Abstract
We have estimated free energies for the binding of nine cyclic carboxylate guest molecules to the octa-acid host in the SAMPL4 blind-test challenge with four different approaches. First, we used standard free-energy perturbation calculations of relative binding affinities, performed at the molecular-mechanics (MM) level with TIP3P waters, the GAFF force field, and two different sets of charges for the host and the guest, obtained either with the restrained electrostatic potential or AM1-BCC methods. Both charge sets give good and nearly identical results, with a mean absolute deviation (MAD) of 4 kJ/mol and a correlation coefficient (R 2) of 0.8 compared to experimental results. Second, we tried to improve these predictions with 28,800 density-functional theory (DFT) calculations for selected snapshots and the non-Boltzmann Bennett acceptance-ratio method, but this led to much worse results, probably because of a too large difference between the MM and DFT potential-energy functions. Third, we tried to calculate absolute affinities using minimised DFT structures. This gave intermediate-quality results with MADs of 5–9 kJ/mol and R 2 = 0.6–0.8, depending on how the structures were obtained. Finally, we tried to improve these results using local coupled-cluster calculations with single and double excitations, and non-iterative perturbative treatment of triple excitations (LCCSD(T0)), employing the polarisable multipole interactions with supermolecular pairs approach. Unfortunately, this only degraded the predictions, probably because of a mismatch between the solvation energies obtained at the DFT and LCCSD(T0) levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the largest challenges of computational chemistry is to predict the binding affinity of a small ligand to a larger receptor molecule, e.g. a drug candidate to its receptor protein [1–6]. Therefore, a large number of methods have been suggested with this aim, ranging from statistical knowledge- and regression-based methods to force-field-based simulations and free-energy perturbation methods. It is well-known that the molecular-mechanics force fields employed in many of these methods have a limited accuracy, e.g. owing to the primitive treatment of electrostatic interactions [7, 8]. Therefore, there has recently been much interest in using quantum mechanical (QM) calculations to improve binding affinities [9–14].
A major problem when estimating binding affinities is the large number of important and to a large extent cancelling interactions, e.g. bonded terms, electrostatics, polarisation, charge transfer, dispersion, exchange repulsion, polar and non-polar solvation, and entropy. For an accurate estimate, all these terms need to be predicted with an accuracy better than 4 kJ/mol, because a difference in the binding constant of one order of magnitude corresponds to a free-energy difference of only 6 kJ/mol. Even worse, the binding might be affected by major changes in the conformation or the protonation state of both the ligand and the receptor. Host–guest systems allow such problems to be studied in a simpler context. In such systems, the binding of small ligands to an organic macrocycle with a few hundred atoms is studied (compared to the tens of thousands of atoms in a biological receptor). Consequently, the configurational freedom as well as the chemical diversity is much smaller.
A good way to test different methods is blind-test challenges, in which the experimental binding affinity is not known when the calculations are performed. Thereby, the results will not be biased against the experimental data and not only success stories will be published. The SAMPL challenges have been leading this development, e.g. providing both protein–ligand and host–guest challenges in the SAMPL3 blind test in 2011 [15].
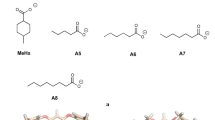
In this paper, we present our efforts to predict binding affinities of the SAMPL4 octa-acid challenge [16]. The octa-acid cavitand (Fig. 1) is a macrocycle of 184 atoms with a four-fold symmetry, twelve benzene rings and eight carboxylic groups [17]. It forms a hydrophobic cavity of ~10 Å depth that can bind various small molecules in aqueous solution [18, 19]. We have tried to predict the binding affinity of the nine carboxylic guest molecules in Fig. 1. The carboxylic group of the guests and the pH of 9.2 during the binding-affinity measurements are chosen to ensure that the complexes remain monomeric and that all carboxylic groups are deprotonated. We selected this test case because the guest molecules are quite rigid and all guests have the same net charge. However, the large negative charge of the host may cause problems in the calculations.
We have performed free-energy perturbations with molecular-mechanics (MM) methods and two sets of charges. Moreover, we have tried to improve these estimates by dispersion-corrected density-functional theory (DFT-D3) methods [20]. In a second approach, we used DFT-D3 optimised structures to estimate the absolute affinities, according to the method recently suggested by Grimme [21]. We have also tried to improve the latter results with density-fitted local coupled cluster calculations (DF-LCCSD(T0)) [22], employing the polarised multipolar interactions with supermolecular pairs (PMISP) approach [8, 23]. The results are of varying quality, providing both the best and the worst predictions (of twelve in total) submitted to the SAMPL4 challenge.
Methods
Force-field parametrisation of the host and guest molecules
All MM calculations employed the general Amber force field (GAFF) [24] for both the host and the guest molecules, and TIP3P for water molecules [25]. Atom types were selected by the antechamber program in the Amber 11 software suite [26]. Two sets of charges were tested. The first was AM1-BCC charges [27, 28], obtained by antechamber (after geometry optimisation at the AM1 level). The second set was standard Amber RESP charges [29]: The molecules were optimised at the AM1 level and then the electrostatic potential was calculated at the Hartree–Fock/6-31G* level in points sampled according to the Merz–Kollman scheme [30], albeit at a higher-than-default density (10 layers with 17 points per unit area, giving ~2,000 points per atom). These calculations were performed with the Gaussian 09 software [31]. Finally, charges were fitted to these points with the restrained electrostatic potential method (RESP) [29] using the antechamber program. For both sets, we ensured that symmetry-equivalent atoms in all molecules had the same charges. The two charge sets will be referred to as BCC and RESP, respectively. A single angle parameter for the host was missing in the GAFF 1.0 force field (ca-c3-h2) and was taken from a similar angle (c3-c3-h2; in fact, the ca-c3-h2 parameter is available in the GAFF 1.4 force field). Amber leap input files for the host and all guest molecules are provided in the supplementary material.
MD simulations
All molecular dynamics (MD) simulations and free-energy perturbation (FEP) calculations were performed with the Amber 11 software [26]. Starting structures for the octa-acid host and the guests were provided by the SAMPL4 organisers and were used without any modification. The host was assumed to always be fully deprotonated with a net charge of −8 e. Likewise, the guests were assumed to be deprotonated with a −1 charge. Starting structures for the complexes of the host and the guests were built manually. No counter ions were employed.
The host and the complexes were solvated in a truncated octahedral box of water molecules extending at least 9 Å from the solute using the leap program in the Amber suite, giving ~3,850 atoms in total. This structure was then subjected to 100 steps of minimisation without any constraints. This was followed by 20 ps constant-volume and 1 ns constant-pressure equilibration. Finally, a 10 ns production simulation was run, during which structures were sampled every 20 ps. In the MD simulations, bonds involving hydrogen atoms were constrained with the SHAKE algorithm [32], allowing for a time-step of 2 fs. In all simulations, the temperature was kept constant at 300 K and the pressure was kept constant at 1 atm using a weak-coupling isotropic algorithm [33] with a relaxation time of 1 ps. Long-range electrostatics were handled by particle-mesh Ewald (PME) summation [34] with a fourth-order B spline interpolation and a tolerance of 10−5. The cut-off for non-bonded interactions was set to 10 Å. For the Bz and EtBz guests (the names of the guests are specified in Fig. 1), ten independent simulations were performed by rotating the complex by a random angle around a random axis before the solvation and employing different starting velocities (the rotations were performed to ensure that the complexes were solvated differently, increasing the difference between the independent simulations [35]).
FEP calculations
The FEP calculations were started from the final structure of these MD simulations (removing water molecules). The complexes were solvated in an octahedral periodic box of TIP3P water molecules extending at least 10 Å from the solute, giving ~4,700 atoms. They were subjected to 500 steps of minimisation and 20 ps equilibration of the water molecules at constant pressure, followed by 2 ns equilibration and 4 ns production simulations without any constraints, during which structures and energies were sampled every 10 ps. In all simulations, the temperature was kept constant at 300 K using a Langevin thermostat with a collision frequency of 2.0 ps−1 [36] and the pressure was kept constant at 1 atm using a weak-coupling isotropic algorithm [33]. The cut-off for non-bonded interactions was set to 8 Å. PME was employed with the same parameters as in the MD simulations. Bonds involving hydrogen atoms were constrained with SHAKE and the time step was 2 fs.
The relative binding free energy (between pairs of guests ∆∆G bind), G1 and G2, was calculated for eight transformations: MeBz → Bz, EtBz → MeBz, pClBz → Bz, mClBz → Bz, Hx → Bz, MeHx → Hx, Hx → Pen, and Hep → Hx. These were estimated using a thermodynamic cycle that relates ∆∆G bind to the free energy of alchemically transforming G1 into G2 when they are either bound to the host, ∆G bound, or are free in solution, ∆G free [37]
∆G bound and ∆G free were estimated by the Bennett acceptance-ratio method [38, 39] (BAR). In this approach, a finite number of λ values is selected between 0 and 1, and for each λ, an MD simulation is run with the potential
where V 0 is the potential with G1 and V 1 is the potential with G2. For each neighbouring pair of λ values, A and B, the free energy difference between the two states is estimated from
where f(x) = (1 + exp(x/kT))−1 is the Fermi function, k is the Boltzmann constant, T is the temperature, and C is a constant. An iterative procedure is applied to find a value of C that makes the first term of the right-hand side of Eq. 3 vanish. Free energies were also calculated by multi-state BAR [40], thermodynamic integration [41], and exponential averaging [42].
We employed 13 states in these calculations (λ = 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 0.95, and 0.99; for technical reasons, AMBER does not allow calculations at λ = 0.00 and 1.00). The electrostatic and van der Waals interactions were transformed simultaneously in the simulation by using soft-core potentials for disappearing atoms and a dual-topology approach [43]. The soft-core potentials were used only for atoms differing between the two guest molecules, i.e. for the transformed CH3 → H or Cl → H groups, or for all atoms in the ring system for the Hx → Bz, Hx → Pen, and Hep → Hx transformations. The V 0 state was always the larger guest molecule. For the calculations with the RESP charges, ten independent simulations were run at each λ value, using different random starting velocities. In a few simulations, SHAKE problems were encountered, which were resolved by removing the SHAKE constraints and decreasing the time step to 0.5 fs. A semi-automatic script used to setup these calculations can be found in http://www.teokem.lu.se/~ulf/Methods/rel_free.html.
Test calculations with nine Na+ counter ions neutralising the charge of the host and the guest were performed for the MeBz → Bz transformation, giving changes in the calculated binding free energy of only 2 kJ/mol compared to calculations without counter ions. Likewise, we tried to include all guest atoms in the perturbed group, treated with soft-core potentials, rather than only the modified Cl or H atom for the pClBz → Bz transformation, and again the effect was only 2 kJ/mol for the calculated binding free energy. Finally, we tried to increase the number of λ values to 27, run twice as long equilibration and production simulations (4 and 8 ns, respectively), half the time step (1 fs), increase the non-bonded cutoff distance (9 Å), and to run separate electrostatic and van der Waals perturbations for the MeBz → Bz transformation. However, none of these five calculations changed the resulting binding free energy by more than 0.5 kJ/mol (the results are collected in Table S1).
DFT-D calculations
All calculations with density-functional theory (DFT) were performed with the Turbomole 6.4 software [44, 45]. The calculations employed either the TPSS [46] or BP86 [47, 48] functionals. Basis sets of four different sizes were employed, def2-SV(P), TZVP, def2-TZVP, and def2-QZVP’ (i.e. the def2-QZVP basis set with discarded f and g-type functions on hydrogen and other atoms, respectively) [49–51]. All DFT calculations were sped up by expanding the Coulomb interactions in auxiliary basis sets, the resolution-of-identity approximation, using the corresponding auxiliary basis sets [52, 53]. The calculations also used the multipole-accelerated resolution-of-identity J approach (MARIJ) [54].
Dispersion effects were included by the DFT-D3 approach [20], either with default damping and no third-order terms, calculated by Turbomole (for geometries) or with Becke–Johnson damping [55] and third-order terms, calculated with the dftd3 program (for single-point energies) [56].
In some of the geometry optimisations, polar solvation effects were estimated by the conductor-like screening model (COSMO) [57, 58]. These calculations were performed with default values for all parameters (implying a water-like probe molecule) and a dielectric constant of 80. For the generation of the cavity, we used the optimised COSMO radii in Turbomole (1.30, 2.00, 1.72, and 2.05 Å for H, C, O, and Cl, respectively) [59].
More accurate solvation energies (including also non-polar effects) were obtained with the COSMO-RS approach [60–62] at 298 K. These calculations were based on two single-point DFT calculations, both at the BP86/TZVP level, either in vacuum or with COSMO continuum solvation with an infinite dielectric constant.
Thermal corrections to the Gibbs free energy at 298 K and 1 atm pressure (G therm; including zero-point vibrational energy, ZPE, entropy, and enthalpy corrections) were calculated by an ideal-gas rigid-rotor harmonic-oscillator approach [63] from vibrational frequencies calculated at the MM level (RESP charges). The rotational contributions were obtained from the DFT (not MM) geometries. Optimised structures of the Bz and pClBz guests have C 2v symmetry and therefore a symmetry number of 2. The other guests have a symmetry number of 1 (C s symmetry for MeBz, EtBz, mClBz, and Hx, C 1 for the others). The octa-acid host has a formal four-fold symmetry, but owing to varying conformations of the propionate side chains, no optimised structure was symmetric. To obtain more stable results, low-lying vibrational modes were treated by the free-rotor approximation, using the interpolation model suggested by Grimme and ω0 = 100 cm−1 [21]. The calculations were performed with the thermo program, kindly provided by Prof. Stefan Grimme. The translational entropy and therefore also the free energy were corrected by 7.9 kJ/mol for the change in the standard state from 1 atm (used in the thermo program) to 1 M (used in the experiments).
Geometries were optimised with the TPSS-D3/def2-SV(P) method. Three different approaches were used for these calculations. One set of structures was obtained in a vacuum and a second set was obtained in the COSMO continuum solvent with a dielectric constant of 80. Both calculations involved only the host and the guest molecules, whereas in a third set of calculations, four water molecules were also included (forming hydrogen bonds to the guest carboxylate group) and the optimisations were performed in the COSMO solvent. These three sets will be called Vac, Cos, and Wat in the following.
After the geometry optimisation, three single-point energy calculations were performed: BP86/TZVP in vacuum and in COSMO (ε = ∞) for the COSMO-RS calculation, and TPSS/def2-QZVP′ (with the finer-than default m5 integration grid). For the Wat structures, the four water molecules were first removed from the complex. The final free-energy estimate:
was the sum of the TPSS/def2-QZVP′ energy (E TPSS), the DFT-D3 dispersion energy, including third-order terms and with Becke–Johnson damping (parameters for the TPSS functional; E DFT-D3), the COSMO-RS solvation free energy (ΔG COSMO-RS), and the entropy, ZPE, and thermal corrections (G therm; specified above). The binding free energy was the difference in this free energy between the complex, host, and guest:
We calculated the binding free energy in two different ways, either using optimised structures for all three species or using rigid structures of the host and guests (i.e. using the complex geometry for the host and the guest in the calculations of G tot(host) and G tot(guest)).
DFT-D FEP calculations
We tried to improve the FEP MM free energies by reweighting selected snapshots from the FEP simulations at the DFT-D level. For the simulations at λ = 0.01, 0.50, and 0.99, ten evenly distributed snapshots were collected (i.e. after 0.4, 0.8, …, 4.0 ns simulation) for each of the ten independent FEP simulations with the RESP charges (i.e. 100 snapshots in total). The snapshots were taken for both the V 0 and V 1 states (i.e. for both guests, which differ only in the coordinates of the perturbed atoms) and from the simulations of both the free guest and the host–guest complex. For each snapshot, a single-point TPSS calculation was performed with the def2-QZVP′ basis set on the guest and the def2-TZVP basis set on the other atoms. The calculations were performed in vacuum with the m5 integration grid, RI, and MARIJ. Three calculations were performed for each snapshot, one with the complex, which was either the host–guest system, including the 36 water molecules closest to the carboxylate C1 atom of the guest, or the guest together with the 91 water molecules closest to any atom of the guest, one with the isolated guest molecule, and one with the other atoms of the complex (host + 36 or 91 water molecules).
We used 36 water molecules, because this was the average number of waters within 7.2 Å of the guest C1 atom (the C1–O11/O12 distances are 1.21 Å) in the 10 ns simulation of the EtBz guest, which had the largest number among the nine guests. The other guests had 29–33 waters on average with standard deviations of 3. Likewise, 91 was the average number of water molecules within 6.0 Å of any atom in the guest molecule in the simulation of the free MeHx guest, which had the largest number among the nine guests; the others had 78–90 waters on average with standard deviations of 2–4. Typical structures are shown in Figure S1. Our previous studies of both ligand binding and enzyme reaction energies have shown that neutral groups (protein residues or water molecules) outside a distance of 4.5–6.0 Å from the ligand or active site have only a very small influence (<1 kJ/mol) on the binding or reaction energy [64–67].
Consequently, 28 800 DFT calculations were performed in total (8 transformations, 3 λ values, 2 states, simulations with either the complex or the free ligand, 100 snapshots, and 3 calculations on each). For each system, a DFT-D3 calculation was also performed, including third-order terms, using parameters for the TPSS functional and Becke–Johnson damping.
These energies were then used to reweight the FEP energy in various ways. BAR cannot be straightforwardly applied, as it would require MD simulations with the QM potential, which is computationally too demanding. We use two types of approaches to get around this problem and still retain the rigorousness. The first type is an endpoint approach, previously applied in reference-potential methods such as QM/FEP, QTCP (QM/MM thermodynamic cycle perturbation), and paradynamics [68–74]. Here, a thermodynamic cycle is used to obtain
where ΔG MM s is the free energy of the whole transformation at the MM level for either the bound or free states (subscript s; i.e. ∆G bound or ∆G free in Eq. 1), obtained by the standard BAR approach for the various subintervals, and the remaining terms are correction terms for going from the MM potential to the QM potential. These corrections have to be evaluated only at the endpoints of the transformation, i.e. for the V 0 state in the λ = 0.01 snapshots and for the V 1 state in the λ = 0.99 snapshots. Each correction term can either be rigorously evaluated using exponential averaging:
or approximated by a plain average (i.e. the first term in the Taylor expansion of the exponential in Eq. 7):
In the second type of approach to calculate a QM free energy, the QM free energy is obtained for each λ interval A → B of the transformation, using the same iterative approach as in BAR. However, the ensemble available for computing the QM energies has been obtained with an MM-based potential; it is as if the ensemble was obtained in a biased simulation, with the bias corresponding to the difference between the approximate (MM) and true (QM) potentials. This situation has been analysed before and has given rise to the non-Boltzmann Bennett acceptance-ratio (NBB) method [75]. In this method, when taking the bias into account, the BAR expression in Eq. (3) has to be modified into
where V bias = V MM − V QM. Thus, this method requires QM and MM evaluations of each of the ligands at each of the snapshots from each of the simulations with different λ values. QM results for intermediate λ values are obtained from two QM calculations of the V 0 and V 1 states, using Eq. 2. This is possible because we performed dual-topology simulations.
All potential energies in Eqs. 7–9 were approximated with the corresponding interaction energies, calculated as in Eq. 5. Moreover, the QM potential energies in Eq. 9 were calculated either only for the isolated QM system (x QM; i.e. the isolated guest with 91 water molecules or the host–guest complex with 36 water molecules) or from the full system in a QM/MM-like fashion:
For the reference-potential methods in Eqs. 7–8, the two approaches give the same result.
LCCSD(T0) PMISP calculations
Finally, we performed local coupled-cluster calculations with single and double excitations, and non-iterative perturbative treatment of triple excitations (LCCSD(T0)) [22]. Full counterpoise correction was used throughout. The cc-pVTZ basis set [76, 77] was applied in these calculations. Density-fitting (DF) approximations [78, 79] were used for both the Hartree–Fock and the correlation part, with the corresponding cc-pVTZ/JKFIT [80] and cc-pVTZ/MP2FIT [81] auxiliary basis sets, respectively. Localized Pipek–Mezey orbitals [82] were used and the orbital domains were determined according to a natural population analysis occupation threshold of TNPA = 0.03 [83]. Pair approximations were applied with the distance criteria rclose = 3 Bohr and rweak = 5 Bohr, and including the amplitudes of close pairs in the coupled-cluster residuals [84]. In order to account for basis-set incompleteness effects, the DF-LCCSD(T0)/cc-pVTZ energies were corrected by estimating the MP2 complete basis-set limit (CBS). Calculations at the DF-MP2/aug-cc-pVTZ and DF-MP2/aug-cc-pVQZ levels of theory were carried out (using the aug-cc-pVnZ/JKFIT and aug-cc-pVnZ/MP2FIT auxiliary basis sets). An n −3 extrapolation of the correlation energy was performed from the two points [85] and added to the aug-cc-pVQZ reference energy (MP2/CBS[3:4]). The final composite energy, including higher-order correlation effects from the local coupled-cluster result and the CBS extrapolation was computed from:
These calculations were performed on the DFT-D3 optimised Cos structures. Interaction energies were calculated with the polarised multipolar interactions with supermolecular pairs (PMISP) approach [8], with extensions to the LCCSD(T0) level, as will be described in detail and verified elsewhere [23]. These LCCSD(T0) PMISP energies replaced the E TPSS and E DFT-D3 interaction energies in Eq. 4. All wave-function calculations were carried out with the Molpro2012.1 program package [10]
Geometric measures
The geometry of the host, guests, and the complexes are described by a number of measures, as is displayed in Fig. 2.
-
r Dm is the closest distance between any guest atom and the average of the coordinates of the four HD atoms (called AD; HD is defined in Fig. 2). It measures how deep the guest is in the host.
-
r DG is the distance between AD and the average coordinate of the guest ring atoms (called AG).
-
r BB1 and r BB2 are the distances between opposite HB atoms in the host (differing by a 90° rotation around the C 4 symmetry axis of the host). The absolute difference, Δr BB = |r BB1 − r BB2|, estimates the distortion of the host.
-
r CO1 and r CO2 are the distances between opposite host CO atoms. They describe the orientation of the carboxybenzyl groups.
-
r 11, r 12, r 13, r 14, r 21, r 22, r 23, and r 24 are the distances between the two guest carboxyl oxygen atoms (O1 and O2) and the four HC atoms of the host. r min is the smallest of these eight distances and r min2 is the second smallest.
-
αt is the angle between the guest C1–C2 and the host AD–AT vectors, where AT is the average coordinate of the four HB atoms. It describes the tilt of the ligand.
-
αr is the angle between the guest O1–O2 vector and one of the host HC–HC vectors. It describes the rotation of the guest in the host.
-
r O1 and r O2 are the distances between the guest O1 and O2 atoms and the average plane defined by the four host CC atoms. They describe how much the guest reaches out of the host.
-
τ11, τ21, τ31, τ41, τ12, τ22, τ32, and τ42 are the inner two C–C–C–C dihedral angles of the four propionate groups, as is shown in Fig. 2.
-
We described hydrogen bonds between the guest carboxylate groups and water molecules by the number of water molecules with a hydrogen atom within 2.5 Å of each of the carboxylate O atoms (n HB1 and n HB2) and the shortest of these distances (r HB1 and r HB2).
-
Finally, n W is the number of water molecules that are inside the host molecule, defined as water molecules with the O atom within 6 Å of all four HM atoms.
Uncertainties and quality estimates
Reported uncertainties are standard errors, i.e. standard deviations divided by the square root of the number of samples. For the BCC charges, a single set of FEP calculations was performed and the uncertainties were obtained by error propagation of the BAR uncertainties for each individual λ value. For the RESP charges, ten independent sets of FEP calculations were performed and the reported uncertainties are the standard error of the net BAR results over these ten sets of calculations divided by \({\sqrt {10} }\). Previous studies have indicated that a single set of calculations underestimates the uncertainty compared to several independent simulations [86] and this is confirmed in the present calculations, which indicate that the BCC uncertainties are underestimated by a factor of 1.0–2.6 compared to the RESP uncertainties. The uncertainties of the free-energy estimates were obtained by non-parametric bootstrap sampling (using 100 samples) of the potential-energy differences in the BAR calculations.
The quality of the binding-affinity estimates compared to experimental data was measured using the mean signed deviation (MSD), the mean absolute deviation (MAD), the MAD after removal of the systematic error (i.e. the MSD), MADtr, the root-mean-squared deviation (RMSD), the correlation coefficient (R 2), Pearlman’s predictive index (PI) [87], and the slope and intercept of the best correlation line. In addition, Kendall’s rank correlation coefficient was calculated, either for all possible pairs of estimates (τ) or after removing pairs for which the predicted or experimental differences in affinities were not significantly different from zero at 95 % significance (τ95) [88]. For the FEP relative affinities, τ was calculated only for the transformations explicitly simulated, τr. The uncertainties of the quality estimates were obtained by a parametric bootstrap (using 500 samples), assuming the estimates are normally distributed with the mean equal to the estimate and the standard deviation equal to the reported uncertainty.
Results and discussion
In this paper, we have tried to estimate the binding affinities of nine small carboxylate guest molecules to the octa-acid host (all shown in Fig. 1). This was a part of the SAMPL4 blind-test challenge. Thus, the experimental affinities were not known during the investigation (besides the one of the Hx guest, which can be found in ref. [18]). Five sets of binding affinities, calculated with different methods, were submitted to SAMPL4. Only at a later stage were the experimental affinities revealed [89]. Additional estimates, which were not submitted, will also be discussed below.
We have worked with both MM and QM (DFT-D3 and LCCSD(T0)) approaches. Our objective was to incorporate as much as possible QM in the calculations. However, this will always come at a cost because the sampling of the conformational space cannot be as thorough as when using fast MM methods. Two main approaches were tested. First, we employed FEP calculations, based on extensive MD simulations at the MM level. This is the only set of results that was obtained solely through MM. Second, we used single minimised DFT-D3 structures with entropy and thermal effects calculated from vibrational frequencies, using a procedure close to that recently suggested by Grimme [21]. For the first approach, we also tried to improve the FEP free energies by reweighting with DFT-D3 methods. For the second approach, we also tried to improve the energies by employing the PMISP approach [8] at the LCCSD(T0) level [22, 23]. All these results will be described in separate sections.
MD simulations
We started by performing MD simulations of the host alone or in complex with the guest molecules. These give us information about the structure and dynamics of the host and the complexes, which can be compared to the optimised DFT structures and can be used to see whether the FEP calculations can be expected to be converged or if there are degrees of freedom that are not adequately sampled during the 6-ns FEP simulations.
The geometry of the free octa-acid host during the MD simulations is described in Table 1 (averages) and Fig. 3 (dynamic variation). There are 1–5 water molecules inside the host, with an average of 3.3–3.4 with both force fields. The host shows a breathing motion with the difference of the two opposite HB–HB distances (Δr BB) varying from 0 to 6 Å with an average of 1.3 Å. The largest fluctuations are found for the propionate groups with C–C–C–C dihedral angles that show transitions between the three stable states with a frequency of 0.1–1.4 ns−1.
The host–guest complexes show a similar dynamics. They still show a breathing motion with an even larger difference in the two HB–HB distances of 1–9 Å. The BCC simulations give 0.2 Å larger HB–HB distances than simulations with the RESP charges. The dihedral angles of the propionate groups also show a similar variation, with transitions between the three minima with a frequency of 0.1–1.4 ns−1. Thus, all minima of the propionate groups will in general not be sampled during a single FEP calculation.
The guest molecules are firmly bound inside the host, with average r Dm distances of 3.4–5.3 Å and a range of ~2 Å between the smallest and largest distances during the simulations (Fig. 4a). The distance is in general smallest for the largest guest EtBz and largest for the smallest guest Pen. The r DG distances are 6.6–7.9 Å with no significant differences between the BCC and RESP charges. The average tilt angle is 12–29°, with variations of ±20° during the simulations (Fig. 4b). The guest rotates quite freely in the host, making 3–8 full rotations during the 10 ns simulations (Fig. 4b). This applies even to the large EtBz guest. The guest essentially always reaches outside the host with its carboxylate atoms (Fig. 4c), with an average 1.7–2.5 Å above the average CC plane.
Dynamics of the octa-acid–Bz complex during the 10 ns simulation (RESP charges): a Variation of the r Dm and distances r DG; b variation of the αt tilt and αr rotation angles; c variation of the r O1 and r O2 out-of-plane distances; d variation in n HB1 and n HB2; e variation in r HB1 and r HB2 distances
The carboxylate O atoms form hydrogen bonds with water molecules: On the average, there are three water molecules within 2.5 Å of each of the carboxylate O atoms (Fig. 4d), with only minimal variations between the various guest molecules. The average shortest distance is always 1.7 Å, with only a restricted dynamics (1.5–2.2 Å; Fig. 4e). There is occasionally a single water molecule inside the host, together with the guest, on average 0–0.14 molecules, highest for the Bz, MeBz, and pClBz guests.
FEP results at the MM level
Relative binding affinities were calculated by FEP, according to Eq. 1. As a preliminary test of the method, we performed FEP calculations for the transformations from hexanoate (C6) to decanoate (C10) in four steps (each transforming a hydrogen atom to a methyl group) with RESP charges. Experimental results for the C6 → C8 and C8 → C10 transformations are available, 4.9 and 3.6 kJ/mol, respectively [18]. Our results (collected in Table S2), 8.0 ± 0.4 and 3.3 ± 0.9 kJ/mol, reproduce these reasonably well, with errors of 3.1 and 0.2 kJ/mol, respectively. This gives some credence to our FEP approach.
The results of the FEP calculations for the nine guest molecules in the SAMPL4 challenge are collected in Table 2. For four of the transformations, the errors are only 0–2 kJ/mol, whereas for the other four transformations, the errors are 5–7 kJ/mol (Fig. 5), giving a mean absolute deviation (MAD) of 4 kJ/mol. Quite unexpectedly, the larger errors are obtained mostly for simple transformations, MeBz → Bz, pClBz → Bz, and MeHx → Hx, but also for the harder Hx → Bz transformation. The correlation between the calculated and experimental values is good (R 2 = 0.8) and the ranking is perfect with τr = 1.0. However, the slope is 1.8–1.9, reflecting that the FEP calculations overestimate the energy difference for all four transformations with large errors.
Relative binding affinities calculated with FEP and the RESP or BCC charges, compared to the experimental results [89]
The two charge sets gave very similar results, with differences of up to 0.2–0.4 kJ/mol for five of the transformations and 1–2 kJ/mol for the remaining three. These differences are within the statistical uncertainty for all except two of the transformations, viz. MeBz → Bz and MeHx → Hx. This reflects that the RESP charges of the methyl groups are much larger in magnitude than the BCC charges, e.g. −0.44 compared to −0.03 e in MeBz and −0.72 and −0.09 e in MeHx. This is compensated by the charges on the methyl hydrogen atoms and the neighbouring C5 atom. However, the two sets agree rather closely, with a mean absolute difference of 0.08 e. The predictions based on the RESP charges are slightly but consistently better than those based on the BCC charges for all the quality estimates, although the improvement is typically not statistically significant.
The precision of the calculated relative affinities is in general excellent: For the FEP calculations with RESP charges, which are based on ten independent simulations, it is 0.05–0.15 kJ/mol for the five simple calculations, involving CH3 → H or Cl → H transformations, 0.3 kJ/mol for Hx → Bz, and 0.7 kJ/mol for Hx → Pen and Hep → Hx. For the BCC simulations, which are based on only a single set of calculations, the standard errors are 1.2–3.1 times larger. As discussed in the “Methods” section, the BCC uncertainty is probably somewhat underestimated.
As shown in Eq. 1, the relative binding free energies are obtained as the difference of estimated free energy differences for the simulations of the guest free in water or bound to the host. As shown in Table S3, these energy components are in general larger than ∆∆G bind, up to 558 kJ/mol for the Hx → Pen transformation. In general, the two simulations give a similar uncertainty, except for the Hep → Hx transformation, for which the uncertainty is more than twice as large in water than in the host with the RESP charges.
We have also calculated the relative binding free-energies with five other methods, thermodynamic integration (TI), TI with a cubic approximation, forward exponential averaging, backward exponential averaging, and multi-state BAR (all using the same simulations). For five of the transformations, the results of the five methods agree within 1 kJ/mol, indicating well-behaving transformations and a proper number of λ values (Table S4). For the Hep → Hx transformation, TI gives a 3 kJ/mol lower results and for the MeHx → Hx and Hx → Pen transformations, there are differences of up to 13–15 kJ/mol, indicating poor overlap between some of the intermediate states and that the calculations could gain from using more λ values or employing separate van der Waals and electrostatic transformations.
We submitted two sets of results to the SAMPL4 challenge, based on the RESP and BCC charges, respectively. These were obtained by recalculating our relative affinities to absolute affinities employing an experimental estimate of −21.1 kJ/mol for the Hx guest [18]. These estimates are listed in Table S5 and gave MADs of 3.2 and 3.7 kJ/mol, for RESP and BCC, respectively, whereas both gave τ95 = 1.0, and R 2 = 0.9. The RESP charges gave the best results among the 12 submissions to this SAMPL4 challenge. Still, these results are somewhat affected by the use of the old experimental estimate for the Hx guest (having an error of 2.4 kJ/mol, compared to the new one). Using instead the new experimental estimate for the affinity for Hx (−23.5 kJ/mol [89]), the MAD decreases to 2.6 or 2.9 kJ/mol, whereas R 2 and τ95 are not affected.
FEP results at the DFT-D3 level
Next, we tried to incorporate DFT-D3 calculations into the FEP. Due to the underlying cost of these methods, a different approach had to be used. We run in total 28 000 DFT-D3 calculations at 100 snapshots and three λ values from the RESP FEP simulations. The DFT calculations were performed on truncated systems with only 36 (complex simulation) or 91 (free guest simulation) water molecules. Several methods were used to estimate the binding free-energies at the DFT-D3 level, as is described in the “Methods” section.
A first estimate of the difference between the QM and MM energies can be obtained by taking the average difference between the QM and MM interaction energy for the endpoints of the perturbation (Eq. 8). This energy difference is sizeable, as can be seen in Table 3, up to 136 kJ/mol. The QM interaction energy is always less favourable than the MM one. The standard error is 2–3 kJ/mol, somewhat lower for the free guest than for the complex. For the Bz, Hx, and MeBz guest molecules, we have several estimates (from different transformations; marked with the same colour in Table 3) and these agree within 4 kJ/mol, except for Hx (11 kJ/mol). There is essentially no difference whether the MM energies are calculated with standard or soft-core van der Waals potentials (differences in the average interaction energies of less than 0.8 kJ/mol, except in one case with 1.6 kJ/mol). These energies could be used as simple extrapolation corrections to the FEP binding free energies, giving sizeable corrections to several transformations, in particular the EtBz → MeBz and Hx → Bz transformations (−43 and −45 kJ/mol) with standard errors of 5 kJ/mol (Table 4, column Plain av.). Unfortunately, this correction leads to a degradation of the computed estimates, increasing MAD to 17 kJ/mol, decreasing R 2 to 0.6, and increasing the range of the predicted affinities to 56 kJ/mol.
Strictly, we should not use plain averages for the energy corrections, but instead exponential averages (Eq. 7), which would give a QTCP-like approach. Similar corrections have been used in other contexts before [68–74]. Unfortunately, such an approach becomes unstable if the differences between the MM and QM potentials are too large, leading to corrections that depend only on a few of the DFT calculations [90]. In the present case, this gives a correction of 35–92 kJ/mol for the host simulations but −51 to −149 kJ/mol in water (Table 3). The uncertainties increase to 3–22 kJ/mol, except for three of the Bz and Hx calculations in water, which gave very large and unreliable results. Disregarding these and using the average value for the identical Bz, MeBz, and Hx simulations, we can arrive at reasonable corrections for all transformations of −75 to 6 kJ/mol, with standard errors of 16–32 kJ/mol (Table 4, Exp. av. column). However, these estimates are slightly worse than the plain averages with MAD = 26 kJ/mol, R 2 = 0.6, and a range of 94 kJ/mol.
The problem with doing QM corrections only at the end points of the transformation is that no error cancellation is obtained between the QM calculations of the two states; thus the statistical noise becomes large if the QM and MM potentials are too different. The NBB method (Eq. 9) [75] provides a more balanced way to treat FEP calculations where the sampling has been performed with a cheap MM potential and energies with the more expensive QM potential. This method makes use of QM calculations of both states on the snapshots sampled on all three λ values. Performing NBB on energies obtained for the truncated QM system give energy corrections of −52 to 16 kJ/mol with standard errors of 5–12 kJ/mol (third column in Table 4). This improves the results somewhat to MAD = 17 kJ/mol, R 2 = 0.7, and a range of 68 kJ/mol.
However, it seems more reasonable to perform NBB on energies containing also an estimate of interaction energy from the remaining water molecules outside the QM system, i.e. by using a standard QM/MM approach (Eq. 10). This changes the NBB energy correction by 1–4 kJ/mol, except for the Hx → Bz transformation (26 kJ/mol). The precision is still 5–13 kJ/mol. Such predictions have a similar accuracy compared to experiments with MAD = 20 kJ/mol and R 2 = 0.6, and a range of 68 kJ/mol (fourth column in Table 4).
There are several differences between how the QM and MM free energies were calculated. First, the MM free energies were based on 13 rather than three λ values. Three λ values have been shown to be enough in a previous investigation, giving errors of less than 2 kJ/mol [91]. However, this of course depends on the specific transformations. We can directly estimate this effect by repeating the BAR calculations for the MM FEP data with only three λ values. For six of the transformations, this changes the relative binding free energy by less than 5 kJ/mol. However, for the Hx → Pen and Hep → Hx transformations, the effect is much larger, 12 and 46 kJ/mol, respectively.
Second, the MM FEP calculations were performed with periodic systems and Ewald summation, whereas the QM/(MM) calculations were performed for finite systems. Third, the QM correction is based on interaction energies, i.e. ignoring the internal energy of the guest, whereas the FEP energies include all energy terms; this can make a difference if the dominant conformation differs between the bound and unbound states. We can correct for all these three effects by performing BAR calculations based on the same MM data as used for the QM correction, i.e. either the truncated QM system (QM) or all atoms (QM/MM). The difference between these results and the original BAR results provides a correction for the differences in the method, which has been added to the columns with Corr. = Yes in Table 4. These correction terms are 6 to −68 (QM) or 1 to −48 kJ/mol (QM/MM), with standard errors of 5–13 kJ/mol. Unfortunately, they give worse results than the uncorrected data, with R 2 = 0.4–0.5, MAD = 26–27 kJ/mol, and ranges of 71–73 kJ/mol. The results of the various DFT-FEP methods are shown in Fig. 6. It can be seen that all DFT-FEP approaches fail for the four transformations with small ∆∆G bind energies, giving a much too negative affinity.
Relative binding affinities calculated with the various DFT-FEP approaches, compared to the experimental results [89]
To the SAMPL4 challenge, we submitted one set of predictions, based on the NBB-QM corrections and recalculated to absolute binding energies using the old experimental data for the Hx guest. Unfortunately, there was a bug in the script calculating the NBB free energies so those predictions were incorrect. Correct data are given in Table 4.
DFT-D3 structures
In our second approach, inspired by a method recently suggested by Grimme [21], structures were optimised at the BP86-D3/def2-SV(P) level. The optimisations were started from the end of the 10 ns MD simulation of the nine complexes with RESP charges. Three different sets of structures were calculated, one in vacuum (Vac), one in the COSMO (ε = 80) continuum solvent (Cos), and another one in the same COSMO solvent, but with four water molecules forming hydrogen bonds to the guest carboxylate group (Wat). The latter four molecules were removed before the energy calculations.
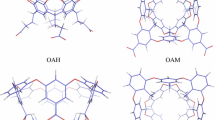
Pictures of the optimised Cos structures are shown in Fig. 7. It can be seen that the general binding pattern of the guest is similar in all structures: It binds inside the host with the carboxylate group just above the rim of the structure, with at least one carboxylate atom 0.4–3.2 Å above the average plane of the four CB atoms (r O1 and r O2 in Table 5). In most of the Vac and Cos structures, each of the two carboxylate oxygen atoms of the guest forms a weak hydrogen bond with one HC atom on the carboxybenzene groups of the host with H–O distances of 1.9–2.3 Å (r min and r min2 in Table 5; the EtBz guest in the Vac structure forms instead a hydrogen bond with the HC atom at 2.6 Å distance). This leads to a tilted orientation of the carboxybenzene group of the guest, forming a tilt angle (αt) of 8–58°. In the Wat structures, the importance of these O–HC interactions is reduced and most of the structures show no or only one such hydrogen bond.
Clearly, this binding mode is somewhat different from what was found in the MD simulations: The guests are more hidden in the host with smaller and more varying r O distances. Moreover, the guest carboxylate groups form hydrogen bonds with the HC atoms, whereas such interactions were rare in the MD simulations. Consequently, the tilt angle is in general larger (typically by ~20°) and more varying in the DFT structures.
The r Dm distances are 2.6–5.9 Å, with an average of 3.9 Å. There is no clear-cut tendency that the larger ligands are closer to the bottom. However, the r DG distances increase for the larger ligands, e.g. Bz < MeBz < EtBz. Thus, there seems to be a major random component in the structures. In particular, we saw in Fig. 3c that the propionate groups can attain many different conformations by rotations around the C–C bonds. In the DFT-optimised structures, one of these conformations is obtained and for the Cos and Wat structures, the obtained structure depends on the starting structure, giving rise to a large variation in these angles. For the Vac structures, the variation is much smaller because the stronger repulsion between the carboxylate groups forces them to be as far from each other as possible (cf. Figure 7).
Likewise, there is a large variation in the distortion of the macrocycle, as is best seen from the top views in Fig. 7d. The Vac structures are nearly symmetric, with Δr BB differences of 0–0.4 Å (again owing to the repulsion of the carboxylate groups; this repulsion also forces the carboxybenzyl to point straight out from the host, giving larger rCO distances; cf. Fig. 7a), whereas for the Cos structures the differences are 1.1–7.2 Å and for the Wat structures, the differences are 0.1–8.5 Å. Again, this is in accordance with the breathing motion of the host seen in the MD simulations: the DFT-D3 optimisations end up in one of many possible conformations of the host. The distortion of the host was even more pronounced in test calculations with the older DFT-D2 dispersion correction [92], which gave strange structures with even larger differences in the r BB distances, cf. Figure S2. Apparently, DFT-D2 overestimates dispersion effects, as has also been observed before [93].
DFT-D3 binding energies
Apparently, the DFT optimisations give one structure of many possible conformations of the host–guest complexes. It is likely that these differences in the structures may affect the calculated affinities in a random way, making the octa-acid system a hard test case for a minimisation-based approach. Unfortunately, the geometry optimisations were quite time consuming (they took typically ~2 weeks on a single processor, but some structures took as long as 2 months; the new statpt optimiser in Turbomole was appreciably faster and more successful than the old relax optimiser). This made it hard to perform an exhaustive investigation of the influence of different conformations on the binding affinities.
An indication of the stability of the calculated energies can be found from the structures of the free host, for which nine independent calculations were run for Vac and 18 for Cos and Wat (which should all give the same structures). The raw BP86-D3/def2-SV(P) + COSMO(ε = 80) energies varied by 7 kJ/mol for the Vac structures and by 34 kJ/mol for the Cos and Wat structures. The reason for the smaller variation in the Vac structures is that the orientation of the propionate groups is more similar in those structures, because of the large repulsion between these groups in vacuum.
As an additional test, we also optimised ten different structures of the Bz complex, starting from different snapshots of the MD simulation. The resulting structures differed by up to 50 kJ/mol in the TPSS-D3/def2-SVP+COSMO(ε=80) energy. On the other hand, ten structures of the isolated guest gave energies that agreed within 0.1 kJ/mol.
Table 6 shows guest binding energies and free energies calculated with various approaches and for the three sets of structures. The first four columns contain the results obtained at the various DFT levels of theory. The raw TPSS-D3/def2-SV(P) results (used for the geometry optimisation) in the first column, show that the binding energies strongly depend on solvation effects: For the Vac structures (with energies also calculated in vacuum), the binding energies are large and positive (818–847 kJ/mol), owing to the Coulombic repulsion between the host and the guests with net charges of −8 and −1 e, respectively. This repulsion is removed in water solution, so that for the Cos and Wat structures (the latter calculated after removal of the water molecules from the complex), the binding energies are negative, −58 to −108 for the Cos structures and −18 to −71 kJ/mol for the Wat structures. The energies give R 2 = 0.2–0.4 and MADtr = 6–15 kJ/mol (the mean absolute deviation after removal of the mean signed error) compared to the experimental data, with the Vac structures giving the best results.
The BP86/TZVP calculations give similar results, although the energies are ~120 kJ/mol more positive, mainly owing to the omitted dispersion energy. It is also notable that the vacuum binding energies for the Cos and Wat structures become more unstable, as is illustrated by a much larger range of the binding energies between the various ligands (e.g. 238 kJ/mol for BP86/TZVP in vacuum, but 50 kJ/mol for the TPSS-D3/def2-SV(P) for the Cos structures), but also by the poor R 2 (−0.4 and −0.1 for the Cos and Wat structures, respectively; a negative sign of R 2 indicates that R is negative) and the large MADtr (78 and 34 kJ/mol). For the Vac structures, the results are slightly better, e.g. R 2 = 0.1 and MADtr = 15 kJ/mol. The BP86/TZVP+COSMO(ε=∞) results are even better, especially for the Wat structures (R 2 = 0.5 and MADtr = 5 kJ/mol).
The TPSS/def2-QZVP′ energies are quite similar to the BP86/TZVP vacuum energies, with differences of up to 12 kJ/mol. The four sets of calculations show that the effects of the basis set and the DFT functional are quite small (but the effect is still important considering that the nine guests have experimental binding free energies that differ by no more than 16 kJ/mol). The DFT integration grid had only a minor effect on the energies, up to 1 kJ/mol (difference between calculations with m3 and m5 grids), so some time could be saved by using only the smaller grid.
Several corrections were added to the raw DFT electronic energies, as shown in Eq. 4. The DFT-D3 dispersion energies were found to be large and significantly contribute to the stability of the complexes (values between −76 to −144 kJ/mol). The polar solvation energy (the difference between the BP86/TZVP COSMO(ε = ∞) and vacuum calculations) is larger, and increases in magnitude from the Vac to the Wat structures (−898, −943, and −961 kJ/mol on average). The COSMO-RS solvation energies (based on these calculations), including also non-polar solvation effects, are 12–43 kJ/mol more negative.
The TPSS/def2-QZVP’, DFT-D3, and COSMO-RS energies can be added to give a binding enthalpy which is negative, −28 to −118 kJ/mol. The Vac and Wat structures show a decent correlation with experimental data (R 2 = 0.5–0.8 and MADtr = 8–9 kJ/mol), whereas the results for the Cos structures are poor, R 2 = 0.0, and MADtr = 21 kJ/mol.
Finally, we have also calculated entropies, ZPEs, and thermal effects. Their sum is positive owing to the loss of translational and rotational entropy. If these terms are added to the enthalpies, we obtain a final binding free-energy estimate. It is 5 to −36 kJ/mol for the Vac structures, 7 to −66 kJ/mol for the Cos structures, and 20 to −22 kJ/mol for the Wat structures. The correlation to experimental results is similar to that of the enthalpies, R 2 = 0.82 and 0.63 and MADtr = 7 kJ/mol for the Vac and Wat structures and R 2 = −0.02 and MADtr = 23 kJ/mol for the Cos structures.
Energies discussed so far are based on the optimised structures for all complexes, guests, and the host. As mentioned above, this may be problematic, because the propionate groups may have different conformations in the various complexes and the host can show different degrees of distortion. Therefore, we also calculated the corresponding energies for rigid structures, i.e. where the structures of the host and the guest were taken from that of the complex, without any optimisation. If this is done for the ten different optimised structures of the Bz guest, the variation in binding energies is reduced from 50 to 20 kJ/mol. Rigid energies for all guest molecules are listed in the four last columns in Table 6.
For the Vac structures, the calculated energies are quite similar to those obtained with the relaxed structures: the various binding energies change by less than 10 kJ/mol, except for the solvation energies (up to 23 kJ/mol). The vacuum BP86 and TPSS, as well as the DFT-D3 binding energies always become more negative, reflecting that the host and ligand are not optimised. On the other hand, the solvation free energies and thermal corrections are always positive. As an effect, a non-systematic variation in the net binding free energies of up to 17 kJ/mol is observed. They reproduce the experimental data about as well as the relaxed data with R 2 = 0.70 and MADtr = 4 kJ/mol.
For the Cos and Wat structure, the differences are much larger, up to 105 kJ/mol, and without any trends regarding the sign. Many of the differences cancel and for the final binding free energies the differences are up to 33 kJ/mol, with a varying sign for the Cos structures, but always negative for the Wat structures. For the latter, there is no change in the performance compared to the experimental data (R 2 = 0.67 and MADtr = 8 kJ/mol), but for the Cos structures, there is a major improvement, giving them a performance similar to that of the other two sets of structures, R 2 = 0.73 and MADtr = 7 kJ/mol.
The effect of using rigid structures comes mainly from the host: The changes for the guests are only up to 6 kJ/mol for the Vac and Cos structures and up to 9 kJ/mol for the Wat structures (owing to the distortion caused by the explicit hydrogen bonds to the four water molecules). The only exception is the solvation term that changes by up to 12 kJ/mol. Therefore, we may reintroduce these terms as a ligand-relaxation energy. Such a correction term, calculated at the TPSS/def2-QZVP’ level, is included in Table 6 (column relax). It amounts to 0 to −6 kJ/mol for the Vac and Cos structures and −4 to −9 kJ/mol for the Wat structures. It has only a minor influence on the results. Adding the remaining terms (DFT-D3, solvation, and thermal effects) has again little influence on the results.
As seen above, the solvation energy is a major correction factor. The systems under study show strong electrostatic interactions and the overall accuracy can be strongly undermined if the continuum solvent is insufficient to capture the solvent effect. We have already observed that the COSMO-RS solvation energies seem to introduce a quite large variation in the calculated values. Therefore, we also tried to calculate final binding free energies, based on the solvation energy from the BP86/TZVP calculations (difference between the COSMO and vacuum calculation) instead of the COSMO-RS energies (thereby including only the electrostatic part of the solvation energy). This gave worse R 2 for the Vac and Cos structures (0.65 and 0.58), but better for the Wat structures (0.67) and better MADtr for all structures (3–6 kJ/mol). However, the estimated affinities were all too low in absolute terms with averages of 7 to −6 kJ/mol.
Finally, we could consider the three sets of structures (Vac, Cos, and Wat) as a minimal sampling of possible structures and simply average the results of the three structures for each guest. This is the consensus estimate shown in Table 7 and Fig. 8, which gave R 2 = 0.77 and MADtr = 6 kJ/mol. However, the range of the estimates is 40 kJ/mol, 2.5 times larger than for the experimental affinities (16 kJ/mol). Consequently, the slope of the best correlation line is 2.0. The τ and τ95 values are 0.67 and 0.71, the same as for the Wat structures, but these measures are slightly larger for the Vac and Cos structures, 0.72 and 0.77.
Absolute binding affinities calculated from the various DFT-optimised structures compared to the experimental results [89]
Only a single set of DFT-D3 energies were submitted to SAMPL4, based on the Cos structures, rigid calculations with guest relaxation. It is also shown in Table 7, but it is not identical to any of the previously discussed entries for several reasons. First, it employed the thermal corrections from the relaxed structures (having only a minor effect, up to 2 kJ/mol). Second, the estimate for the Bz guest was (by mistake) a mixture of the ΔG therm terms for the correct structure and the other energy terms for another structure that is 1 kJ/mol higher in the optimised TPSS-D3/def2-SV(P)+COSMO(ε=80) energy, giving a 1 kJ/mol more negative binding energy. Third, the correction for the change in the standard state for the entropy was missing (making the energies 7.9 kJ/mol too negative). Fourth and most important, the result for the Pen guest was an outlier (in the submission we wrote: “results of ligand 8 seem to be wrong, but we have not had time to spot the error”). The problem could be traced to the structure (shown in Figure S3 in the supplementary material and also included in Table 5): The ligand binds deeper inside the host, with only one O–HC hydrogen bonds. Reoptimisation gave the structure in Fig. 7b, which was 10 kJ/mol more stable at the optimised TPSS-D3/def2-SV(P)+COSMO(ε=80) level and 106 kJ/mol more stable at the TPSS/def2-QZVP’ level. Consequently, the latter structure has been employed throughout this paper.
The group of Grimme also submitted a prediction for this test case with similar methods [94]. Their calculations differed in several aspects:
-
1.
They neutralised the host by protonation and added corrections for this afterwards
-
2.
They performed a scan at the HF3c level [95] of 28 structures for each complex, varying the tilt and rotation of the guest in the host. The final prediction was based on the one with the lowest free energy of binding.
-
3.
They employed optimised structures for the binding energies, but started the optimisation of the free host from the structure of the complex.
-
4.
Structures were optimised at the TPSS-D3(BJ)/def2-TZVP+COSMO(ε=78) level and single-point energies were calculated at the TPSS-D3(BJ)/def2-TZVP level.
-
5.
Frequencies were obtained at the HF3c level [95].
Their submitted results were in general slightly worse than our results in Table 7, with MADtr = 12 kJ/mol, R 2 = 0.3, and τ = 0.3. It is not fully clear why this is the case. Undoubtedly, their systematic search of the most favourable structure should be better than our more random selection of one (or three) structures, but the MD simulations show that the rotation and tilt of the guest are not the most important sources of conformational variation in the octa-acid structures. Frequencies at the HF3c level are also expected to be better. We used a smaller basis set for the geometries but a slightly better one for the energies, but this is not expected to have a major influence on the results. Instead, we believe that our use of rigid structures for the energy calculations may be the most important reason of our improved results. The effect of the neutralisation of the host is hard to judge, although the results indicate that it is no disadvantage to employ a fully charged host.
LCCSD(T0) PMISP binding energies
In a last set of calculations, we tried to compute the host–guest interactions at the coupled cluster level of theory. CCSD(T) is often considered a gold standard in quantum chemical methods. The use of local approximations allows the latter method to be routinely used for systems of up to ~60 atoms, but the octa-acid complexes contain 198–207 atoms. Therefore, we developed a PMISP approach [8, 23] to calculate the (rigid) ligand-binding energies at the LCCSD(T0) level (with an MP2 estimate to CBS), employing a total of 60 fragment and conjugate capping calculations. The calculations were based on the Cos structures and the LCCSD(T0)-PMISP binding energies simply replaced the E TPSS + E DFT-D3 energies in Eq. 4. The results are included in Table 7.
Unfortunately, the LCCSD(T0)-PMISP corrections make the results significantly worse with R 2 = 0.28 and MADtr = 14 kJ/mol. In particular, the range of the calculated affinities increases to 55 kJ/mol and the estimates become ~37 kJ/mol too negative. A likely explanation for this is that the solvation energies, which we took directly from the DFT calculations, are not accurate at the CCSD(T0) level for these highly charged complexes, leading to the observed exaggerated variations between the guest molecules (Fig. 9). This will be tested and further discussed elsewhere [23]. Another possible reason for the over-binding of both the LCCSD(T0) and DFT results is that guests bind deeper in the host in the DFT structures than in the MD snapshots (cf. Tables 1, 5) and that minimised structures are used rather than a thermodynamic ensemble of properly sampled structures.
Absolute binding affinities before and after the LCCSD(T0) correction of the Cos optimised structures compared to the experimental results [89]
Of course, the original submission had the same problems as for the DFT-D3 results, discussed above, with the extra complication that the LCCSD(T0) calculations for the Bz guest was based on a structure taken before it was fully converged (0.4 kJ/mol difference at the TPSS-D3/def2-SV(P)+COSMO(ε=80) level). Also for this submission, the Pen guest was noted as an outlier.
Conclusions
In this study, we have calculated ligand-binding free energies with four different approaches in the SAMPL4 octa-acid host–guest blind challenge. The results were of varying quality.
The best predictions were obtained with standard MM FEP calculations and energies obtained with BAR. They gave relative affinities for the eight studied transformations of reasonable quality, with a correlation coefficient (R 2) of 0.8 and a perfect τr, thereby providing the best prediction among the 12 submissions. However, the MAD of 4 kJ/mol and the fact that half of the transformations gave errors of 5–9 kJ/mol was somewhat disappointing, considering the simplicity of the studied system. The results are well converged with standard errors of 0.05–0.7 kJ/mol using ten independent sets of calculations. Interestingly, there were only minor differences between the two charge methods tested, RESP and BCC, less than 2 kJ/mol.
Our attempt to improve these results with 28 800 DFT-D3 calculations using systems with 36 or 91 water molecules (287–312 atoms) and the NBB approach was unfortunately a failure. The reason for this is most likely the poor precision of the corrections (5–13 kJ/mol), caused by the large difference between the MM and QM potentials [90]. Of course, this could be improved by running more DFT calculations, but the results indicate that 25–169 times more calculations are needed to bring the uncertainty down to 1 kJ/mol. It is probably more economic to perform DFT-D3 calculations for more λ values, especially as two of the transformations gave very poor results with only three λ values.
Third, we calculated absolute affinities with single minimised DFT-D3 structures, using the approach recently suggested by Grimme [21]. This gave reasonable predictions with R 2 = 0.6–0.8 and τ95 = 0.7–0.8. However, the large MADtr (5–9 kJ/mol) and the great slope of the best correlation line (1.7–2.1) indicate that this approach exaggerates the differences between the ligands. The key problem is the use of single structures, which ignores the flexibility of the molecules. This is a major problem for the flexible octa-acid host molecule with its breathing motion and the movements of the propionate side chains. The problem can partly be solved by using rigid interaction energies or performing the optimisations in vacuum, but we doubt that really accurate results can be obtained with an approach based on only minimised structures.
Finally, we tried to improve the DFT-D3 affinities by using LCCSD(T0) calculations with the PMISP approach. Unfortunately, this did not improve the predictions, probably owing to problems with the solvation energies. However, it shows that it is possible to calculate binding affinities at the LCCSD(T0)/CBS level for systems of this size.
It should be pointed out that the octa-acid test case is not fully ideal for these calculations. In particular, the high charge of the host gave severe problems in the calculations, e.g. numerous positive orbital energies and extremely high solvation energies. We have chosen not to neutralise the complexes, because this would lead to additional problems with many possible conformations of the counter ions or protons (hydrogen bonds). An alternative solution could be to simply delete the propionate side chains, which cause major problems with their large flexibility and many conformations.
In conclusion, the best approach to obtain relative binding free energies for this octa-acid host–guest system is FEP calculations at the MM level. Currently, there is no computational protocol for QM approaches that give improved predictions at a reasonable computational effort. However, this is likely to change in the future, owing to the continuing increase in computational resources and the development of improved approaches. In particular, we need to find effective ways to apply QM corrections in systems with large fluctuations in the conformational space. Otherwise, the impact of the bias introduced may be larger than the benefit.
References
Gohlke H, Klebe G (2002) Angew Chem Int Ed 41:2644
Jorgensen WL (2009) Acc Chem Res 42:724
Zhou H-X, Gilson MK (2009) Chem Rev 109:4092
Michel J, Essex JW (2010) J Comput Aided Mol Des 24:639
Christ CD, Mark AE, van Gunsteren WF (2010) J Comput Chem 31:1569
Wereszczynski J, McCammon JA (2012) Quart Rev Biophys 45:1
Halgren TA, Damm W (2001) Curr Opin Struct Biol 11:236
Söderhjelm P, Ryde U (2009) J Phys Chem A 113:617
Cavalli A, Carloni P, Recanatini M (2006) Chem Rev 106:3497
Werner H.-J, Knowles P. J, Knizia G, Manby F. R, Schütz M et al (2012) MOLPRO,, version 2012.1, a package of ab initio programs. see http://www.molpro.net
Raha K, Peters MB, Wang B, Yu N, Wollacott AM, Westerhoff LM, Merz KM (2007) Drug Discov Today 12:725
Söderhjelm P, Kongsted J, Genheden S, Ryde U (2010) Interdiscip Sci Comput Life Sci 2:21–37
Söderhjelm P, Genheden S, Ryde U (2012) Protein–ligand interactions. In: Gohlke H (ed) Methods and principles in medicinal chemistry, vol 53. Wiley-VCH, Weinheim, pp 121–143
Antony J, Grimme S (2012) J Comput Chem 33:1730
Muddana HS, Varnado CD, Bielawski CW, Urbach AR, Isaacs L, Geballe MT, Gilson MK (2012) J Comput Aided Mol Des 26:475
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) Blind prediction of the host–guest binding affinities from the SAMPL4 challenge. J Comput-Aided Mol Design (in press)
Gibb CLD, Gibb BCJ (2004) Am Chem Soc 126:11408
Sun H, Gibb CLD, Gibb BC (2008) Supramol Chem 20:141
Gibb CLD, Gibb BC (2009) Tetrahedron 65:7240
Grimme S, Antony J, Ehrlich S, Krieg H (2010) J Chem Phys 132:154104
Grimme S (2012) Chem Eur J 18:9955
Hampel C, Werner H-J (1996) J Chem Phys 104:6286
Andrejić M, Mata RA, Ryde U, Söderhjelm P (2014) Chem Phys Chem (submitted)
Wang JM, Wolf RM, Caldwell KW, Kollman PA, Case DA (2004) J Comput Chem 25:1157–1174
Jorgensen WL, Chandrasekhar J, Madura JD, Impley RW, Klein ML (1983) J Chem Phys 79:926–935
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts BP, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye Q, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2010) AMBER 11. University of California, San Francisco
Jakalian A, Bush BL, Jack DB, Bayly CI (2000) J Comput Chem 21:132–146
Jakalian A, Jack DB, Bayly CI (2002) J Comput Chem 23:1623–1641
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) J Phys Chem 97:10269–10280
Besler BH, Merz KM, Kollman PA (1990) J Comput Chem 11:431–439
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, revision A02. Gaussian Inc, Wallingford CT
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) J Comput Phys 23:327–341
Berendsen HJC, Postma JPM, Van Gunsteren WF, Dinola A, Haak JR (1984) J Chem Phys 81:3684–3690
Darden T, York D, Pedersen L (1993) J Chem Phys 98:10089–10092
Genheden S, Ryde U (2011) J Comput Chem 32:187
Wu X, Brooks BR (2003) Chem Phys Lett 381:512–518
Tembe BL, McCammon JA (1984) Comp Chem 8:281–283
Bennett CH (1976) J Comput Phys 22:245–268
Shirts MR, Pande VS (2005) J Chem Phys 122:144107
Shirts MR, Chodera JD (2008) J Chem Phys 129:124105
Kirkwood JG (1935) J Chem Phys 3:300–313
Zwanzig RWJ (1954) Chem Phys 22:1420–1426
Steinbrecher T, Mobley DL, Case DA (2007) J Chem Phys 127:214108
Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C (1989) Chem Phys Lett 162:165
Treutler O, Ahlrichs RJ (1995) Chem Phys 102:346
Tao J, Perdew JP, Staroverov VN, Scuseria GE (2003) Phys Rev Lett 91:146401
Becke AD (1988) Phys Rev A 38:3098–3100
Perdew JP (1986) Phys Rev B 33:8822–8824
Schäfer A, Huber C, Ahlrichs R (1994) J Chem Phys 100:5829
Weigend F, Ahlrichs R (2005) Phys Chem Chem Phys 7:3297–3305
Weigend F, Furche F, Ahlrichs R (2003) J Chem Phys 119:12753
Eichkorn K, Treutler O, Öhm H, Häser M, Ahlrichs R (1995) Chem Phys Lett 240:283–290
Eichkorn K, Weigend F, Treutler O, Ahlrichs R (1997) Theor Chem Acc 97:119–126
Sierka M, Hogekamp A, Ahlrichs R (2003) J Chem Phys 118:9136
Grimme S, Ehrlich S, Goerigk L (2011) J Comput Chem 32:1456–1465
http://www.thch.uni-bonn.de/tc/index.php?section=downloads&subsection=getd3
Klamt A, Schüürmann J (1993) J Chem Soc Perkin Trans 2:799–805
Schäfer A, Klamt A, Sattel D, Lohrenz JCW, Eckert F (2000) Phys Chem Chem Phys 2:2187–2193
Klamt A, Jonas V, Bürger T, Lohrenz JCW (1998) J Phys Chem 102:5074–5085
Klamt A (1995) J Phys Chem 99:2224
Eckert F, Klamt A (2002) AIChE J 48:369
Eckert F, Klamt A (2010) COSMOtherm, version C30, release 1301. COSMOlogic GmbH & Co KG, Leverkusen
Jensen F (1999) Introduction to computational chemistry. Wiley, Chichester, pp 298–303
Kaukonen M, Söderhjelm P, Heimdal J, Ryde U (2008) J Chem Theory Comput 4:985
Söderhjelm P, Husberg C, Strambi A, Olivucci M, Ryde U (2009) J Chem Theory Comput 5:649
Hu L, Eliasson J, Heimdal J, Ryde U (2009) J Phys Chem A 113:11793
Genheden S, Ryde U (2012) J Chem Theory Comput 8:1449
Wesolowski T, Warshel A (1994) J Phys Chem 98:5183–5187
Olsson MH, Hong G, Warshel A (2003) J Am Chem Soc 125:5025–5039
Wood RH, Yezdimer EM, Sakane S, Barriocanal JA, Doren DJJ (1999) Chem Phys 110:1329
Rod TH, Ryde U (2005) Phys Rev Lett 94:138302
Plotnikov NV, Kamerlin SCL, Warshel A (2011) J Phys Chem B 115:7950–7962
Woods CJ, Manby FR, Mulholland AJ (2008) J Chem Phys 128:014109
Beierlein FR, Michel J, Essex JW (2011) J Phys Chem B 115:4911–4926
König G, Boresch S (2011) J Comput Chem 32:1082
Dunning TH (1989) J Chem Phys 90:1007
Woon DE, Dunning TH (1993) J Chem Phys 98:1358
Polly R, Werner H-J, Manby FR, Knowles PJ (2004) Mol Phys 102:2311
Werner H-J, Manby FR, Knowles PJ (2003) J Chem Phys 118:8149
Weigend F (2002) Phys Chem Chem Phys 4:4285
Weigend F, Köhn A, Hättig C (2002) J Chem Phys 116:3175
Pipek J, Mezey PG (1989) J Chem Phys 90:4916–4926
Mata RA, Werner H-J (2007) Mol Phys 105:2753–2761
Dieterich JM, Werner H-J, Mata RA, Metz S, Thiel W (2010) J Chem Phys 132:035101
Helgaker T, Klopper W, Koch H, Noga J (1997) J Chem Phys 106:9639
Genheden S, Ryde U (2010) J Comput Chem 31:837–846
Pearlman DA, Charifson PS (2001) J Med Chem 44:3417
Mikulskis P, Genheden S, Rydberg P, Sandberg L, Olsen L, Ryde U (2012) J Comput-Aided Mol Design 26:527–554
Gibb CLD, Gibb B (2013) J Comput-Aided Mol Design. doi:10.1007/s10822-013-9690-2
Heimdal J, Ryde U (2012) Phys Chem Chem Phys 14:12592–12604
Genheden S, Nilsson I, Ryde U (2011) J Chem Inf Model 51:947–958
Grimme S (2006) J Comput Chem 27:1787–1799
Ryde U, Mata RA, Grimme S (2011) Dalton Trans 40:11176
Sure R, Antony J, Grimme S (2014) J Phys Chem B (submitted)
Sure R, Grimme S (2013) J Comput Chem 34:1672–1685
Acknowledgments
This investigation has been supported by Grants from the Swedish research council (project 2010-5025). The computations were performed on computer resources provided by the Swedish National Infrastructure for Computing (SNIC) at Lunarc at Lund University and HPC2N at Umeå University. The collaboration between the Universities of Lund and Göttingen has been carried out within the framework of the International Research Training Group 1422 Metal Sites in Biomolecules—Structures, Regulation, Mechanisms and M. A. is supported through a Ph.D. scholarship in this International Research Training Group. D. C. thanks FEBS for a short-term fellowship. We are grateful to Prof. Stefan Grimme for providing us with the thermo program.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mikulskis, P., Cioloboc, D., Andrejić, M. et al. Free-energy perturbation and quantum mechanical study of SAMPL4 octa-acid host–guest binding energies. J Comput Aided Mol Des 28, 375–400 (2014). https://doi.org/10.1007/s10822-014-9739-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9739-x