Abstract
Plants recognize biotic challengers and respond with the appropriate defense by utilizing phytohormone signaling and crosstalk. Despite this, microbes and insects have evolved mechanisms that compromise the plant surveillance system and specific defenses, thus ensuring successful colonization. In nature, plants do not experience insect herbivores and microbes in isolation, but in combination. Over time, relationships have developed between insects and microbes, varying on a continuum from no-relationship to obligate relationships that are required for both organisms to survive. While many reviews have examined plant-insect and plant-microbe interactions and the mechanisms of plant defense, few have considered the interface where microbes and insects may overlap, and synergies may develop. In this review, we critically evaluate the requirements for insect-associated microbes to develop synergistic relationships with their hosts, and we mechanistically discuss how some of these insect-associated microbes can target or modify host plant defenses. Finally, by using bioinformatics and the recent literature, we review evidence for synergies in insect-microbe relationships at the interface of plant-insect defenses. Insect-associated microbes can influence host-plant detection and/or signaling through phytohormone synthesis, conserved microbial patterns, and effectors, however, microbes associated with insects must be maintained in the environment and located in opportunistic positions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants experience an array of environmental insults including attack by pathogenic microorganisms and insect herbivores. Plants succeed despite these challenges, largely due to sophisticated defense systems that utilize morphological, biochemical, and molecular mechanisms (Howe and Jander 2008; Jones and Dangl 2006). For example, phytohormone signaling and crosstalk play a major role in the perception of invaders, and in the initiation of the appropriate defense response against the attacker (Erb et al. 2012; Pieterse et al. 2012). Nevertheless, pathogens and insects still successfully colonize plants by actively compromising plant perception and/or defense responses. While many reviews on plant-insect and plant-microbe interactions have described the general trends of defensive signaling in response to these biotic challenges (Howe and Jander 2008; Jones and Dangl 2006; Stout et al. 2006; Walling 2008, 2009), few have considered the interface where microbes and insects may overlap and where synergies may develop.
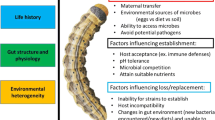
Recent efforts to catalog and characterize microbial diversity have increased public awareness of how microbe communities live in and on nearly all plant and animal species (Engel and Moran 2013; Hansen and Moran 2013; McFall-Ngai et al. 2013; Ng et al. 2011; Roossinck et al. 2010; Stobbe and Roossinck 2014; Vorholt 2012). As a result, it is increasingly clear that insect herbivores are not alone, but in fact harbor various viruses, fungi, and bacteria in their bodies, guts, saliva, and/or on the surface of their exoskeleton (Fig. 1). Plants too are colonized by numerous microbes both above and below ground, and in their phyllosphere and rhizosphere, respectively (Vorholt 2012). How plants effectively detect and respond to both microbe and herbivore natural enemies, and ignore microbes and herbivores that do not decrease plant fitness, is largely still unknown. In this review, we focus on insect herbivore-associated bacteria, and evaluate if these microbes are important for the ecology and evolution of insect–plant interactions. We evaluate the requirements for insect-associated bacteria to develop synergistic relationships with their hosts and mechanistically discuss how some of these microbes can target or modify host plant defenses. Finally, by using bioinformatics and the recent literature, we review evidence for synergies in insect-microbe relationships at the interface of plant-insect interactions. Recently, several papers and reviews examining the impact of virus infections of plant-insect interactions have been published and are highly relevant to this review, and we point readers to these for additional details (Casteel et al. 2014; Casteel and Jander 2013; Mauck et al. 2010, 2012; Stafford et al. 2011).
The insect microbiome is composed of environmental, gut, and intra/extracellular microbes. Microbes can easily deliver proteins and chemicals to the host plant surface through the insect’s fecal or oral material, and on the insect’s exoskeleton or eggs. In more stable associations, insects may introduce microbes intra- or inter-cellularly into host plants when feeding, chewing, or probing, directly impacting host plant regulatory networks, including phytohormones and related defenses. Mechanisms for manipulation by insect-associated microbes include phytohormone synthesis, Pathogen Associated Molecular Patterns (PAMPs), and effectors
Insect Herbivores Threaten Plants, Plants Respond Actively
Plants and insects are two of the most abundant macro-terrestrial taxa, making interactions almost unavoidable. Herbivory is perhaps the most important shared interaction between plants and insects, as it is the main route for the sun’s energy to enter the rest of the food web. The intimate associations that have developed between insects and plants have helped define and shape their evolution over time. Plants respond to herbivory with an active immune system that has the ability to recognize mechanical pressure, foreign molecules, and damaged cells (Boller and Felix 2009; Howe and Jander 2008; Pieterse et al. 2012). Defensive compounds produced can directly affect feeding, growth, or survival of herbivores (Mithofer and Boland 2012). In addition, some plants have indirect defense; recruiting the natural enemies of herbivores by releasing volatiles upon herbivore damage (Baldwin 2010; Hare 2011). These strategies either act independently, or in conjunction with one another depending on the timing of the attack, the attacker, and the ecological context.
Plants utilize phytohormones that coordinate recognition of individual attackers, and that determine the most appropriate defensive response. The phytohormones jasmonic acid (JA), salicylic acid (SA) and ethylene (ET) have major roles in plant defense (Bari and Jones 2009; Erb et al. 2012; Pieterse et al. 2012). However, recently the role of abscisic acid (ABA), giberellins (GA), auxins, cytokinins (CKs), and brassinosteroids in host plant signaling and herbivore defense also has been recognized (Bari and Jones 2009; McSteen and Zhao 2008; Robert-Seilaniantz et al. 2007). In general, the production of SA is critical for defensive responses to biotrophic pathogens, which obtain nutrients from living tissue (Glazebrook 2005; Stout et al. 2006). The production of JA and ET are important signals in the wound response to chewing herbivores, some phloem feeding herbivores, and in response to necrotrophic pathogens, which obtain nutrients from dead tissue (Glazebrook 2005; Howe and Jander 2008). Modulation in hormone composition, timing, and concentrations specify plant responses to an attack (Mur et al. 2006; Verhage et al. 2010). For example, the SA- and JA-signaling pathways often negatively influence each other (Doares et al. 1995; Leon-Reyes et al. 2010b; Mur et al. 2006). However if SA is induced prior to JA, JA inhibition is prevented (Koornneef et al. 2008). Additionally, induction of ET signaling can make the JA pathway insensitive to SA-mediated suppression (Leon-Reyes et al. 2010a). For a more comprehensive review on insect-plant interactions, and phytohormone signaling and crosstalk, see Erb et al. (2012), Howe and Jander (2008), McSteen and Zhao (2008), Pieterse et al. (2012), or Walling (2009).
Insects are not Alone: Which Microbes Matter
Maintenance of microbial associates in an herbivore population is necessary if the microbe (s) are to influence the insect’s fitness, and ultimately shape its ecology and evolution with its host-plant. Insect herbivores are widely associated with fungal, bacterial, and viral microbiomes that can be located externally on the insect’s exoskeleton or within the insect’s gut, or inside the insect’s body, including the salivary glands, and within or between insect cells (Brault et al. 2010; Engel and Moran 2013; Hansen and Moran 2013; Oliver et al. 2010; Roossinck 2012) (Fig. 1). Intracellular bacterial symbionts that are necessary for insect survival are prevalent among most sap-feeding insects, as they provide the essential nutrients that are in low abundance or completely absent from the insect’s sap diet (Hansen and Moran 2013). Most obligate symbionts are stably inherited through maternal transmission, and all insect individuals of a species harbor the same bacterial symbiont taxa. Many non-obligate endosymbionts (facultative endosymbionts), which include several plant pathogens, also are inherited maternally, however, they generally do not infect all individuals within a population, and many strains may exist (Casteel et al. 2012; Oliver et al. 2010). Even though facultative endosymbionts are not required for insect survival, specific strains are known to confer beneficial effects on their insect host’s phenotype depending on the environmental context (Oliver et al. 2010, 2013). Another subset of insect-associated microbes is insect-vectored plant pathogens, which can form highly specialized and often beneficial relationships with their insect vectors. Some of these plant pathogens, can be vertically transmitted, host specific, and persistently maintained in insect populations (reviewed in Hansen and Moran 2013).
In contrast to intracellular endosymbionts and some insect-vectored plant pathogens, gut and environmental microbes generally do not have stable associations with insect herbivores (Colman et al. 2012; Hansen and Moran 2013). Exceptions exist, and these stable gut and environmental microbes generally are maintained either by maternal transmission, selective host acquisition, and/or the insect’s social behavior (Engel and Moran 2013; Hansen and Moran 2013). It currently is unclear if this lack of evidence for microbiome stability of insect gut and environmental microbes is related to insufficient sampling, or simply because stable core microbiomes are not commonly associated with most insect herbivores. Disentangling the microbes that constitute the core insect and core plant microbiomes will be challenging. For example, Vorholt (2012) have shown that some plant species harbor core microbiomes in their phyllospheres, which are independent of the plant’s geography. Thus, a microbiome specific to a plant species can falsely be identified as a persistent gut microbiome of an herbivore specialist, when in reality it is just microbially contaminated host plant material passing through the digestive tract of the insect. More landscape and population level insect and plant microbiome studies will be needed in the future to dissect these relationships.
Another phenomenon that further complicates studying the interface where insect microbiomes and plants intersect, is the evolutionary process of horizontal gene transfer (HGT). This process of gene exchange can occur among microbes (Ochman et al. 2005), or remarkably between a microbe species and the insect or plant (Armijos Jaramillo et al. 2013; Husnik et al. 2013; Nikolaidis et al. 2014; Sloan et al. 2014). For example, individual microbes that encode gene cassettes that modulate host-plant interactions (e.g., secretion systems, toxins) can be transferred horizontally to unrelated microbes. Recipient microbes then can take on a similar functional role with the host-plant. As such, screening for 16S ribosomal RNA similarity alone will not provide direct functional evidence of how an individual microbe species interacts with a given host plant.
Although highly specific gene cassettes are critical for some insect-microbe- interactions (Dale et al. 2001; Hansen et al. 2012; Oliver et al. 2009), we predict that more generalized interactions also may result from broadly conserved bacterial genes in insect-plant interactions. In these situations, the identity of a specific bacterial strain may not necessarily be important for inducing a particular host-plant interaction, but instead the maintenance of any bacterium may be sufficient. Future metagenomic studies of microbial populations associated with insect-plant interactions have the capability to identify the microbe-encoded proteins involved in highly specialized, and/or generalized plant responses. This distinction may be critical because highly specialized interactions may require the insect to maintain a particular microbe (or gene set) leading to impacts on the insect’s ecology and evolution, whereas generalized interactions most likely result in more transient and diffuse relationships between the insect host and microbe.
In addition to the stability of a particular microbe (s) within insect populations, and/or the fidelity of a specific microbe-encoded gene set, the location of the microbe in/on the insect is expected to influence the microbe’s potential role in insect-host-plant interactions. For example, physical proximity of the microbe to the host plant can influence the relative amount of microbial proteins and/or active enzymes that can be successfully delivered to the plant tissue. An environmental or gut microbe (s) would be in a more likely position to deliver large quantities of microbial compounds to the plant, compared to an endosymbiont. The location of the microbe in/on the insect also affects the microbe’s genomic architecture and evolution. For example, when a microbe transitions from a free-living lifestyle, as a gut or environmental microbe, to a maternally inherited lifestyle as an endosymbiont, several hallmarks of endosymbiont genome evolution result (McCutcheon and Moran 2012). Genome size becomes remarkably reduced, mutational AT bias generally occurs, and in the most extreme cases, which is seen in most obligate symbionts, DNA repair machinery and genes involved in horizontal gene transfer are lost (McCutcheon and Moran 2012). In turn, obligate endosymbiont genomes generally only retain a small subset of core genes required for their symbiotic lifestyle, and are not able to incorporate novel genes, in contrast to the dynamic genomes of gut, environmental, and some facultative symbionts (McCutcheon and Moran 2012).
Mechanisms and Evidence: Manipulation of Phytohormones and Plant Defense
Insect microbes can influence host-plant detection and/or signaling, if they are stably maintained and located in opportunistic positions in the environment, gut, or salivary glands. These microbes can easily deliver proteins to the host plant surface through the insect’s fecal or oral material, and on the insect’s exoskeleton or eggs. In more stable associations, insects may introduce microbes intra- or inter-cellularly into host plants when feeding, chewing, or probing, directly impacting host plant regulatory networks, including phytohormones and related defenses. Below, we discuss mechanisms by which insect-associated microbes may manipulate phytohormones and related defenses (Fig. 1), and the specific evidence from the literature if available (Table 1).
I. Mechanisms: Phytohormone Synthesis, Microbe Associated Molecular Patterns, and Effectors
Phytohormone Synthesis
Plant hormones regulate growth, development, and plant responses to the environment including attack by insects and pathogens (Bari and Jones 2009; Durbak et al. 2012; Erb et al. 2012; Pieterse et al. 2012; Veit 2009). Many microbes take advantage of these control points to manipulate plant development and defenses by synthesizing phytohormones or their functional mimics. This results in inappropriate signals and defense responses in the plant (Robert-Seilaniantz et al. 2007). One example of this is in Pseudomonas syringae, a pathogenic bacterium that produces coronatine, a JA-IIe mimic, the bioactive form of JA. Coronatine triggers JA defense responses when introduced into the plant leading to the inhibition of SA-dependent defenses and increased susceptibility to the pathogen (Cui et al. 2005). Another example of phytohormone synthesis by microbes is in the gall-forming bacteria, Agrobacterium tumefaciens that produces auxin and cytokinins (Patten and Glick 1996; Pertry et al. 2009; Spaepen and Vanderleyden 2011). Auxin and cytokinins modulate plant development and growth (Benjamins and Scheres 2008; Durbak et al. 2012; Werner and Schmulling 2009). Production of these phytohormones allows the bacterium to modulate cell proliferation at the site of infection, contributing to gall formation and disease establishment (Hwang et al. 2010; Stes et al. 2011). In addition, auxin and cytokinins have been shown to have an inhibitory impact on SA signaling (Choi et al. 2010), and certain free-living biotrophic pathogens exploit this by synthesizing these phytohormones that suppress SA and increase susceptibility of the host (Chen et al. 2007). Many root-colonizing bacteria also have been shown to produce large quantities of phytohormones, altering root architecture and nutrient acquisition (Chen et al. 2007). The ability of microbes to synthesize phytohormones or functional mimics is widespread (Glickmann et al. 1998; Jameson 2000).
Recent studies have demonstrated that insects have evolved ways to manipulate plant signaling for their own benefit by modulating hormone pathways (Weech et al. 2008; Zarate et al. 2007; Zhang et al. 2011, 2013). For example, immature whiteflies induce SA signaling, thus preventing the activation of JA–dependent host defenses against the insects (Zarate et al. 2007). Further, it has been demonstrated that Myzus persicae secrete salivary proteins into their host plant during feeding, which subsequently alters plant defense responses (Pitino and Hogenhout 2012). Insects and their microbe associates may act together and manipulate plant signaling and defense, potentially increasing the palatability of the plant for the insect (Fig. 1). Examples of phytohormone or phytohormone-mimic synthesis have not yet been documented in insect-associated-bacteria. However, multiple phytohormones have been identified in aphid honeydew including salicylic acid, auxin, giberillic acid, and cytokinins (Cleland and Ajami 1974; Hussain et al. 1974), and recently non-plant based cytokinins have been identified in lepidopteran bodies (Body et al. 2013). Insect-associated bacteria may be producing these phytohormones (Fig. 1), or alternatively the insect itself may be sequestering them from the plant during feeding. The growing availability of microbe and insect genome sequencing projects will shed light on this possibility, allowing the detection of phytohormone synthesis genes, and experimentally testing their functionalities.
Microbe Associated Molecular Patterns and Effectors
Upon plant entry, common residues from microbes and damaged cells are recognized, indicating infection/invasion to the plant. Recognized signals generally are conserved across bacteria and known as Microbe/Pathogen Associated Molecular Patterns (MAMPs/PAMPs). MAMPs/PAMPs are general elicitors of non-specific plant immunity (PAMP-triggered immunity (PTI)) (Jones and Dangl 2006). Activation of PTI triggers resistance mechanisms that are effective across a broad range of pathogens/microbes. However, microbes have evolved ways that overcome PTI (Bardoel and Strijp 2011; Cui et al. 2009). Successful microbes produce effectors, which can prevent detection of their PAMPs or suppress PTI. In response to the microbe’s effectors, plants have evolved mechanisms that recognize pathogen effectors by activating effector-triggered immunity (ETI) (Cui et al. 2009). A significant amount of research has examined various mechanisms utilized by microbes that overcome and manipulate plant defense. However, these studies largely focus on non-insect-associated bacteria and pathogens (Fu and Dong 2013; Henry et al. 2013; Pieterse et al. 2012; Stout et al. 2006). Many facultative insect symbionts, insect vectored pathogens, and most likely many environmental and gut microbes, also encode PAMPs and virulence factors, similar to plant and animal pathogens, such as TypeI or TypeIII secretion systems, iron, amino acid, carbon transporters, and/or modifiers of eukaryotic hormonal pathways (Dale et al. 2002; Dale and Moran 2006; Ochman and Moran 2001; Pontes et al. 2011). Consequently, it is highly probable that some insect-associated bacteria have the genetic capability to produce PAMPs and effectors that impact insect-host plant interactions (Fig. 1).
PAMPs from intra/extra-cellular, gut, or environmental insect bacteria may be introduced onto the host plant through oral secretions, or through excreta (honeydew/frass) (Leroy et al. 2011; Sabri et al. 2013) (Fig. 1). A panel of known PAMPs was used to investigate the diversity and distribution of conserved PAMP homologs encoded in insect herbivore-associated microbial genomes (from Mukhtar et al. 2011). PAMPs from six fully sequenced plant and animal microbe-associates (pathogens, commensals, and symbionts) were used as blastp queries to identify putative homologous PAMPs from 45 insect-associated bacterial genomes, which were each used as blastp databases. These 45 bacterial genomes represent the only insect herbivore-associated bacterial genomes that are fully sequenced or near complete in draft form, and that are publicly available from the Joint Genome Institute (JGI), Integrated Microbial Genomes/Expert Review (IMG/ER) database as of 05/2014 (please see Supplementary file 1 for more details on methods and analysis). As expected, most of these insect-associated bacteria encode conserved PAMPs, such as flagellar genes, lipopolysaccharide biosynthesis genes, and the Elongation factor Tu, with the exception of a few obligate endosymbionts with tiny genomes (McCutcheon and Moran 2012) (Supplementary file 1). Facultative and obligate endosymbionts produce proteins inside of the insect’s tissues or cells, and therefore, delivery of PAMPs to the leaf surface is expected to be less likely. Surprisingly, a recent proteomic study of pea aphid honeydew revealed that almost half of the identified proteins in aphid honeydew were not only homologous to several gut microbes, but to a facultative endosymbiont, and to a lesser extent the obligate endosymbiont, Buchnera aphidicola (Sabri et al. 2013). Another recent proteomic study identified an obligate endosymbiont bacterial protein, GroEL in aphid saliva (Vandermoten et al. 2014). GroEL is a highly conserved and highly expressed heat shock chaperone in bacteria, and has been retained in nearly every insect endosymbiont (Filichkin et al. 1997). In turn, GroEL may act as a PAMP when introduced into host plants, potentially inducing plant defenses. Consistent with this hypothesis, a recent study identified GroEL in aphid saliva (Macrosiphum euphorbiae) and infiltrating GroEL into plant leaves induced an oxidative burst and marker transcripts of PTI (Chaudhary et al. 2014). Additionally, transgenic plants engineered to express GroEL decreased aphid fecundity compared to control plants (Chaudhary et al. 2014). In another study, Elzinga et al. 2014 found similar impacts of GroEL on aphid fecundity. Additionally, expression of PR1, a marker transcript of SA defenses, also was induced in transgenic plants expressing GroEL compared to controls (Elzinga et al. 2014). These data suggest that PAMPs of the insect microbiome may have the potential to influence insect-plant interactions. However, plant specific microbes also are commonly present on the leaf surface (Vorholt 2012), and therefore, how and if the plant can distinguish between different PAMPs and respond accordingly, remains unclear.
Effectors from the insect microbiome can be delivered into the host-plant through insect feeding behavior (Fig. 1). As with the PAMPs, we conducted a genomic analysis to explore if known plant pathogen effector homologs also are encoded in insect-associated bacterial genomes from insect herbivores (for methods see Supplementary file 1). From our analysis, no known plant pathogen effectors were identified in any of the obligate endosymbionts. However, two core microbial genes that are conserved in bacteria, the molecular chaperone protein DnaJ, and the 16S ribosomal RNA methyltransferase (RsmE), display significant homology to the domains in the plant effectors Hopl1 and Candidate 4480, respectively, from Pseudomonas syringae (Supplementary file 1). Most likely, plant effectors Hopl1 and Candidate 4480 are paralogs of core genes DnaJ and RsmE, and therefore, they encode different functions for the microbe. Interestingly, two facultative endosymbionts Regiella insecticola R5.15 and Hamiltonella defense MED, which are harbored intra and inter-cellularly in aphids and whiteflies, respectively, encode genes that are homologous to four plant pathogen effectors (Supplementary file 1). However, based on protein sequence similarity, we suggest that these effector candidates in R. insecticola R5.15 likely target insects not plants because they are more similar to mcf- toxins, rtx-toxins, and Invasion proteins known to target animals (Daborn et al. 2002; Satchell 2011). In addition, this Regiella strain (R5.15) was shown to confer protection toward aphids against an insect parasitoid potentially by utilizing these virulence factors (Hansen et al. 2012).
II. Evidence: Environmental, Gut, and Intra/Extracellular Microbes
Environmental and Gut Microbes
Herbivores possess diverse microbes in their digestive systems, and recent research has demonstrated that these gut microbes can modify plant–insect interactions. For example, during host-plant feeding, the Colorado potato beetle (Leptinotarsa decemlineata) introduces environmental/gut bacteria into the plant, inducing transcripts related to JA and SA signaling in tomato (Chung et al. 2013) (Table 1). When beetles are cured of bacteria using antibiotics, transcripts related to JA signaling are strongly induced compared to feeding by untreated beetles. Bacteria introduced into the plant in isolation of insects, elicit marker genes of SA-signaling and inhibit JA related transcripts and defense responses (Chung et al. 2013).This suggests that JA induction by beetle feeding is suppressed when they secrete their environmental/gut microbes into the host-plant. Additionally, re-introducing the bacteria to antibiotic-treated larvae restores the insect’s ability to suppress defenses (Chung et al. 2013). As SA signaling often inhibits jasmonate signaling (Leon-Reyes et al. 2010b; Mur et al. 2006), larvae may exploit bacteria in their oral secretions and suppress plant defenses (Table 1). The authors hypothesized that bacteria are manipulating signaling and defenses through PAMPs introduced into the host plant during insect feeding. Mechanistically, they demonstrated a flagellin protein which is encoded in a Pseudomonas species isolated from the beetle’s oral secretions was able to suppress plant defenses. (Chung et al. 2013). It would be of interest to investigate how generalized or specific this host plant response is toward other flagellin proteins encoded in other microbes associated with the host-plant and beetle microbiomes.
In another recent study, Chu et al. (2013) revealed that the gut microbiome of the western corn rootworm, Diabrotica virgifera virgifera, differed between beetle populations that fed primarily on corn compared to populations that were adapted to soybean (Table 1). They demonstrated that the insects’ guts from the soybean-adapted insects had higher protease activity compared to antibiotic-treated insects. The authors suggested that the gut bacteria may be important for maintaining protease activity and tolerating plant protease inhibitors introduced in the insect's soybean diet (Chu et al. 2013). The direct impact of the bacteria on protease activity or the ability of microbes to detoxify protease inhibitors was not tested in Chu et al. (2013). Moreover, antibiotics used to treat insects in this study previously have been shown to directly suppress protease enzyme activity (Castro and Tanus-Santos 2013; Hirakata et al. 1992). For both Chu et al. 2013 and Chung et al. 2013, it is unclear if a specific gut microbe (s) and the specialized gene cassettes they encode may facilitate these altered host-plant interactions. If so, determining if these specific strains are persistently maintained in beetle's populations, and thus, can ultimately impact the insect’s ecology and evolution will be required. Alternatively, these gut/environmental microbes may have more of a generalized host-plant response through PAMPs, and therefore, these insect-microbe interactions may be more diffuse resulting in dynamic and transient host-plant interactions in nature.
Intra/Extracellular Microbes
One of the best examples illustrating the effect of insect microbes on plant signaling and metabolism was conducted on Arabidopsis thaliana and the leafhopper Macrosteles quadrilineatus, which vectors Aster Yellows phytoplasma, strain Witches’ Broom (AY-WB) (Sugio et al. 2011) (Table 1). AY-WB is obtained by the leafhopper during feeding on infected-plants. After an incubation period in the insect’s body, AY-WB moves to the salivary gland where it can be injected back into a host plant during feeding. Leafhoppers feeding on AY-WB-infected plants have higher fecundity compared to uninfected plants, primarily due to the phytoplasma’s ability to inhibit JA. Further Sugio et al. (2011) found that the phytoplasma’s effector protein (SAP11) is responsible for JA inhibition, by destabilizing two Arabidopsis transcription factors (Sugio et al. 2011) (Table 1).
In another specialized insect–plant pathogen system, the tomato psyllid Bactericera cockerelli vectors the alphaproteobacterium, Liberibacter psyllaurous (same as Liberibacter solanacearum), into solanaceous host plants (Casteel et al. 2012; Hansen et al. 2008). Liberibacter psyllaurous is associated with the plant disease, psyllid yellows, and is vertically transmitted and maintained throughout psyllid development (Casteel et al. 2012; Hansen et al. 2008). When L. psyllaurous is inoculated into tomato (Solanum lycopersicum) alone without the insect, tomato genes induced by JA and SA signaling are suppressed (Casteel et al. 2012). Similarly, when infected psyllids feed on tomato plants, defensive transcript induction is dampened for psyllid life stages that harbor the highest concentrations of the bacterium (Casteel et al. 2012) (Table 1). Further studies are needed in this system to dissect the molecular mechanisms mediating psyllid interactions with host plants in isolation of the microbe. Collectively, these results suggest that the bacterium can suppress defense transcripts and may play a role in suppressing insect-related host plant defenses when L. psyllaurous is vectored into tomato by the psyllid (Table 1).
Liberibacter species also are implicated in manipulating plant signaling and psyllid–host plant interactions through volatile production (Davis et al. 2012; Mann et al. 2012) (Table 1). Plants respond to herbivore attack and pathogen infection by releasing volatiles. Release of volatiles can prevent further colonization by conspecific insects, attract vectors to infected plants, and recruit natural enemies of the herbivores to the plant (Baldwin 2010). In one system, L. asiaticus, infected citrus plants are initially more attractive to adult psyllids (Diaphorina citri) than non-infected plants; however, psyllids that subsequently disperse prefer to settle on non-infected plants rather than infected plants (Mann et al. 2012). Initial attraction to infected plants by psyllids did not depend on infection status of the insect, suggesting changes in insect behavior are mediated through the host plant. Mann et al. determined that levels of methyl-salicylate were elevated in Liberibacter-infected plants, and that psyllids are attracted to this compound (Mann et al. 2012). Methyl salicylate is a derivative of SA and has been implicated as the mobile signal in systemic acquired resistance, an inducible resistance that is triggered in systemic healthy tissue of infected plants (Vlot et al. 2008).
In a similar system, L. psyllaurous infection of potatoes also initially attracts psyllid vectors, but ultimately adults move from infected plants and prefer to settle on healthy potato plants (Solanum tuberosum) (Davis et al. 2012) (Table 1). The role of the bacteria in potato-psyllid manipulations is not clear. Infected potatoes had increased levels of β–caryophyllene, but attraction to the compound was not demonstrated. β-Caryophyllene emissions inhibit bacterial growth in Arabidopsis, and may be a direct defense response to L. psyllaurous (Huang et al. 2012). β-Caryophyllene also may be a direct defense to the insect, as it decreases the growth and survival of cotton herbivores (Langenheim 1994), and serves as an indirect defense attracting natural enemies and parasitic wasps to herbivores in maize (Kollner et al. 2008; Rasmann et al. 2005). Thus, β-caryophyllene induction by the bacteria may not be responsible for attraction, but instead the deterrence observed in the system (Davis et al. 2012).
Cytokinins are plant hormones involved in growth and cell division, nutrient mobilization, and inhibition of senescence (Werner and Schmulling 2009). Recently, cytokinins have emerged as potential players in a variety of insect–plant and microbe–plant interactions as well (Choi et al. 2011; Perilli et al. 2010; Werner and Schmulling 2009) (Table 1). The leaf-miner Phyllonorycter blancardella and a facultative symbiont Wolbachia, survives in senescing apple leaves by feeding within photosynthetically active green patches on the leaf. The green patches and insects contain increased levels of cytokinines (Body et al. 2013; Kaiser et al. 2010). After treatment with antibiotics, the leaf miner cannot produce green patches in senescing leaves, and cytokinin concentrations decline in herbivore leaves and insect bodies. In contrast, untreated leaf miners maintain Wolbachia infections, cytokinin levels, produce green patches, and display higher fitness (Body et al. 2013; Kaiser et al. 2010). Therefore, symbiont presence in the leaf miner corresponds to green patches and higher cytokinin concentrations (Table 1). In this system, it is not clear if and how Wolbachia is involved in green-patch production, as the moth or symbiont could be producing cytokinin, and the phenomenon may be caused by a different microbe that is present and that can also be eliminated during antibiotic treatments.
In another system, the western corn rootworm, harbors Wolbachia as an endosymbiont. When infected beetles feed on corn, defense-related genes are down-regulated in the host plant compared to when antibiotic-treated beetles feed (Barr et al. 2010) (Table 1). It was not tested if Wolbachia alone could suppress plant defense transcripts, can infect plant tissue, or if Wolbachia proteins are injected into the host plant. However, the suppression of plant defense transcripts and mRNAs could not be reproduced in another study (Robert et al. 2013), and this suggests that findings may be context dependent (Table 1).
Conclusions and Future Directions
Recent literature on manipulation of phytohormones and plant defenses by insect-associated bacteria is rapidly growing, however, there are still many unanswered questions and gaps of knowledge that need to be addressed (see below). Few studies have actually dissected the molecular mechanisms mediating hormone and defense manipulation in these systems (Sugio et al. 2011). In particular, how insect-associated bacteria alter plant signaling either through specialized interactions or generalized responses needs to be addressed (Table 1, Box 1). In addition, these studies often frame relationships as beneficial to one or both players suggesting mutualisms or commensalisms. However, the impact of these relationships often are not fully elucidated, and the selfish benefit of the microbe itself generally has been ignored (Table 1). To better understand plant-insect relationships, and the functional role of microbes in them, additional studies must examine the ecological relevance for both players. Finally, this increased interest in the literature will certainly continue and be aided by increased use of molecular genetics and genomics in identifying potential players and mechanisms. Bacteria, fungi, and viruses largely have been studied in relation to their ability to cause disease in agriculturally important crops. However, it is becoming increasingly apparent in natural systems that viruses, bacteria, and fungi exist often without visible symptoms of disease in the plant and animal host. Microbiomes, including viruses, fungi, and bacteria, will be identified on and in virtually every organism and in every environment, and as scientists our challenge will be to determine if these microbial associates are ecologically and evolutionarily relevant to the plant-insect interaction.
Future questions to address on the effect of the insect microbiome on insect-plant interactions:
-
1)
What plant signaling and defense mechanisms are altered by insect-associated bacteria?
-
2)
How does plant phyllosphere-dwelling organisms influence insect-plant interactions?
-
3)
How does the insect microbiome impact the plant phyllosphere?
-
4)
How do insect-associated bacteria manipulate plant hormone and defense signaling? Are putative microbe genes specialized and present in specific microbe strains, or are they conserved and present in most bacterial species?
-
5)
What impact do insect associations have on the microbe? Are they ecologically and evolutionarily important for the microbe?
References
Armijos Jaramillo VD, Vargas WA, Sukno SA, Thon MR (2013) New insights into the evolution and structure of Colletotrichum Plant-Like Subtilisins (CPLSs). Comm & Integ Biol 6:e25727. doi:10.4161/cib.25727
Baldwin IT (2010) Plant volatiles. Curr Biol 20:392–397. doi:10.1016/j.cub.2010.02.052
Bardoel BW, Strijp JA (2011) Molecular battle between host and bacterium: recognition in innate immunity. J Mol Recogn 24:1077–1086. doi:10.1002/jmr.1156
Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488. doi:10.1007/s11103-008-9435-0
Barr KL, Hearne LB, Briesacher S, Clark TL, Davis GE (2010) Microbial symbionts in insects influence down-regulation of defense genes in maize. PLoS One 5:e11339. doi:10.1371/journal.pone.0011339
Benjamins R, Scheres B (2008) Auxin: the looping star in plant development. Annu Rev Plant Biol 59:443–465. doi:10.1146/annurev.arplant.58.032806.103805
Body M, Kaiser W, Dubreuil G, Casas J, Giron D (2013) Leaf-miners co-opt microorganisms to enhance their nutritional environment. J Chem Ecol 39:969–977. doi:10.1007/s10886-013-0307-y
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406. doi:10.1146/annurev.arplant.57.032905.105346
Brault V, Uzest M, Monsion B, Jacquot E, Blanc S (2010) Aphids as transport devices for plant viruses. C R Biol 333(6–7):524–538. doi:10.1016/j.crvi.2010.04.001
Casteel CL, Jander G (2013) New synthesis: investigating mutualisms in virus-vector interactions. J Chem Ecol 39:809
Casteel CL, Hansen AK, Walling LL, Paine TD (2012) Manipulation of plant defense responses by the tomato psyllid (Bactericerca cockerelli) and its associated endosymbiont Candidatus Liberibacter psyllaurous. PLoS One 7:e35191. doi:10.1371/journal.pone.0035191
Casteel CL, Yang C, Nanduri AC, Whitham SA, Jander G (2014) The Nia-Pro protein of turnip mosaic virus improves growth and reproduction of the aphid vector, Myzus persicae (green peach aphid). Plant J 77:653–663
Castro MM, Tanus-Santos JE (2013) Inhibition of matrix metalloproteinases (MMPs) as a potential strategy to ameliorate hypertension-induced cardiovascular alterations. Curr Drug Targets 14:335–343
Chaudhary R, Atamian HS, Shen Z, Briggs SP, Kaloshian I (2014) GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc Natl Acad Sci U S A 111(24):8919–8924
Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci U S A 104:20131–20136. doi:10.1073/pnas.0704901104
Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I (2010) The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell 19:284–295. doi:10.1016/j.devcel.2010.07.011
Choi J, Choi D, Lee S, Ryu CM, Hwang I (2011) Cytokinins and plant immunity: old foes or new friends? Trends Plant Sci 16:388–394. doi:10.1016/j.tplants.2011.03.003
Chu C-C, Spencer JL, Curzi MJ, Zavala JA, Seufferheld MJ (2013) Gut bacteria facilitate adaptation to crop rotation in the western corn rootworm. Proc Natl Acad Sci U S A 110:11917–11922. doi:10.1073/pnas1301886110
Chung SH, Rosa C, Scully ED, Peiffer M, Tooker JF, Hoover K, Luthe DS, Felton GW (2013) Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc Natl Acad Sci U S A 110:15728–15733. doi:10.1073/pnas.1308867110
Cleland CF, Ajami A (1974) Identification of the flower-inducing factor isolated from aphid honeydew as being salicylic acid. Plant Physiol 54:904–906
Colman DR, Toolson EC, Takacs-Vesbach CD (2012) Do diet and taxonomy influence insect gut bacterial communities? Mol Ecol 21:5124–5137. doi:10.1111/j.1365-294x.2012.05752.x
Cui H, Bahrami AK, Pringle EG, Hernadez-Guzman G, Bender CL, Pierce NE, Ausubel F (2005) Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci U S A 102:1791–1796
Cui H, Xiang T, Zhou JM (2009) Plant immunity: a lesson from pathogenic bacterial effector proteins. Cellu Microbiol 11:1453–1461. doi:10.1111/j.1462-5822.2009.01359.x
Daborn PJ, Waterfield N, Silva CP, Au C, Sharma S, Ffrench-Constant RH (2002) A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc Natl Acad Sci U S A 99:10742–10747. doi:10.1073/pnas.10268099
Dale C, Moran NA (2006) Molecular interactions between bacterial symbionts and their hosts. Cell 126:453–465. doi:10.1016/j.cell.2006.07.014
Dale C, Young SA, Haydon DT, Welburn SC (2001) The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci U S A 13(98):1883–1888
Dale C, Plague GR, Wang B, Ochman H, Moran NA (2002) Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci U S A 99:12397–12402. doi:10.1073/pnas.182213299
Davis TS, Horton DR, Munyaneza JE, Landolt PJ (2012) Experimental infection of plants with an herbivore-associated bacterial endosymbiont influences herbivore host selection behavior. PLoS One 7:e49330. doi:10.1371/journal.pone.0049330
Doares SH, Narvaez-Vasquez J, Conconi A, Ryan CA (1995) Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol 108(4):1741–1746
Durbak A, Yao H, McSteen P (2012) Hormone signaling in plant development. Curr Opin Plant Biol 15(1):92–96. doi:10.1016/j.pbi.2011.12.004
Elzinga DA, De Vos M, Jander G (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol Plant Microbe Interact 27:747–756
Engel P, Moran NA (2013) The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi:10.1111/1574-6976.12025
Erb M, Meldau S, Howe GA (2012) Role of phytohormones in insect-specific plant reactions. Trends Plant Sci 17:250–259. doi:10.1016/j.tplants.2012.01.003
Filichkin SA, Brumfield S, Filichkin TP, Young MJ (1997) In vitro interactions of the aphid endosymbiotic SymL chaperonin with barley yellow dwarf virus. J Virol 71:569–577
Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64:839–863. doi:10.1146/annurev-arplant-042811-105606
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopath 43:205–227
Glickmann E, Garden L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y (1998) Auxin produciton is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 11:156–162
Hansen AK, Moran NA (2013) The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol 23:1473–1496. doi:10.1111/mec.12421
Hansen AK, Trumble JT, Stouthamer R, Paine TD (2008) A new huanglongbing species, “Candidatus Liberibacter psyllaurous” found to infect tomato and potato, is vectored by the psyllid Bactericera cockerelli (Sulc). App Environ Microbiol 74:5862–5865. doi:10.1128/aem.01268-08
Hansen AK, Vorburger C, Moran NA (2012) Genomic basis of endosymbiont-conferred protection against an insect parasitoid. Genome Res 22:106–114. doi:10.1101/gr.125351.111
Hare JD (2011) Ecological role of volatiles produced by plants in response to damage by herbivorous insects. Annu Rev Entomol 56:161–180. doi:10.1146/annurev-ento-120709-144753
Henry E, Yadeta KA, Coaker G (2013) Recognition of bacterial plant pathogens: local, systemic and transgenerational immunity. New Phytol 199:908–915. doi:10.1111/nph.12214
Hirakata Y, Kaku M, Mizukane R, Ishida K, Furuya N, Matsumoto T, Tateda K, Yamaguchi K (1992) Potential effects of erythromycin on host defense systems and virulence of Pseudomonas aeruginosa. Antimicrob Agents Chemother 36:1922–1927
Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59:41–66. doi:10.1146/annurev.arplant.59.032607.092825
Huang M, Sanchez-Moreiras AM, Abel C, Sohrabi R, Lee S, Gershenzon J, Tholl D (2012) The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-beta-caryophyllene, is a defense against a bacterial pathogen. New Phytol 193:997–1008. doi:10.1111/j.1469-8137.2011.04001.x
Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M, Tanaka M, Satoh N, Bachtrog D, Wilson ACC, von Dohlen CD, Fukatsu T, McCutcheon JP (2013) Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153:1567–1578
Hussain A, Forrest JM, Dixon AF (1974) Sugar, organic acid, phenolic acid and plant growth regulator content of extracts of honeydew of the aphid Myzus persicae and of its host plant, Raphanus sativus. Ann Appl Biol 78:65–73
Hwang HH, Wang MH, Lee YL, Tsai YL, Li YH, Yang FJ, Liao YC, Lin SK, Lai EM (2010) Agrobacterium-produced and exogenous cytokinin-modulated Agrobacterium-mediated plant transformation. Mol Plant Pathol 11:677–690. doi:10.1111/j.1364-3703.2010.00637.x
Jameson P (2000) Cytokinins and auxins in plant-pathogen interactions - an overview. Plant Growth Regul 32:369–380
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329. doi:10.1038/nature05286
Kaiser W, Huguet E, Casas J, Commin C, Giron D (2010) Plant green-island phenotype induced by leaf-miners is mediated by bacterial symbionts. Proc Biol Sci 277:2311–2319. doi:10.1098/rspb.2010.0214
Kollner TG, Held M, Lenk C, Hiltpold I, Turlings TC, Gershenzon J, Degenhardt J (2008) A maize (E)-beta-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell 20:482–494. doi:10.1105/tpc.107.051672
Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, Van Loon LC, Pieterse CM (2008) Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol 147:1358–1368. doi:10.1104/pp. 108.121392
Langenheim JH (1994) Higher plant terpenoids: a phytocentric overview of their ecological roles. J Chem Ecol 20:1223–1280. doi:10.1007/bf02059809
Leon-Reyes A, Du Y, Koornneef A, Proietti S, Korbes AP, Memelink J, Pieterse CM, Ritsema T (2010a) Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol Plant Microbe Interact 23:187–197. doi:10.1094/mpmi-23-2-0187
Leon-Reyes A, Van der Does D, De Lange ES, Delker C, Wasternack C, Van Wees SCM, Ritsema T, Pieterse CMJ (2010b) Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta (Berlin) 232:1423–1432. doi:10.1007/s00425-010-1265-z
Leroy PD, Sabri A, Heuskin S, Thonart P, Lognay G, Verheggen FJ, Francis F, Brostaux Y, Felton GW, Haubruge E (2011) Microorganisms from aphid honeydew attract and enhance the efficacy of natural enemies. Nat Commun 2:348. doi:10.1038/ncomms1347
Mann RS, Ali JG, Hermann SL, Tiwari S, Pelz-Stelinski KS, Alborn HT, Stelinski LL (2012) Induced release of a plant-defense volatile ‘deceptively’ attracts insect vectors to plants infected with a bacterial pathogen. PLoS Pathog 8:e1002610. doi:10.1371/journal.ppat.1002610
Mauck KE, De Moraes CM, Mescher MC (2010) Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc Natl Acad Sci U S A 107:3600–3605
Mauck K, Bosque-Pérez NA, Eigenbrode SD et al (2012) Transmission mechanisms shape pathogen effects on host–vector interactions: evidence from plant viruses. Funct Ecol 26:1162–1175
McCutcheon JP, Moran NA (2012) Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi:10.1038/nrmicro2670
McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci 110:3229–3236. doi:10.1073/pnas.1218525110
McSteen P, Zhao Y (2008) Plant hormones and signaling: common themes and new developments. Dev Cell 14:467–473. doi:10.1016/j.devcel.2008.03.013
Mithofer A, Boland W (2012) Plant defense against herbivores: chemical aspects. Annu Rev Plant Biol 63:431–450. doi:10.1146/annurev-arplant-042110-103854
Mukhtar MS, Carvunis A-R, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, Pevzner SJ, Donovan SE, Ghamsari L, Santhanam B, Romero V, Poulin MM, Gebreab F, Gutierrez BJ, Tam S, Monachello D, Boxem M, Harbort CJ, McDonald N, Gai L, Chen H, He Y, Consortium EUE, Vandenhaute J, Roth FP, Hill DE, Ecker JR, Vidal M, Beynon J, Braun P, Dangl JL (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333:596–601. doi:10.1126/science.1203659
Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C (2006) The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140:249–262
Ng TF, Duffy S, Polston JE et al (2011) Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS One 6:e19050
Nikolaidis N, Doran N, Cosgrove DJ (2014) Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion. Mol Biol Evol 31:376–386. doi:10.1093/molbev/mst206
Ochman H, Moran NA (2001) Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096–1099. doi:10.1126/science.1058543
Ochman H, Lerat E, Daubin V (2005) Examining bacterial species under the specter of gene transfer and exchange. Proc Natl Acad Sci USA 1:6595-65959
Oliver KM, Degnan PH, Hunter MS, Moran NA (2009) Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325:992–994. doi:10.1126/science.1174463
Oliver KM, Degnan PH, Burke GR, Moran NA (2010) Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. doi:10.1146/annurev-ento-112408-085305
Oliver KM, Smith AH, Russell JA (2013) Defensive symbiosis in the real world – advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol 28:341–355. doi:10.1111/1365-2435.12133
Patten CL, Glick BR (1996) Bacterial biosynthesis of indole-3-acetic acid. Canad J Microbiol 42:207–220
Perilli S, Moubayidin L, Sabatini S (2010) The molecular basis of cytokinin function. Curr Opin Plant Biol 13:21–26. doi:10.1016/j.pbi.2009.09.018
Pertry I, Vaclavikova K, Depuydt S, Galuszka P, Spichal L, Temmerman W, Stes E, Schmulling T, Kakimoto T, Van Montagu MC, Strnad M, Holsters M, Tarkowski P, Vereecke D (2009) Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc Natl Acad Sci U S A 106:929–934. doi:10.1073/pnas.0811683106
Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521. doi:10.1146/annurev-cellbio-092910-154055
Pitino M, Hogenhout SA (2012) Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol Plant Microbe Interact 26:130–139. doi:10.1094/MPMI-07-12-0172-FI
Pontes MH, Smith KL, De Vooght L, Van den Abbeele J, Dale C (2011) Attenuation of the sensing capabilities of PhoQ in transition to obligate insect-bacterial association. PLoS Genet 7:e1002349–e1002349. doi:10.1371/journal.pgen.1002349
Rasmann S, Kollner TG, Degenhardt J, Hiltpold I, Toepfer S, Kuhlmann U, Gershenzon J, Turlings TC (2005) Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434(7034):732–737. doi:10.1038/nature03451
Robert CA, Frank DL, Leach KA, Turlings TC, Hibbard BE, Erb M (2013) Direct and indirect plant defenses are not suppressed by endosymbionts of a specialist root herbivore. J Chem Ecol 39:507–515. doi:10.1007/s10886-013-0264-5
Robert-Seilaniantz A, Navarro L, Bari R, Jones JD (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10:372–379. doi:10.1016/j.pbi.2007.06.003
Roossinck MJ (2012) Plant virus metagenomics: biodiversity and ecology. Annu Rev Gen 46:359–369. doi:10.1146/annurev-genet-110711-155600
Roossinck MJ, Saha P, Wiley GB et al (2010) Ecogenomics: using massively parallel pyrosequencing to understand virus ecology. Mol Ecol 19(Suppl 1):81–88
Sabri A, Vandermoten S, Leroy PD, Haubruge E, Hance T, Thonart P, De Pauw E, Francis F (2013) Proteomic investigation of aphid honeydew reveals an unexpected diversity of proteins. PLoS One 8:e74656. doi:10.1371/journal.pone.0074656
Satchell KJFK (2011) Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol 65:71–90. doi:10.1146/annurev-micro-090110-102943
Sloan DB, Nakabachi A, Richards S, Murali SC, Gibbs RA, Moran NA (2014) Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol. Biol. Evol.: Oxford University Press. pp 857–871
Spaepen S, Vanderleyden J (2011) Auxin and plant-microbe interactions. Cold Spring Harb Perspect in Biol 3. doi:10.1101/cshperspect.a001438
Stafford CA, Walker GP, Ullman DE (2011) Infection with a plant virus modifies vector feeding behavior. Proc Natl Acad Sci 108:9350–9355
Stes E, Vandeputte OM, El Jaziri M, Holsters M, Vereecke D (2011) A successful bacterial coup d’etat: how Rhodococcus fascians redirects plant development. Annu Rev Phytopathol 49:69–86. doi:10.1146/annurev-phyto-072910-095217
Stobbe AH, Roossinck MJ (2014) Plant virus metagenomics: what we know and why we need to know more. Front Plant Sci 5:150
Stout MJ, Thaler JS, Thomma BP (2006) Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu Rev Entomol 51:663–689. doi:10.1146/annurev.ento.51.110104.151117
Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA (2011) Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc Natl Acad Sci USA 1081254–1263. doi:10.1073/pnas.1105664108.
Vandermoten S, Harmel N, Mazzucchelli G, De Pauw E, Haubruge E, Francis F (2014) Comparative analyses of salivary proteins from three aphid species. Insect Mol Biol 23:67–77
Veit B (2009) Hormone mediated regulation of the shoot apical meristem. Plant Mol Biol 69:397–408. doi:10.1007/s11103-008-9396-3
Verhage A, van Wees SCM, Pieterse CMJ (2010) Plant immunity: it’s the hormones talking, but what do they say? Plant Physiol 154:536–540. doi:10.1104/pp. 110.161570
Vlot AC, Klessig DF, Park SW (2008) Systemic acquired resistance: the elusive signal (s). Curr Opin Plant Biol 11:436–442. doi:10.1016/j.pbi.2008.05.003
Vorholt JA (2012) Microbial life in the phyllosphere. Nature Publishing Gr 10:828–840. doi:10.1038/nrmicro2910
Walling LL (2008) Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol 146:859–866. doi:10.1104/pp. 107.113142
Walling LL (2009) Adaptive defense responses to pathogens and insects. In: VanLoon LC (ed) Plant innate immunity, vol 51. Advances in Botanical Research, p 551–612. doi:10.1016/s0065-2296(09)51013-0
Weech MH, Chapleau M, Pan L, Ide C, Bede JC (2008) Caterpillar saliva interferes with induced Arabidopsis thaliana defence responses via the systemic acquired resistance pathway. J Exp Bot 59:2437–2448. doi:10.1093/jxb/ern108
Werner T, Schmulling T (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12:527–538. doi:10.1016/j.pbi.2009.07.002
Zarate SI, Kempema LA, Walling LL (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143:866–875
Zhang P, Zhu X, Huang F, Liu Y, Zhang J, Lu Y, Ruan Y (2011) Suppression of jasmonic acid-dependent defense in cotton plant by the Mealybug Phenacoccus solenopsis. PLoS One 6:e22378. doi:10.1371/journal.pone.0022378
Zhang PJ, Li WD, Huang F, Zhang JM, Xu FC, Lu YB (2013) Feeding by whiteflies suppresses downstream jasmonic acid signaling by eliciting salicylic acid signaling. J Chem Ecol 39:612–619. doi:10.1007/s10886-013-0283-2
Acknowledgments
We thank Dezi Elzinga and Patrick Degnan for comments on earlier versions of the manuscript. We also thank Andrew Jang for help with formatting figures and tables. This publication was supported by the United States Department of Agriculture award 2013-2013-03265 to CLC and 2011-67012-30707 to AKH, and by University of Illinois at Urbana-Champaign start-up funds to AKH.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 140 kb)
Rights and permissions
About this article
Cite this article
Casteel, C.L., Hansen, A.K. Evaluating Insect-Microbiomes at the Plant-Insect Interface. J Chem Ecol 40, 836–847 (2014). https://doi.org/10.1007/s10886-014-0475-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10886-014-0475-4