Abstract
Objectives This study aimed to characterize the surface properties of experimental resin polymers consisting of monomers differing in functionality and chain length, and to evaluate differences in Streptococcus mutans adhesion. Material and Methods Six resins were prepared (70/30 ratio UDMA/monomer); camphorquinone and ethyl-4-dimethylaminebenzoate were added for light activation. A conventional composite was used as a control. Surface free energy was determined prior and after saliva exposition (2 h, 37 °C). After saliva incubation (2 h, 37 °C), specimens were incubated with Streptococcus mutans NCTC 10449 for 2.5 h at 37 °C. Adherent bacteria were quantified by determining the relative substratum area covered by bacteria using SEM analysis, and by using a fluorometric assay for viable cell quantification. Results No statistically significant differences in total surface free energies were found for uncoated specimens (mean total surface free energies ranging from 39.79 to 49.73 mJ/m−2); after saliva coating, statistically significant differences were observed for some of the polymers (mean total surface free energies ranging from 44.13 to 65.81 mJ/m−2). Few differences were observed between SEM and fluorescence quantification, finding statistically significant differences in streptococcal adhesion to the experimental polymers. Median bacteria surface coverage ranged from 1.4% for UDMA mixed with 1,10-decandiol dimethacrylate to 16.2% for the control composite material; lowest fluorescence intensities indicating lowest adhesion of bacteria were found for UDMA mixed with 1,10-decandiol dimethacrylate (median 712), and highest values indicating highest adhesion of bacteria were found for UDMA mixed with polyethyleneglycol (600) dimethacrylate (median 11974). ConclusionStreptococcus mutans adhesion appears to be different on polymers differing in monomer mixtures, yet correlations between substratum surface free energy and streptococcal adhesion were poor. Further studies are necessary to evaluate additional substratum surface properties and pellicle distribution and composition more thoroughly.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The first step in dental plaque formation is the adsorption of salivary constituents to teeth and restorative surfaces; this layer of salivary proteins, lipids and carbohydrates has been labelled salivary pellicle [1], and its formation is followed by the initial adhesion of oral bacteria such as streptococci and actinomyces species [2]. Dental plaque has been correlated with the occurrence of dental and oral diseases such as caries or parodontopathia [3], and extensive plaque formation on dental restoratives may contribute to secondary caries or periodontal inflammation. Thus, it is wishful that dental materials reveal low susceptibility to adhere oral microorganisms. Composite materials are commonly used for direct and indirect tooth restoration as well as for cementation of crowns and bridges, and the scope of this material class is still broadening. Modern dental composites consist of organic, hydrophobic matrix monomers and predominantly inorganic, rather hydrophilic fillers that are linked by silane coupling agents, and influences of composite constituents such as monomers or fillers on pellicle and plaque formation have been reported. For example, researchers observed that resin monomers such as triethyleneglycol dimethacrylate or methacrylic acid modulate bacterial growth [4], and Gröger and co-workers [5] revealed accumulation of salivary pellicle on resin fillers and in the matrix substance between resin and fillers in vivo, which might be a hint towards material deterioration, filler loss or degradation of the filler-matrix bonding.
Furthermore, numerous in vivo and in vitro studies concluded that the adhesion of oral microorganisms is significantly influenced by substratum surface properties [6–13]. Substratum surface roughness and surface free energy have been counted among the most significant factors influencing the adhesion of bacteria [14]. High substratum surface roughness has been found to promote bacterial adhesion to composite surfaces [15]. It has been observed that substrata featuring high surface free energies harbour significantly more plaque than substrata with lower surface free energies in in vivo as well as in vitro studies [14]. The surface properties of the bacteria themselves have been found to play a decisive role in this thermodynamical adhesion approach, too: high surface free energy strains preferentially adhere to substrata with high surface free energy, whereas strains featuring lower surface free energy adhere preferentially to substrata with lower surface free energy [16, 17]. Little information is available on the influence of surface charge of dental materials on the adhesion of oral bacteria; however, Reynolds and co-workers [18] found a decrease in bacterial adhesion to hydroxyapatite discs precoated with acidic proteins, which implicated a more negative surface charge. Similar results are reported by Jansen and co-workers [19], using experimental polymers modified by glow discharge and Streptococcus epidermidis. The adsorption of salivary proteins to substratum surfaces has been found to influence substrata’s surface properties indirectly, too, as the salivary pellicle masks differences in surface free energy of various substrata and thus changes conditions for streptococcal adhesion [20]. Some researchers, however, maintain that a distinct influence of original substratum surface properties remains despite a salivary protein coating, which has been named shine-through effect [18, 21–23].
To date, there is no study that assesses potential influences of organic composite resin matrix on both physicochemical surface properties of the corresponding polymer and the adhesion of oral bacteria. In modern composite technology, resin monomers are frequently modified in terms of functionality and chain length for providing resins with less polymerization shrinkage [24, 25] and improved mechanical properties [26] by increased cross-linking. As substratum surface properties such as surface free energy and its polar and disperse components may change with alterations in number and kind of functional resin monomer groups, influences of monomer chemistry on resin surface properties and the initial adhesion of oral bacteria cannot be excluded. Thus, the aim of this study was to evaluate the impact of various resin matrix polymers prepared from experimental resin matrix methacrylate monomers differing in chain length or the number of functional groups on the physicochemical surface properties of the polymer (1), and on the initial adhesion of a cariogenic strain of the bacterium Streptococcus mutans (2).
2 Material and methods
2.1 Material
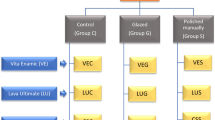
Six experimental methacrylate resins (obtained from Ivoclar Vivadent (Ivoclar Vivadent, Schaan, FL)) were used in this study (cf. Fig. 1): urethane dimethacrylate (C23H38N2O3, 470.63 g/mol), 1,10-decandiol dimethacrylate (C18H30O4, 310.48 g/mol), 1,1,1-trimethylolpropane trimethacrylate (C18H26O6, 338,44 g/mol), ditrimethylpropane tetramethacrylate (C28H42O9, 522.7 g/mol), polyethylenglycol (400) dimethacrylate (C26H46O12, 550.7 g/mol), and polyethylenglycol (600) dimethacrylate (C46H71O17, 770 g/mol).
For preparation of test specimens, blends were mixed (w/w) in 70/30% ratio urethane dimethacrylate (UDMA)/methacrylate resin monomer, and camphorquinone (0.25% w/w) and ethyl-4-dimethylaminebenzoate (EPD, 0.25% w/w) were added for light activation. Six test specimens of each mixture (diameter 10 mm, 2 mm thick) were prepared by filling resin in moulds and subsequent light curing (2 × 90 s, Ivomat, Ivoclar Vivadent, Schaan, FL). Prior to polymerization, specimens were covered with a transparent plastic film to prevent the formation of an oxygen inhibited layer.
A veneering composite (Sinfony, 3 M Espe, Seefeld, G) served as control, and was cured according to the manufacturerer’s instructions using Visio Alfa and Visio Beta (3 M ESPE, Seefeld, G).
All test specimens were polished using silicone carbide grinding paper grit 1000 and 4000 (Buehler, Düsseldorf, Germany) and a rotating grinding disk apparatus (Motopol 8, Buehler Ltd., Coventry, UK). Peak-to-valley surface roughness was determined at three spots of each sample (one in central position, two at the margins) using a profilometric contact surface measurement device (Perthometer S6P, Feinprüf-Perthen, Göttingen, Germany). Roughness values up to 0.08 μm were tolerated, and specimens with higher roughness values were rejected for keeping surface roughness far below the threshold value at 0.2 μm [27].
After manufacturing, all resin samples were stored in distilled water for six days after manufacturing to minimize potential influences of residual monomer of resin samples on the viability of bacteria.
Surface free energy of the various resin substrata was calculated from contact angle measurements using an automated contact angle measurement device equipped with a video camera and an image analyzer (OCA 15 plus, Dataphysics Instruments GmbH, Filderstadt, Germany). Uncoated and saliva coated specimens were examined. For each substratum (uncoated and saliva coated), three liquids differing in hydrophobicity were used: deionized water, diiodomethane (Sigma-Aldrich, St. Louis, USA) and ethylene glycol (Merck KgaA, Darmstadt, Germany). Nine drops for each liquid (1 μL) and substratum were examined, and left ands right contact angles of each drop were averaged. The surface free energy of the substrata and its polar and disperse components were calculated according to the Owens, Wendt, Rabel and Kaelble (OWRK) method.
2.2 Bacteria
The strain Streptococcus mutans NCTC 10449 (DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) was used in this research. After a frozen (−60 °C) preculture was established bacteria were exposed on an agar plate and incubated at 37 °C for 48 h. A single colony was incubated with sterile DSMZ-medium 92 (Trypticase Soy Yeast Extract Medium, containing 30 g tryptic soy broth (Becton Dickinson Microbiology Systems, Sparks, USA) and 3 g yeast extract (Sigma-Aldrich, St. Louis, USA)). This mixture was incubated at 37 °C for 16 h in a thermo shaking device (OrbitalShaker, ThermoForma, Marietta, USA) and subsequently kept at 4 °C. The day before the experiment 1 mL of bacteria suspension was inoculated with 250 mL of sterile DSMZ-medium 92 and incubated for 12 h as described above. Cells were harvested by centrifugation (2,200 rpm, 19 °C, 5 min; Hettich Rotixa P, Tuttlingen, Germany), washed twice with PBS and resuspended in the same buffer. The cell suspension was subsequently subjected to low intensity ultrasonic energy in order to disperse bacterial chains [28], and the optical density was adjusted to 0.3 at 550 nm (Genesys 10-S, Thermo Spectronic, Rochester, USA), which corresponds to a microbial concentration of 3.65 × 108 cells/mL [29].
2.3 Saliva preparation
Unstimulated human whole saliva was collected by expectoration from one healthy, 35 year old donor without active carious lesions or periodontal diseases. Saliva was sterilized using single-use filtration devices with pore sizes 0.45 and 0.2 μm (Vacuflo PV050/2 and PV050/3, Schleicher & Schüll Microscience GmbH, Dassel, Germany).
2.4 Test assay and Streptococccus mutans quantification
For quantification of adherent Streptococcus mutans, scanning electron microscopic (SEM) analysis and a fluorometric assay (Resazurin reduction, Alamar Blue) were used.
For SEM quantification, six test specimens of each experimental resin were equilibrated with ethanol for removing traces of proteins and lipids and transferred to 12 well cell clusters (Tissue Culture Clusters, costar, Cambridge, USA). Pellicle formation was initiated by incubating test specimens with 1 mL of human whole saliva for 2 h in a thermo shaking device (OrbitalShaker, ThermoForma, Marietta, USA). After 2 h, samples were carefully rinsed with phosphate buffered saline solution (PBS, Sigma-Aldrich, St. Louis, USA) and incubated with 1 mL of Streptococcus mutans suspension in a thermo shaking device. After 2.5 h, samples were gently rinsed twice with PBS to remove unbound bacteria, and subsequently dried and glued to holders. In order to subject the resin specimens to scanning electrom microscopy (Steroscan S240, Cambridge, Nußloch, G), they were sputtered with a thin layer of gold (BALTEC, Walluf, G). Five SEM micrographs (×2000) of randomly selected areas of each resin specimens were taken, and the relative area covered by Streptococcus mutans was determined using the image analysis program Optimas 6.2 (Media Cybernetics, Bethesda, USA).
Resazurin reduction was carried out as described previously [30]. In brief, this cell quantification method is based on the reduction of the blue, non fluorescent redox indicator Resazurin (maximum absorbance at 605 nm) into the violet, fluorescent pigment Resorufin (maximum absorbance at 573 nm) by metabolically active, viable cells [31]. Fifteen samples of each experimental resin were used, and prior saliva and bacteria incubation auto-fluorescence of the resin specimens was determined using an automated multi-detection reader (Fluostar Optima, BMG Labtech, Offenburg, G). Similarly to SEM quantification, samples were equilibrated with ethanol, transferred to 48 well cell clusters (48 Well Cell Culture Cluster, Corning Inc., Corning, USA) and subsequently incubated with 1 mL whole saliva for pellicle formation. After 2 h, samples were carefully rinsed with PBS and incubated with 1 mL of Streptococcus mutans suspension and 15 μL Resazurin (Resazurin, Sigma-Aldrich, St. Louis, USA). After 2.5 h, samples were gently rinsed twice with PBS for removing unbound cells. Relative fluorescence intensities were measured using the automated multidetection reader, and auto-fluorescence values were subtracted from these values. The remaining values for fluorescence intensity correlate linearly with the number of adherent bacteria [32].
2.5 Statistics
For evaluating differences in surface free energies, means and standard deviations were calculated. Statistical analysis was performed using one-way analysis of variance (ANOVA) and Tukey’s HSD test (α < 0.05). For evaluating differences in the adhesion of Streptococcus mutans to the various substrata, medians and 25/75% percentiles were calculated. Statistical analysis was performed using the Mann–Whitney U-Test (α < 0.05). All calculations were performed using SPSS 13.0 for Windows (SPSS Inc., Chicago, USA).
3 Results
3.1 Contact angle measurements and surface free energy
Contact angle measurements (cf. Table 1) of the uncoated as well as the saliva coated resin specimens differed among the various resin mixtures. After saliva-coating, broad variations in measured contact angles were observed for some of the various polymers and liquids.
Prior saliva coating, one-way ANOVA revealed no statistically significant differences in total surface free energy (cf. Table 2) between the various specimens (p = 0.053). Lowest values were calculated for UDMA mixed with 1,10-decanediol dimethacrylate (39.79 ± 2.29 mJ/m−2), and highest values were measured for UDMA mixed with polyethylenglycol (600) dimethacrylate (49.73 ± 1.92 mJ/m−2). Statistically significant differences between the various substrata were found for the contribution of the polar (ANOVA, p = 0.000) and disperse (ANOVA, p = 0.003) components to total surface free energy. Tukey’s HSD revealed a significantly higher proportion of the polar components of the surface free energy of UDMA mixed with polyethylenglycol (400) dimethacrylate (10.46 ± 3.17 mJ/m−2) than for all other substrata, and the disperse components of the total surface free energy of UDMA mixed with polyethylenglycol (400) dimethacrylate (34.01 ± 3.49 mJ/m−2) were significantly lower than for the veneering composite (42.08 ± 0.91 mJ/m−2; p = 0.032), UDMA mixed with 1,1,1-trimethylolpropane trimethacrylate (41.62 ± 3.42 mJ/m−2; p = 0.047) and UDMA mixed with polyethylenglycol (600) dimethacrylate (46.35 ± 1.39 mJ/m−2; p = 0.001).
After saliva coating, the total surface free energy of the veneering composite and UDMA mixed with ditrimethylpropane tetramethacrylate increased significantly (from 42.91 ± 1.06 to 58.75 ± 2.30 mJ/m−2, and from 42.57 ± 1.41 to 65.81 ± 2.90 mJ/m−2, respectively; p = 0.000, respectively).
For the other resin mixtures no statistically significant differences were observed concerning total surface free energy. The polar components of the surface free energy increased significantly for the veneering composite (from 0.83 ± 0.54 mJ/m−2 prior saliva coating to 26.73 ± 1.89 mJ/m−2 after saliva coating; p = 0.000), UDMA mixed with 1,10-decanediol dimethacrylate (from 2.04 ± 0.94 to 14.09 ± 2.30 mJ/m−2; p = 0.000), UDMA mixed with 1,1,1-trimethylolpropane trimethacrylate (from 3.84 ± 1.91 to 11.50 ± 3.49 mJ/m−2; p = 0.005) and UDMA mixed with dimethylpropane tetramethacrylate (from 1.80 ± 0.54 to 28.14 ± 2.19 mJ/m−2; p = 0.000). For the veneering composite, the proportion of the disperse components of the surface free energy diminished significantly from 42.08 ± 0.91 to 32.02 ± 1.32 mJ/m−2 (p = 0.002), too.
ANOVA revealed statistically significant differences in total surface free energy among the various resin polymers after saliva coating (p = 0.000). Highest surface free energies were calculated for UDMA mixed with ditrimethylpropane tetramethacrylate (65.81 ± 2.90 mJ/m−2) and the veneering composite (58.75 ± 2.30 mJ/m−2), which were significantly higher than for all other resin mixtures that did not differ significantly from each other in terms of total surface free energy. The polar components of the surface free energy of the saliva coated specimens were significantly higher for the veneering composite (26.73 ± 1.89 mJ/m−2) and UDMA mixed with ditrimethylpropane tetramethacrylate (28.14 ± 2.19 mJ/m−2) than for all other substrata. Significantly lowest values for the polar components of the surface free energy were calculated for pure UDMA (2.87 ± 1.34 mJ/m−2), UDMA mixed with polyethylenglycol (400) dimethacrylate (5.13 ± 2.67 mJ/m−2) and polyethylenglycol (600) dimethacrylate (1.98 ± 1.80 mJ/m−2). The highest proportions of the disperse components after saliva coating were found for pure UDMA resin (45.66 ± 2.38 mJ/m−2) and UDMA mixed with polyethylenglycol (600) dimethacrylate (44.57 ± 2.53 mJ/m−2), which were significantly higher than for all other resin mixtures. Lowest values for the disperse components of the surface free energy were found for the veneering composite (32.02 ± 1.32 mJ/m−2) and UDMA mixed with 1,10-decanediol dimethacrylate (31.69 ± 2.19 mJ/m−2), but values were similar compared with the other resins.
3.2 Adherence of Streptococcus mutans
SEM analysis revealed equal distribution of Streptococcus mutans on the various resin specimens, and cell quantification and statistical analysis revealed significant differences between the various resin specimens concerning streptococcal adhesion. With the exception of the veneering composite control, few bacteria were discovered on the resin specimens’ surfaces (cf. Fig. 2). Highest values were observed for the control composite material (median surface coverage: 16.2%), which were significantly higher (p = 0.016 and p = 0.033, respectively) than values measured for urethane dimethacrylate mixed with 1,10-decanediol dimethacrylate (median 1.4%) or polyethyleneglycol (600) dimethacrylate (median 3.8%), but not the other mixtures. Lowest adhesion of Streptococcus mutans was noticed for urethane dimethacrylate mixed with 1,10-decanediol dimethacrylate (median 1.4%). Significantly higher adhesion was found on urethane dimethacrylate mixed with 1,1,1-trimethylolpropane trimethacrylate (median 3.3%; p = 0.018), and tendentially higher adhesion of Streptococcus mutans was found for urethane dimethacrylate mixed with ditrimethylolpropane tetramethacrylate (median 4.6%; p = 0.251). On urethane dimethacrylate mixed with polyethyleneglycol (600) dimethacrylate (median 3.8%) significantly fewer bacteria adhered than on urethane dimethacrylate mixed with polyethyleneglycol (400) dimethacrylate (median 7.5%; p = 0.006). On pure urethane dimethacrylate resin monomer adhesion of Streptococcus mutans was significantly higher (median 6.4%; p = 0.047) compared with urethane dimethacrylate mixed with 1,10-decanediol dimethacrylate, but was similar to all other resin monomers and the reference.
The experiments were repeated for validation using a Resazurin reduction assay for viable cell quantification. Similarly to SEM quantification, lowest fluorescence intensities (cf. Fig. 3) indicating lowest adhesion of Streptococcus mutans were found for UDMA mixed with 1,10-decandiol dimethacrylate (median fluorescence intensity 712), which were significantly lower than for UDMA mixed with ditrimethylolpropane tetramethacrylate (1683, p = 0.001) and UDMA mixed with polyethylenglycol (400) dimethacrylate (median 2299, p = 0.000) and tendentially lower than for UDMA mixed with 1,1,1-trimethylolpropane trimethacrylate (1561, p = 0.063). Highest fluorescence intensities correlating with highest adhesion of streptococci were found for UDMA mixed with polyethylenglycol (600) dimethacrylate (11974); values were significantly higher than for all other substrata evaluated (p = 0.000). The veneering composite (1382) and pure UDMA resin (2041) did not differ significantly from each other and from the other substrata in terms of fluorescence intensity.
4 Discussion
In this study, focus was set on the influence of composite matrix monomers on the surface properties of experimental polymers, and on the initial adhesion of Streptococcus mutans to these experimental substrata. Urethane dimethacrylate was chosen as basic resin as it is one of the most commonly used and most important resin monomers in current dental technology and science [33]. In order to provide low shrinking dental composites, resin monomer chain length is frequently extended to reduce shrinkage during polymerization [34]; for simulating this aspect, the monomers polyethyleneglycol (400) dimethacrylate and polyethyleneglycol (600) dimethacrylate have been used, which differ from each other in its number of ethylene functional groups. 1,10-decanediol dimethacrylate is frequently used in dental composite technology as a diluent [35], and 1,1,1-trimethylpropane trimethacrylate and ditrimethylpropane tetramethacrylate have been used to investigate the impact of ramifications in molecular structure and functional groups on surface properties and streptococcal adhesion.
Dental restorations are usually polished to high gloss prior or after insertion for minimizing in situ plaque formation; thus, resin specimens were subjected to intense polishing prior to measurements. As excessive surface roughness has been found to promote bacterial adhesion [15] and influence contact angle measurements [36], specimens with higher roughness values than 0.08 μm were rejected. Bollen and co-workers [27] found a threshold value at 0.2 μm, insisting that roughness values lower than 0.2 μm do not influence bacterial adhesion.
Numerous studies agree that coating dental materials with a salivary pellicle affects bacterial adhesion due to the masking properties of the salivary pellicle, which may cause a levelling of originally distinct surface free energies, for instance [37–39]. In this study, few differences in surface free energy were found among the various uncoated resin specimens, and differences concerning the contribution of the polar and disperse components to surface free energy were poor with the exception of polyethyleneglycol (400) dimethacrylate. Thus, it may be concluded that the various monomers only sparsely influenced the surface properties of the uncoated specimens. The extraordinarily high proportion of the polar components of UDMA mixed polyethyleneglycol (400) dimethacrylate was surprising, as UDMA mixed with polyethylenglycol (600) dimethacrylate, differing only in its number of ethylene functional groups from polyethylenglycol (400) dimethacrylate (seven versus twelve), featured similar values as the other resin mixtures; only slightly lower values have been expected for UDMA mixed with polyethylenglycol (600) dimethacrylate. Surprisingly, similar values for the total surface free energy and its components were found for the various pure resin mixtures and the veneering composite despite of its content of about 50% hydrophilic fillers. Carlèn and co-workers, however, found similarly high disperse components of the surface free energy for polished composite resin [40], which supports the values calculated in this research.
In contrast to other studies, no levelling effect of the salivary pellicle concerning substrata’s surface properties was observed in this research, yet it has to be borne in mind that most of the other studies dealing with this topic used experimental substrata or substrata clearly differing in their original physicochemical surface properties; in this study, already original total surface free energies were similar. For the veneering composite and UDMA mixed with decanediol dimethacrylate, 1,10-decandiol dimethacrylate, 1,1,1-trimethylolpropane trimethacrylate or ditrimethylpropane tetramethacrylate, total surface free energy increased after saliva coating, and particularly the proportion of the polar components incremented decisively. For pure UDMA, UDMA mixed with polyethyleneglycol (400) dimethacrylate, and UDMA mixed with polyethyleneglycol (600) dimethacrylate either total surface free energy or its polar component decreased.
Despite of different functional groups and chain length, the correlations between monomer structure and surface properties such as surface free energy were poor. Similar total surface free energies for UDMA mixed with polyethylenglycol (400) dimethacrylate and UDMA mixed with polyethylenglycol (600) dimethacrylate were found, and after saliva coating total surface free energy as well as its polar components decreased for both of the resins. For the other resin mixtures, similar surface free energies were found before saliva coating, which indicates that the influence of increasing monomer functionality on physicochemical surface properties of the experimental polymers was rather poor. After saliva coating, however, significant differences in both surface free energy and its polar and disperse components were found between some of the polymers, which hints that protein adsorption was different on the various polymers.
Concerning alterations of surface free energies or its components of dental materials after saliva coating, no consistent information can be found in the literature. Evaluating surface free energy of various denture base materials, Sipahi and co-workers [41] observed a decrease of the disperse Lifshitz–van der Waals surface free energy components as well as of total surface free energy after saliva coating, whereas the polar Lewis-base components of total surface free energy increased. On ceramic specimens, Milleding and co-workers [42] did, however, not detect significant changes in total surface free energy after saliva coating, but the polar Lewis-acid and Lewis-base components decreased. These divergent data, along with the rather diverse results of this study, indicate that further studies are necessary to clarify the influence of substratum surface properties on salivary protein adsorption and surface free energy more thoroughly. Surprisingly, broad variations in contact angles were observed particularly on saliva coated specimens, which indicates that salivary proteins were not equally distributed on the various surfaces. Although numerous studies found that salivary protein adsorption is dependent on substratum surface properties such as surface free energy [43, 44], Hannig and co-workers [45] observed that substratum surface free energy correlates with the rate of pellicle formation rather than the total amount of salivary proteins adsorbed to the substratum surfaces. Moreover, Carlèn and co-workers observed that protein adsorption to dental biomaterials appears to be particularly dependent on substratum surface roughness [40]. Judging from these divergent findings, it is not totally clear whether total protein amount adsorbed to the various specimens might serve as an explanation for differences in surface free energy. Investigating plasma protein adsorption to ceramics, Rosengren and co-workers [46] found that about 70% of the surface were not covered by proteins at all; this phenomenon might, however, serve as a valid explanation for variations in contact angles and surface free energies calculated in this study. Thus, these findings indicate that further studies are necessary to investigate adsorption patterns and distribution of salivary proteins more thoroughly by means of fluoroscence imaging in order to correlate substratum surface properties and protein adsorption.
Streptococcus mutans has been chosen as a representative oral bacterium as it is considered as one of the most abundant microorganisms in the oral cavity [47]. Moreover, the bacterium has been discovered in early dental plaque, and is regarded as one of the major causative agents for dental caries [48]. Most studies dealing with the adherence of bacteria to biomaterials do not incorporate saliva in the test assay, yet in the oral cavity, dental restorations are covered with the salivary pellicle within minutes [49]. This phenomenon justifies pellicle formation prior to simulation of streptococcal adhesion, as in vivo bacteria adhere to the salivary pellicle on the restorative surface rather than the mere surface itself. Oral shear stress has been simulated by employing semistatic incubation conditions for both pellicle formation and bacteria adhesion. Although there are broad interindividual variations, about 108–109 microrganisms/mL saliva have been found [50, 51], which justifies the microbial concentration used in this study. As it might be argued that SEM quantification revealed too low values for the relative substratum areas covered by Streptococcus mutans for allowing proper statistical analysis, streptococcal adherence has been assessed with a fluorometric assay in addition to SEM analysis. The Resazurin reduction assay employed in this study offers the accessory advantage that only viable cells are subjected to quantification, as merely metabolically active bacteria reduce Resazurin to its fluorescent form, Resorufin.
Both assays revealed rather low adhesion of Streptococcus mutans. Using SEM quantification, highest adhesion of Streptococcus mutans was found on the control samples; surprisingly, using the fluorometric assay, significantly highest adhesion was found to UDMA mixed with polyethylenglycol (600) dimethacrylate, and the control samples featured only intermediate values for Streptococcus mutans adhesion. In contrast, SEM analysis found only low values of adherent streptococci on UDMA mixed with polyethylenglycol (600) dimethacrylate. Measurements were repeated twice because of these divergent data, but repetitions still yielded similar results. Possibly, data differ as by SEM quantification only randomly selected areas of the specimens can be analyzed, whereas bacteria on the entire surface of the specimen are subject to quantification when using the fluorometric assay. Moreover, using SEM analysis it cannot be distinguished between viable and dead cells, whereas only viable cells are quantified by fluorescence analysis. However, both assays found lowest adhesion to UDMA mixed with 1,10-decanediol dimethacrylate, and revealed significantly or at least tendentially higher adhesion of streptococci to UDMA mixed with 1,1,1-trimethylolpropane trimethacrylate and ditrimethylpropane tetramethacrylate. Mostly similar adhesion of streptococci to pure UDMA compared with the experimental polymers was found regardless of quantification method.
Previous studies found a correlation between bacterium surface properties and the surface properties of the substratum, which has still been observed even after saliva coating of the specimens. Weerkamp and co-workers [52] found high surface free energies for numerous Streptococcus mutans strains; in accordance with the established thermodynamic models, increased adhesion of Streptococcus mutans to substrata with high total surface free energy was expected in this study. However, no correlation between total surface free energy uncoated specimens and the relative adherence of Streptococcus mutans could be established, which might, in part, be due to the fact that prior saliva coating samples featured rather similar values for total surface free energy and its polar and disperse components. After saliva coating, significantly highest values for total surface free energy were calculated UDMA mixed with ditrimethylpropane tetramethacrylate, yet only intermediate adhesion of Streptococcus mutans to this substratum was observed regardless of quantification method applied. These findings correspond to studies by Sardin and co-workers [53], who evaluated the adhesion of several strains of oral streptococci to saliva coated prosthetic materials. However, Sardin and co-workers [53] found a significant correlation between the number of adherent bacteria and the values of the non-polar component of the surface free energy of saliva coated substrata, yet the findings of this study do not correpond to this approach, too. Highest values for the non-polar components of total surface free energy after saliva-coating were found for pure UDMA, yet only intermediate Streptococcus mutans adhesion was found.
It is obvious that our results cannot be explained sufficiently by the thermodynamic approach. Significant differences in streptococcal adhesion to various experimental polymers were found, yet surface free energy determination revealed almost similar values for uncoated resins. Moreover, alterations in surface free energy after saliva coating did not follow a clear pattern, and no correlation between surface free energy or its components after saliva coating could be established neither with SEM nor fluorescence quantification. These findings hint that other substratum properties such as surface charge might play a role in the adhesion of Streptococcus mutans to saliva coated resin polymers. Protein adsorption to experimental polymer surfaces should be studied extensively, too, as differences in protein distribution and pellicle composition cannot be excluded. Further studies are necessary to investigate the properties of experimental polymers and their influence on protein adsorption and streptococcal adherence more thoroughly.
References
C. DAWES, G. N. JENKINS and C. H. TONGE, Br. Dent. J. 16 (1963) 65
P. E. KOLENBRANDER, R. N. ANDERSEN, D. S. BLEHERT, P. G. EGLAND, J. S. FOSTER and R. J. PALMER Jr, Microbiol. Mol. Biol. Rev. 66 (2002) 486
P. AXELSSON and J. LINDHE, J. Clin. Periodontol. 5 (1978) 131
P. KHALICHI, D. G. CVITKOVITCH and J. P. SANTERRE, Biomaterials 25 (2004) 5467
G. GRÖGER, M. ROSENTRITT, M. BEHR, J. SCHRÖDER and G. HANDEL, J. Mater. Sci. Mater. Med. 17 (2006) 825
D. R. ABSOLOM, F. V. LAMBERTI, Z. POLICOVA, W. ZINGG, C. J. VAN OSS and A. W. NEUMANN, Appl. Environ. Microbiol. 46 (1983) 90
J. M. SCHAKENRAAD, H. J. BUSSCHER, C. R. H. WILDEVUUR and J. ARENDS, J. Biomed. Mater. Res. 20 (1986) 773
L. J. VAN DIJK, F. HERKSTRÖTER, H. J. BUSSCHER, A. H. WEERKAMP, H. W. B. JANSEN and J. ARENDS, J. Clin. Periodontol. 14 (1987) 300
M. QUIRYNEN, M. MARECHAL, H. J. BUSSCHER, A. H. WEERKAMP, J. ARENDS, P. L. DARIUS and D. VAN STEENBERGHE, J. Dent. Res. 68 (1989) 796
J. M. SCHAKENRAAD, H. J. BUSSCHER, C. R. WILDEVUUR and J. ARENDS, J. Biomed. Mater. Res. 20 (1986) 773
J. M. SCHAKENRAAD, J. ARENDS, H. J. BUSSCHER, F. DIJK, P. B. VAN WACHEM and C. R. H. WILDEVUUR, Biomaterials 10 (1989) 43
S. EICK, E. GLOCKMANN, B. BRANDL and W. PFISTER, J. Oral Rehabil. 31 (2004) 278
L. MONTANARO, D. CAMPOCCIA, S. RIZZI, M. E. DONTATI, L. BRESCHI, C. PRATI and C. R. ARCIOLA, Biomaterials 25 (2004) 4467
W. TEUGHELS, N. VAN ASSCHE, I. SLIEPEN and M. QUIRYNEN, Clin. Oral Implants Res. 17(Suppl 2) (2004) 68
K. KAWAI and M. URANO, Oper. Dent. 26 (2001) 396
A. H. WEERKAMP, H. M. UYEN and H. J. BUSSCHER, J. Dent. Res. 67 (1988) 1483
F. MABBAUX, L. PONSONNET, J. J. MORRIER, N. JAFFREZIC and O. BARSOTTI, Colloids Surf. B Biointerfaces 39 (2004) 199
E. C. REYNOLDS and A. WONG, Infect. Immun. 39 (1983) 1285
B. JANSEN and W. KOHNEN, J. Ind. Microbiol. 15 (1995) 391
J. SATOU, A. FUKUNAGA, A. MORIKAWA, I. MATSUMAE, N. SATOU and H. SHINTANI, J. Oral Rehabil. 18 (1991) 421
M. FLETCHER, J. Gen. Microbiol. 94 (1976) 400
A. ABBOTT, P. R. RUTTER and R. C. W. BERKLEY, J. Gen. Microbiol. 129 (1983) 439
I. H. PRATT-TERPSTRA, A. H. WEERKAMP and H. J. BUSSCHER, J. Gen. Microbiol. 133 (1987) 3199
M. ATAI, D. C. WATTS and Z. ATAI, Biomaterials 26 (2005) 5015
A. ELLAKWA, N. CHO and I. B. LEE, Dent. Mater. 23 (2007) 1229
J. TANAKA, T. HASHIMOTO, J. W. STANSBURY, J. M. ANTONUCCI and K. SUZUKI, Dent. Mater. J. 20 (2001) 206
C. M. L. BOLLEN, P. LAMBRECHTS and M. QUIRYNEN, Dent. Mater. 13 (1997) 258
M. W. STINSON, M. J. LEVINE, J. M. CAVESE, A. PRAKOBPHOL, P. A. MURRAY, L. A. TABAK and M. S. REDDY, J. Dent. Res. 61 (1982) 1390
J. SATOU, H. FUKUNAGA, N. SATOU, H. SHINTANI and K. OKUDA, J. Dent. Res. 67 (1988) 588
M. ROSENTRITT, S. HAHNEL, G. GRÖGER, B. MÜHLFRIEDEL, R. BÜRGERS and G. HANDEL, J. Biomed. Mater. Res. Part B. Appl. Biomater. (2007). doi: 10.1002/jbm.b.30985
G. R. NAKAYAMA, M. C. CATON, M. P. NOVA and Z. PARANDOOSH, J. Immunol. Methods 204 (1997) 205
J. O’BRIEN, I. WILSON, T. ORTON and F. POGNAN, Eur. J. Biochem. 267 (2000) 5421
I. D. SIDERIDOU, M. M. KARABELA and D. N. BIKIARIS, Dent. Mater. 23 (2007) 1142
B. DARVELL, in “Materials Science for Dentistry” (Hong Kong, 2002) p. 133
N. MOSZNER, in “Die Geheimnisse von Kompositen” (Ivoclar-Vivadent Report Nr. 18, Ivoclar Vivadent, Schaan, 2007) p. 10
A. A. ADAMSON and A. P. GAST, in “Physical Chemistry of Surfaces” (Wiley-Interscience, New York, 1997) p. 358
M. A. JENDRESEN and P. O. GLANTZ, Acta Odontol. Scand. 38 (1980) 379
L. J. Van DIJK, R. GOLDSWEER and H. J. BUSSCHER, Biofouling 1 (1988) 19
S. MORGE, E. ADAMCZAK and L. A. LINDEN, Arch. Oral. Biol. 34 (1989) 669
A. CARLÈN, K. NIKDEL, A. WENNERBERG, K. HOLMBERG and J. OLSSON, Biomaterials 22 (2001) 481
C. SIPAHI, N. ANIL and E. BAYRAMLI, J. Dent. 29 (2001) 197
P. MILLEDING, S. GERDES, K. HOLMBERG and S. KARLSSON, Eur. J. Oral Sci. 107 (1999) 384
D. R. ABSOLOM, W. ZINGG and A. W. NEUMANN, J. Biomed. Mater. Res. 21 (1987) 161
L. LINDH, Swed. Dent. J. Suppl. 152 (2002) 1
M. HANNIG, A. DÖBERT, R. STIGLER, U. MÜLLER and S. A. PROKHOROVA, J. Nanosci. Nanotechnol. 4 (2004) 532
A. ROSENGREN, E. PAVLOVIC, S. OSCARSSON, A. KRAJEWSKI, A. RAVAGLIOLI and A. PIANCASTELLI, Biomaterials 23 (2002) 1237
M. KAWASHIMA, M. HANADA, T. HAMADA, J. TAGAMI and H. SENPUKU, Oral Microbiol. Immunol. 18 (2003) 220
B. NYVAD and M. KILIAN, Caries Res. 24 (1990) 267
M. HANNIG, Eur. J. Oral Sci. 107 (1999) 55
M. A. LISTGARTEN, in “Dental Plaque Revisited. Oral Biofilms in Health and Disease”, edited by H. N. NEWMAN and M. WILSON (Bioline, Cardiff, 1999) p. 189
M. V. MARSH and P. MARTIN, in “Orale Mikrobiologie” (Thieme, Stuttgart/New York, 2003) p. 49
A. H. WEERKAMP, H. C. van der MEI and H. J. BUSSCHER, J. Dent. Res. 64 (1985) 1204
S. SARDIN, J. J. MORRIER, G. BENAY and O. BARSOTTI, J. Oral Rehabil. 31 (2004) 140
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hahnel, S., Rosentritt, M., Bürgers, R. et al. Surface properties and in vitro Streptococcus mutans adhesion to dental resin polymers. J Mater Sci: Mater Med 19, 2619–2627 (2008). https://doi.org/10.1007/s10856-007-3352-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10856-007-3352-7