu Q, Yang PJ, Luan NN, Wang LF, Liu YN (2006) DNA Binding and cleavage activity of Ni(II) complex with all-trans retinoic acid. J Inorg Biochem 100(10):1685–1691
Wilson WD, Ratmeyer L, Zhao M, Stekowshi L, Boykin D (1993) The search for structure-specific nucleic acid-interactive drugs: effects of compound structure on RNA versus DNA interaction strength. Biochemistry 32(15):4098–4104
Xu H, Zheng KC, Lin LJ, Li H, Gao Y, Ji LN (2004) Coordination modes vs. antitumor activity: synthesis and antitumor activity of novel platinum(II) complexes of N-substituted amino dicarboxylic acids. J Inorg Biochem 98(1):87–97
Lepecq JB, Paoletti C (1967) A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol 27(1):87–106
Lee M, Rhodes AL, Wyatt MD, Forrow S, Harltley JA (1993) GC base sequence recognition by oligoimidazolecarboxamide and C-terminus-modified analogs of distamycin deduced from circular dichroism, proton nuclear magnetic resonance, and methidiumpropylethylenediaminetetraacetate-iron(II) footprinting studies. Biochemistry 32(16):4237–4245
Anbu S, Kandaswamy M, Suthakaran P, Murugan V, Varghese B (2009) Structural, magnetic, electrochemical, catalytic, DNA binding and cleavage studies of new macrocyclic binuclear copper(II) complexes. J Inorg Biochem 103(3):401–410
Lerman LS (1961) Structural considerations in the interaction of deoxyribonucleic acid and acridines. J Mol Biol 3(1):18–30
Satyanarayana S, Dabrowiak JC, Chaires JB (1993) Tris(phenanthroline)ruthenium(II) enantiomer interactions with DNA: mode and specificity of binding. Biochem 32(10):2573–2584
Liu JG, Zhang QL, Shi XF, Ji LN (2001) Interaction of [Ru(dmp)2(dppz)]2+ and [Ru(dmb)2(dppz)]2+ with DNA: effects of the ancillary ligands on the DNA-binding behaviors. Inorg Chem 40(19):5045–5050
Liu JG, Ye BH, Li H, Zhen QX, Ji LN, Fu YH (1999) Polypyridyl ruthenium(II) complexes containing intramolecular hydrogen-bond ligand: syntheses, characterization, and DNA-binding properties. Inorg Biochem 76(3–4):265–271
Towyz RM (2004) Reactive oxygen species, vascular oxidative stress, and redox signaling in hypertension. Hypertension 44(3):248–252
Fujimori T, Yamada S, Yasui H, Sakurai H, In Y, Ishida T (2005) Orally active antioxidative copper(II) aspirinate: synthesis, structure characterization, superoxide scavenging activity, and in vitro and in vivo antioxidative evaluations. J Biol Inorg Chem 10(8):831–841
Leonard SS, Keil D, Mehlmanb T, Proper S, Shi X, Harris GK (2006) Essiac tea: scavenging of reactive oxygen species and effects on DNA damage. J Ethnopharmacol 103(2):288–296
Li Y, Yang ZY, Li TR (2008) Synthesis, characterization, antioxidative activity and DNA binding properties of the Copper(II), Zinc(II), Nickel(II) complexes with 1, 2-Di(4′-iminonaringenin)ethane. Chem Pharm Bull 56(11):1528–1534
Xing RE, Yu HH, Liu S, Zhang WW, Zhang QB, Li ZE, Li PC (2005) Antioxidant activity of differently regioselective chitosan sulfates in vitro. Bioorg Med Chem 13(4):1387–1392
Acknowledgements
This work is supported by the National Natural Science Foundation of China (20975046) and Gansu NSF (0710RJZA012).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, Y., Yang, ZY. & Wang, MF. Synthesis, Characterization, DNA Binding Properties, Fluorescence Studies and Antioxidant Activity of Transition Metal Complexes with Hesperetin-2-hydroxy Benzoyl Hydrazone. J Fluoresc 20, 891–905 (2010). https://doi.org/10.1007/s10895-010-0635-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-010-0635-z