Abstract
We present 19F longitudinal and transverse relaxation studies for four differently fluorosubstituted l-tryptophans, which carry single F atoms in the indole ring, both in the context of the free amino acid and when located in the cyclophilin A protein. For the free 4F-, 5F-, 6F-, 7F-l-Trp, satisfactory agreement between experimentally measured and calculated relaxation rates was obtained, suggesting that the parameters used for calculating the rates for the indole frame are sufficiently accurate. We also measured and calculated relaxation rates for four differently 19F-tryptophan labeled cyclophilin A proteins, transferring the parameters from the free amino acid to the protein-bound moiety. Our results suggest that 19F relaxation data of the large and rigid indole ring in Trp are only moderately affected by protein motions and provide critical reference points for evaluating fluorine NMR relaxation in the future, especially in fluorotryptophan labeled proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Relaxation rates are influenced by the physical properties of the molecule in its specific environment and can provide important information about these properties. The fluorine spins relax by dipole–dipole interactions (DD) with the proton spins that surround them and chemical shift anisotropy (CSA) (Gerig 2001). The dipolar interactions result in 19F–1H and 1H–19F nuclear Overhauser effects (NOEs) (Noggle and Schirmer 1971) that provide information about internuclear distances similar to 1H–1H NOEs, which are vital structural parameters in biomolecular structure determinations. In principle, relaxation rate constants yield quantitative estimates about molecular motions. However, in practice, application of 19F to assess protein dynamics in solution is fraught with complications: (i) the relative orientations of 19F–1H dipolar interactions with surrounding protons differ for the different sites of F incorporation; (ii) the orientations of dipolar interactions and CSA may not be colinear in the molecular frame (Mehring 1983; Peng 2001); and (iii) DD-CSA cross correlation may affect the observed rate (Goldman 1984; Kay et al. 1992). These confounding factors contribute to the limited number of applications of 19F-relaxation to probe dynamics in biomolecules in solution (Peng 2001; Luck et al. 1996; Hull and Sykes 1976; Hoang and Prosser 2014; Shi et al. 2011), although 19F NMR has been widely used to study folding and interactions of biomolecules (Dalvit and Piotto 2017; Mishra et al. 2014; Aramini et al. 2014; Matei et al. 2013; Sharaf and Gronenborn 2015).
In this article, we aim to reduce some of the above barriers by investigating a judiciously selected system, using a pragmatic approach: (1) we chose an amino acid that has a large, rigid molecular frame, the bulky Trp; (2) a single F atom was introduced next to a proximal proton at a fixed distance at four different sites in the indole ring. This reduces complexity in calculating the 19F–1H DD contribution and permitted assessment of other parameters for extracting intra-amino acid relaxation rates; (3) we utilized the results from individual fluorosubstituted tryptophans, for which 19F relaxation is predominantly influenced by intra-amino acid interactions. This allowed us to realistically evaluate the difference between the experimental and calculated relaxation rates for the Trp sidechain in a protein, assuming that no significant internal motion is present in the indole ring.
For our study, we selected singly fluorosubstituted tryptophans, namely, 4F-, 5F-, 6F-, 7F-l-Trp, both, the free amino acids and when incorporated into the human cyclophilin A (CypA) protein (Fig. 1) (Nigro et al. 2013; Howard et al. 2003). CypA possesses only one tryptophan at a position 121, close to its active site. We measured 19F longitudinal and transverse relaxation rates (R1 and R2, respectively) and assessed factors that affect relaxation rates for the amino acid systems. For the protein case, we experimentally determined 19F R1 and R2 for four CypA protein samples, each with a differently fluorine substituted Trp. Calculations of relaxation rates were carried out using the basic parameters from the individual amino acid calculations and assuming overall isotropic tumbling of the protein molecule without internal motion. Our data for the protein 19F relaxation show that (1) any apparent effect of DD-CSA cross correlation has to be small due to fast proton spin-flips (Kay et al. 1992), (2) intra-residue DD and CSA is dominating in all four 19F-Trp labeled protein data sets, and (3) comparison of the experimental with the calculated rates shows good agreement, although 15–30% larger values for the calculated R2 values are noted.
a Molecular structures of 4F-l-Trp, 5F-l-Trp, 6F-l-Trp, and 7F-l-Trp, illustrating the position of fluorine atom, the ellipsoidal chemical shielding surface for fluorine (magenta), and the principal axes of the CSA for each fluorinated tryptophan, displayed using TensorView (Young et al. 2019). Individual CSA values are provided in Table S1. b Schematic illustration of fluorotryptophan-labeled CypA [PDB 3K0N (Fraser et al. 2009)]. The fluorine atom is represented by gray (position 4), magenta (position 5), green (position 6), and blue (position 7) spheres. A detailed view of the local environment is shown in the expansion on the left. All sidechains within a 6 Å radius of any of the four fluorine atoms are shown in light cyan
Most importantly, our results provide critical reference data for evaluating fluorine NMR relaxation in future studies, especially for fluorotryptophan labeled proteins.
Experimental
Sample preparation
19F-Trp,15N CypA proteins (F-Trp-CypA) were expressed in E. coli Rosetta 2 (DE3), cultured in modified M9 medium, containing 4 g/L U-12C6-glucose, 1 g/L 15NH4Cl, and 20 mg/L 4, 5, or 7-fluoroindole as carbon, nitrogen, and fluorine sources (Sharaf and Gronenborn 2015; Crowley et al. 2012; Gakh et al. 2000). In the case of 6F-Trp 15N-CypA expression, 100 mg/L 6-l-fluorotryptophan, along with 100 mg/L phenylalanine, 100 mg/L tyrosine, and 1 g/L glyphosate was used instead of fluoroindole to improve the percentage of fluorine labeling. Cultures were grown to 1.0–1.2 OD and induced with 0.5 mM IPTG for protein expression at 18 °C for 16 h. CypA was purified using the same protocol as reported previously (Liu et al. 2016; Lu et al. 2015). Cells were harvested by centrifugation at 4000×g for 25 min at 4 °C, resuspended in 25 mM sodium phosphate buffer (pH 7.0), and ruptured by microfluidization. Cell debris was removed by centrifugation at 27,000×g for 1 h at 4 °C. The pH of the supernatant was adjusted to 5.8 with acetic acid, and the conductivity was reduced to below 2.5 ms/cm with deionized water. Following another centrifugation at 27,000×g for 1 h at 4 °C, the final supernatant was loaded onto a cation exchange column (HiTrap SP HP, 5 mL) and eluted with a 0–1 M NaCl gradient in buffer containing 25 mM sodium phosphate (pH 5.8), 1 mM DTT, 0.02% NaN3. Concentrated protein fractions were further purified using a size-exclusion column (HiLoad 26/600 Superdex 75), equilibrated in 25 mM sodium phosphate buffer (pH 6.5), 1 mM DTT, 0.02% NaN3. Final samples contained ~ 100 μM protein in 25 mM sodium phosphate buffer, pH 6.5, 1 mM TCEP, 0.02% NaN3, 7% D2O.
4F-l-tryptophan and 7F-l-tryptophan were purchased from Advanced ChemBlocks Inc. 5F-l-tryptophan and 6F-l-tryptophan were purchased from Sigma-Aldrich. Samples of ~ 100 μM 4, 5, 6, or 7F-l-Trp in 25 mM sodium phosphate buffer, pH 6.5, 1 mM TCEP, 0.02% NaN3, 7% D2O were used for recording spectra.
NMR spectroscopy
NMR measurements were performed on a 14.1 T Bruker AVANCE spectrometer, equipped with a CP TXO F/C–H–D triple-resonance, z-axis gradient cryoprobe. The Larmor frequencies of 1H and 19F are 600.1 and 564.6 MHz, respectively. The temperature in all NMR experiments was maintained at 298 K using the Bruker temperature controller. 1H and 19F chemical shifts were referenced with respect to DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) and TFA (trifluoroacetic acid). 19F longitudinal relaxation rates, R1, were measured using inversion-recovery (Vold 1972), with a recycle delay of 10 s for F-Trp samples and 6 s for F-Trp-CypA samples. Measurements were repeated twice to estimate the experimental error. 19F transverse relaxation rates, R2, were measured using a CPMG pulse scheme with a half duration between the CPMG pulses of 20 ms for F-Trp samples and 0.5 ms for F-Trp-CypA samples (Carr and Purcell 1954; Meiboom and Gill 1958). All data were processed and analyzed with Topspin 3.1. Relaxation rates were obtained by fitting the experimental data points to single exponential functions.
Calculation of 19F relaxation rates
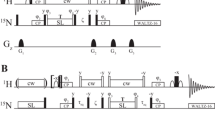
19F longitudinal relaxation rates (R1) and transverse relaxation rates (R2) were calculated based on the following Eqs. (1), (2), (3), and (4) (Gerig 2001; Ernst et al. 1988; Abragam 1961). Eqs. (1) and (2) describe the influence of DD on the longitudinal and transverse relaxation, whereas Eqs. (3) and (4) describe the influence of CSA on the longitudinal and transverse relaxation.
γF and γH, respectively, are the gyromagnetic ratios of fluorine and hydrogen, ℏ is the reduced Planck’s constant, rFH is the fluorine–proton distance, τc is the overall rotational correlation time, ωF and ωH are the resonance frequencies for fluorine and hydrogen, respectively.
with δσ and η the reduced anisotropy and the asymmetry parameters of the 19F chemical shift tensor, as defined in Haeberlen (1976) convention given by Eqs. (5)–(8).
Two sets of relaxation rate calculations were performed. In the first, only a single nearest proton was considered for DD. In the second, any proton within a 3 Å radius around the fluorine atom was taken into account (Table S2). For both cases, (i) single isotropic rotational correlation times (τc) were used, 8.2 ns and 55 ps for F-Trp-CypA (Ottiger et al. 1997) and F-Trp, respectively, and no internal motion was taken into account; (ii) 19F CSA values, previously determined by solid-state magic angle spinning (MAS) NMR for 4, 5, 6, or 7-fluorotryptophans were used in the calculations for both free F-Trp and F-Trp-CypA calculations (Fig. 1, Table S1) (Lu et al. 2018). All calculations were performed in MATLAB R2019a. In addition, time courses of 19F R1 and R2 magnetization decays were simulated using the Bloch–Redfield–Wangsness relaxation theory as implemented in the Spinach (Hogben et al. 2011) program, which accounts for all cross-relaxation and cross-correlations in multi-spin systems. In the time course simulations, the same τc and CSA values as listed above were used and all protons that are located within a 3 Å radius around the fluorine atom, were taken into account. Cross correlation terms were extracted using the Spinach (Hogben et al. 2011) program.
Results and discussion
19F NMR R1 and R2 values of fluorotryptophans in solution were determined at a field strength of 14.1 T (Fig. 2a, c). Relaxation rates (Table 1) were obtained by fitting all peak intensities to single-exponential functions. As can be appreciated from the data presented, the location of the fluorine atom in the indole ring (Fig. 1) clearly influences the R1 and R2 values. Comparing 4-, 5-, 6- and 7-F-l-Trp in the free amino acid, 4F-l-Trp possesses the largest R1 value of 0.99 s−1, followed by 7F-l-Trp with a R1 value of 0.87 s−1. The values for 5F-l-Trp and 6F-l-Trp are somewhat smaller at 0.75 and 0.67 s−1. For small molecules, such as an amino acid, it is expected that only minor differences exist between R1 and R2 values (Fig. S1). This is borne out by our data. All R2 values are slightly larger than their R1 counterparts for the same molecule. The R2 values for 4F-l-Trp and 7F-l-Trp are 1.38 and 1.03 s−1, respectively, and 5F-l-Trp and 6F-l-Trp possess R2 values of 0.89 and 0.78 s−1.
Experimental 19F longitudinal relaxation curves (R1; a) and transverse relaxation curves (R2; c) of 4-, 5-, 6-, or 7-fluorotryptophan and those simulated including all protons within a 3 Å radius around the F atom (b, d, respectively). Curves are color coded according to the different fluorine position: 4 (black), 5 (magenta), 6 (green), and 7 (blue). The entire simulated curves are shown in the insets
We previously measured fluorine chemical shift tensors by solid-state NMR on several fluorinated tryptophans and found that all of the tensors are rhombic with asymmetry parameters ranging from 0.5 to 0.9 (Lu et al. 2018) (Fig. 1, Table S1). Therefore, we used our experimentally measured CSA values for 4, 5, 6, or 7F-tryptophan in the R1 and R2 calculations reported here. In the free amino acid, the closest proton(s) to the different fluorines on the indole ring of Trp are 2.6 Å away, with the 5F-Trp and 6F-Trp positions flanked by two H atoms at this distance, while 4F-Trp and 7F-Trp have only one flanking H on the aromatic ring. 4F-Trp may have the NH2 group close and in 7F-Trp the NεH is located at a distance of 2.9 Å (Table S2). Gratifyingly, the experimental values for 5F-l-Trp and 6F-l-Trp are very similar, reflecting their close structural correspondence. Both have two H atoms as neighbors in an identical geometric arrangement. Not unexpectedly, the values calculated using the experimental CSA and the nearest proton DD are very similar (10% smaller) to those calculated including all protons within 3 Å (Table 1). For 5F-l-Trp and 6F-l-Trp, the R1 and R2 values calculated with the CSA and a single nearest proton DD are smaller than those for 4F-l-Trp and 7F-l-Trp, reflecting the smaller CSAs. The simulated curves also exhibit similar features, i.e., rates for 5F-l-Trp and 6F-l-Trp were slower than R1 and R2 for 4F-l-Trp and 7F-l-Trp (Fig. 2b, d). Although differences between the experimental relaxation curves for 5F-l-Trp and 6F-l-Trp are still discernable, the difference is very small (0.1 s−1; Fig. 2a, b).
Somewhat less good agreement between experimental and calculated R1 and R2 values is noted for 4F-l-Trp and 7F-l-Trp; this is even more pronounced when all protons within a 3 Å radius are included in the calculation (Table 1). This may be a result of flexibility around the χ1 and χ2 angles and/or the presence of close non-carbon bound protons, such as the NεH (close to the 7F position) and the NH2 amino group (potentially close to the 4F position; Fig. 1).
Overall, however, the data obtained here for the free amino acids indicate that using the experimental CSA values in our analysis allowed us to adequately calculate 19F relaxation at each site.
We next measured R1 and R2 values of fluorotryptophans incorporated into the CypA protein (Fig. 3a, c). For the Trp sidechain in the 18.3 kDa CypA protein [τc = 8.2 ns (Ottiger et al. 1997)], 5F, 6F, and 7F-Trp-labeled CypA exhibit similar R1 values of ~ 1.2 s−1, respectively, whereas 4F-Trp-labeled CypA possesses a larger R1 value of ~ 2.0 s−1. With regard to transverse relaxation, we observe that the 4F- and 7F-l-Trp containing proteins exhibit similar experimental R2 values of ~ 110 s−1, and, likewise, the R2 values of 5F- and 6F-l-Trp containing CypA are 65 and 63 s−1, respectively (Table 2). This grouping into two similar sets is also apparent in the experimentally measured linewidths at half height for all the different F-l-Trp CypA variants: 5F-CypA and 6F-CypA exhibit ∆1/2 of ~ 31 Hz, while 4F-CypA and 7F-CypA exhibit broader lines with ∆1/2 of ~ 40 Hz. Most importantly, this grouping into two similar sets is consistent with the observation for the free Trp amino acid data (Table 1), suggesting that the intra-amino acid contributions to 19F R2 are essentially the same for the free amino acid and the amino acid sidechain in a protein.
Experimental 19F longitudinal relaxation curves (R1; a) and transverse relaxation curves (R2; c) of 4, 5, 6, or 7-fluorotryptophan CypA and those simulated including all protons within a 3 Å radius around the F atom (b, d, respectively). Curves are color coded according to the different fluorine position: 4 (black), 5 (magenta), 6 (green), and 7 (blue). The entire simulated curves are shown in the insets
Interestingly, while the experimental 19F R1 curves for all the four F-Trp-CypA proteins fit a single-exponential decay function, multi-exponential decays were observed in the simulations (Fig. 3b), where multi-spin models were used. The fast and slow decay components, captured by simulations, are caused by DD-CSA cross correlation, and can be significantly averaged by the fast proton spin-flip in the case of proteins (Kay et al. 1992), thus resulting in an approximately single-exponential decay. The experimental 19F R2 curves for all the four F-Trp-CypA also fit a single-exponential decay function. Since CSA is the dominant source in transverse relaxation, this suggests that the cross-term cannot be large and only contributes 10–25% to the total R2 relaxation rate (Table S3), depending on the magnitude of the CSA and the relative orientation of the DD to CSA. Thus, an approximately single-exponential behavior is also observed in the simulated R2 curves (Fig. 3d).
Although using a single-exponential model is an approximation for extracting R2 values, good agreement between experimentally determined and calculated R2 values is obtained when all protons within a 3 Å radius around the F-atom are used in the calculation (Table 2). In the CypA protein [PDB 3K0N (Fraser et al. 2009)], like in the free amino acid, the 5F and 6F positions possess two H atoms 2.6 Å away, while 4F and 7F, in addition to the flanking H on the aromatic ring, have other H atoms close by: 4F has the amide NH proton at a distance of 2.3 Å, the Hβ at a distance of 2.6 Å and the Glu120 Hγ at 2.8 Å, while 7F has the NεH at 2.9 Å distance and the Hε proton on the aromatic ring of a neighboring Phe60 sidechain at 2.6 Å (Fig. 1, Table S2).
The calculation results for the proteins clearly show that the calculated values for R1 are too small when only a single nearest proton is taken into account (the calculated values are 30–40% of the measured ones) while they get closer to the experimental values when the calculations consider all protons within a 3 Å radius for the dipole–dipole contributions (60–90% of the experimental R1 values; Table 1). Given that spin-flips affect the R1 rate itself (Fig. 3b), the observed agreement clearly is remarkable. For example, R1 for 4F-Trp CypA, is ~ 2.0 s−1, and clearly larger than R1 values for the other F-Trp-CypAs, which are ~ 1.0 s−1 (Table 2). This feature is faithfully reproduced in the calculations.
In contrast to R1, the calculated R2 values essentially reproduced the experimental values, irrespective of whether a single close proton or all protons within a sphere of 3 Å radius around the F atom were considered (Table 2). This is noteworthy, since the calculations are clearly employing simple approximations, such as using the crystal structure of the non-F-Trp CypA protein and neglecting any potential internal motions. If a motionally active group were in proximity to the F atom, an effect on R2 would be expected and this would not be correctly modelled using a static X-ray structure.
At this juncture it may be instructive to consider how similar or different the analysis of protein motions from NMR relaxation data has to be when different nuclei are involved. Amide relaxation in proteins is commonly analyzed using the model-free formalism, and this approach is traditionally employed to assess backbone or sidechain motions of proteins in solution (Kay et al. 1992; Lipari and Szabo 1982a, b; Mandel et al. 1995). However, such analyses cannot be simply transferred to 19F-labeled proteins since CSA and DD may be affected by motions in different orientations and thus sensitive to anisotropic internal dynamics. Given the uncertainties in calculating R1 and R2 for a fluorine atom on a protein in an accurate manner, we decided to take a pragmatic approach and to experimentally measure fluorine R1 and R2 values for very similar molecular structures in the context of a small molecule and of the same amino acid in the macromolecular system (Figs. 2, 3).
Our choice of tryptophan was a deliberate one: the indole ring is a rigid scaffold and motions within the amino acid will be limited to those around χ1 and χ2 angles. Furthermore, whereas phenylalanine or tyrosine sidechains commonly undergo ring flipping or other rapid motions within proteins (Khan et al. 2006; Boeszoermenyi et al. 2019; Wagner et al. 1976, 1979), the much bigger tryptophan sidechain does not (Munro et al. 1979). Therefore, it is reasonable to assume that a fluorine atom on a fluorotryptophan indole ring, which is a rigid moiety, will rotate with the same rotational correlation time as the whole protein. This assumption is borne out by the data presented here: the protein 19F R1 and R2 values can be very well recapitulated by simple calculations, assuming an approximate CSA and including protons within a sphere of 3 Å radius around the F atom and an overall single rotational correlation time for the protein, neglecting internal motions.
For the free amino acids, our simple calculations of R1 and R2 values also exhibited good agreement with the experimental values, even though it would be simplistic to assume that no motions around the χ1 and χ2 occur in solution. We reason that the good agreement between experimental and calculated values is a reflection of the fact that 19F relaxation at 4F-, 5F-, 6F-, 7F- positions in Trp is mainly determined by the 19F CSA and 19F–1H dipolar interactions within the indole ring. This is supported by previous studies (Peng 2001). Using model CSA parameters of a fluoro-phenyl ring from Hiyama (Hiyama et al. 1986), a field strength of 11.7 T (500 MHz for 1H), and an internuclear distance of rFH of 2.6 Å, Peng showed that transverse relaxation of a fluorine in a six-membered aromatic ring is dominated by the CSA mechanism and that the high-frequency spectral densities JDD(ωH), JDD(ωH ± ωF) of the 19F–1H dipole–dipole interaction can be neglected over a wide range of correlation times τc (Peng 2001). More recently, Dalvit and Piotto (Dalvit and Piotto 2017) reported calculations of 19F R1 and R2 values for 5F-Trp at two field strengths, 9.4 T and 18.8 T (19F Larmor frequencies of 376 MHz and 752 MHz). In their calculations they also used the two interacting spin approximation at a 2.6 Å distance, and a literature value for the CSA of ∆σ of 76.8 ppm with ηCSA = 0 (Durr et al. 2008).
Although we believe our data and analysis are an important first step, it still uses a qualitative approach, particularly for evaluating R1, which is affected by the proton-spin flip effect. It is hoped that more data sets on proteins will be accumulated in the future, which will enable a more systematic treatment, either using a 19F relaxation database approach or more rigorous calculation strategies.
Conclusions
Here, we systematically investigated fluorine relaxation in fluorosubstituted tryptophan amino acids and in fluorotryptophan-labeled CypA proteins in solution. Distinct longitudinal and transverse relaxation rates were observed for fluorine atoms at different positions in the indole ring. Experimentally measured 19F R1 and R2 values are generally in good agreement with calculated values. Overall, our results demonstrate that both dipole–dipole and CSA relaxation mechanisms play important parts in determining the 19F R1 and R2 relaxation rates in fluorotryptophans as free amino acids, and that the parameters for calculating relaxation the free amino acid can be transferred to the protein-bound moiety. The data reported here are a critical benchmark for evaluating fluorine NMR relaxation of fluorotryptophan-labeled proteins in the future.
References
Abragam A (1961) The principles of nuclear magnetism. Clarendon Press, Oxford
Aramini JM, Hamilton K, Ma L-C, Swapna GVT, Leonard PG, Ladbury JE, Krug RM, Montelione GT (2014) 19F NMR reveals multiple conformations at the dimer interface of the nonstructural protein 1 effector domain from influenza A virus. Structure 22:515–525
Boeszoermenyi A, Chhabra S, Dubey A, Radeva DL, Burdzhiev NT, Chanev CD, Petrov OI, Gelev VM, Zhang M, Anklin C, Kovacs H, Wagner G, Kuprov I, Takeuchi K, Arthanari H (2019) Aromatic 19F–13C TROSY: a background-free approach to probe biomolecular structure, function, and dynamics. Nat Methods 16:333–340
Carr HY, Purcell EM (1954) Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys Rev 94:630–638
Crowley PB, Kyne C, Monteith WB (2012) Simple and inexpensive incorporation of 19F-tryptophan for protein NMR spectroscopy. Chem Commun 48:10681–10683
Dalvit C, Piotto M (2017) 19F NMR transverse and longitudinal relaxation filter experiments for screening: a theoretical and experimental analysis. Magn Reson Chem 55:106–114
Durr UHN, Grage SL, Witter R, Ulrich AS (2008) Solid state 19F NMR parameters of fluorine-labeled amino acids. Part I: aromatic substituents. J Magn Reson 191:7–15
Ernst R, Bodenhausen G, Wokaun A (1988) Principles of nuclear magnetic resonance in one and two dimensions. Clarendon Press, Oxford
Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T (2009) Hidden alternative structures of proline isomerase essential for catalysis. Nature 462:669–673
Gakh YG, Gakh AA, Gronenborn AM (2000) Fluorine as an NMR probe for structural studies of chemical and biological systems. Magn Reson Chem 38:551–558
Gerig J (2001) Fluorine NMR. Published online, http://www.biophysics.org/portals/1/pdfs/education/gerig.pdf
Goldman M (1984) Interference effects in the relaxation of a pair of unlike spin-12 nuclei. J Magn Reson 60:437–452
Haeberlen U (1976) High resolution NMR in solids selective averaging. Academic Press, New York
Hiyama Y, Silverton JV, Torchia DA, Gerig JT, Hammond SJ (1986) Molecular-structure and dynamics of crystalline para-fluoro-D, l-phenylalanine—a combined X-ray NMR investigation. J Am Chem Soc 108:2715–2723
Hoang J, Prosser RS (2014) Conformational selection and functional dynamics of calmodulin: a 19F nuclear magnetic resonance study. Biochemistry 53:5727–5736
Hogben HJ, Krzystyniak M, Charnock GTP, Hore PJ, Kuprov I (2011) Spinach—a software library for simulation of spin dynamics in large spin systems. J Magn Reson 208:179–194
Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP (2003) Structural insights into the catalytic mechanism of cyclophilin A. Nat Struct Biol 10:475–481
Hull WE, Sykes BD (1976) Fluorine-19 nuclear magnetic resonance study of fluorotyrosine alkaline phosphatase: the influence of zinc on protein structure and a conformational change induced by phosphate binding. Biochemistry 15:1535–1546
Kay LE, Nicholson LK, Delaglio F, Bax A, Torchia DA (1992) Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J Magn Reson 97:359–375
Khan F, Kuprov I, Craggs TD, Hore PJ, Jackson SE (2006) 19F NMR studies of the native and denatured states of green fluorescent protein. J Am Chem Soc 128:10729–10737
Lipari G, Szabo A (1982a) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc 104:4546–4559
Lipari G, Szabo A (1982b) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc 104:4559–4570
Liu C, Perilla JR, Ning JY, Lu MM, Hou GJ, Ramalho R, Himes BA, Zhao GP, Bedwell GJ, Byeon IJ, Ahn J, Gronenborn AM, Prevelige PE, Rousso I, Aiken C, Polenova T, Schulten K, Zhang PJ (2016) Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat Commun 7:10714
Lu M, Hou G, Zhang H, Suiter CL, Ahn J, Byeon IJL, Perilla JR, Langmead CJ, Hung I, Gor’kov PL, Gan ZH, Brey W, Aiken C, Zhang PJ, Schulten K, Gronenborn AM, Polenova T (2015) Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proc Natl Acad Sci USA 112:14617–14622
Lu M, Sarkar S, Wang M, Kraus J, Fritz M, Quinn CM, Bai S, Holmes ST, Dybowski C, Yap GPA, Struppe J, Sergeyev IV, Maas W, Gronenborn AM, Polenova T (2018) 19F magic angle spinning NMR spectroscopy and density functional theory calculations of fluorosubstituted tryptophans: integrating experiment and theory for accurate determination of chemical shift tensors. J Phys Chem B 122:6148–6155
Luck LA, Vance JE, OConnell TM, London RE (1996) 19F NMR relaxation studies on 5-fluorotryptophan- and tetradeutero-5-fluorotryptophan-labeled E. coli glucose/galactose receptor. J Biomol NMR 7:261–272
Mandel AM, Akke M, Palmer AG III (1995) Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol 246:144–163
Matei E, André S, Glinschert A, Infantino AS, Oscarson S, Gabius H-J, Gronenborn AM (2013) Fluorinated carbohydrates as lectin ligands: dissecting glycan–cyanovirin interactions by using 19F NMR spectroscopy. Chem Eur J 19:5364–5374
Mehring M (1983) Principles of high resolution NMR in solids. Springer, Berlin
Meiboom S, Gill D (1958) Modified spin-echo method for measuring nuclear relaxation times. Rev Sci Instrum 29:688–691
Mishra NK, Urick AK, Ember SWJ, Schönbrunn E, Pomerantz WC (2014) Fluorinated aromatic amino acids are sensitive 19F NMR probes for bromodomain–ligand interactions. ACS Chem Biol 9:2755–2760
Munro I, Pecht I, Stryer L (1979) Sub-nanosecond motions of tryptophan residues in proteins. Proc Natl Acad Sci USA 76:56–60
Nigro P, Pompilio G, Capogrossi MC (2013) Cyclophilin A: a key player for human disease. Cell Death Dis 4:e888
Noggle JH, Schirmer RE (1971) The nuclear Overhauser effect. Academic Press, New York
Ottiger M, Zerbe O, Guntert P, Wuthrich K (1997) The NMR solution conformation of unligated human cyclophilin A. J Mol Biol 272:64–81
Peng JW (2001) Cross-correlated 19F relaxation measurements for the study of fluorinated ligand-receptor interactions. J Magn Reson 153:32–47
Sharaf NG, Gronenborn AM (2015) 19F-modified proteins and 19F-containing ligands as tools in solution NMR studies of protein interactions. Methods Enzymol 565:67–95
Shi P, Wang H, Xi Z, Shi C, Xiong Y, Tian C (2011) Site-specific 19F NMR chemical shift and side chain relaxation analysis of a membrane protein labeled with an unnatural amino acid. Protein Sci 20:224–228
Vold RL (1972) Measurement of transverse relaxation rates in complex spin systems. J Chem Phys 56:28–31
Wagner G, DeMarco A, Wüthrich K (1976) Dynamics of the aromatic amino acid residues in the globular conformation of the basic pancreatic trypsin inhibitor (BPTI). Biophys Struct Mech 2:139–158
Wagner G, Tschesche H, Wuthrich K (1979) Influence of localized chemical modifications of the basic pancreatic trypsin-inhibitor on static and dynamic aspects of the molecular-conformation in solution. Eur J Biochem 95:239–248
Young RP, Lewis CR, Yang C, Wang L, Harper JK, Mueller LJ (2019) TensorView: a software tool for displaying NMR tensors. Magn Reson Chem 57:211–223
Acknowledgements
This work was supported by the National Science Foundation (CHE-1708773), the National Institutes of Health (P50 GM082251) and is a contribution from the Pittsburgh Center for HIV Protein Interactions. Mike Delk is acknowledged for NMR technical support and Teresa Brosentisch for editorial help.
Author information
Authors and Affiliations
Contributions
AMG conceived the project and AMG and TP guided the work. ML prepared the samples, performed NMR experiments and analyzed the experimental data. RI conducted further in-depth data analysis. All authors discussed the results and contributed to the manuscript preparation.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, M., Ishima, R., Polenova, T. et al. 19F NMR relaxation studies of fluorosubstituted tryptophans. J Biomol NMR 73, 401–409 (2019). https://doi.org/10.1007/s10858-019-00268-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-019-00268-y