Abstract
The success of molecular fragment-based design depends critically on the ability to make predictions of binding poses and of affinity ranking for compounds assembled by linking fragments. The SAMPL3 Challenge provides a unique opportunity to evaluate the performance of a state-of-the-art fragment-based design methodology with respect to these requirements. In this article, we present results derived from linking fragments to predict affinity and pose in the SAMPL3 Challenge. The goal is to demonstrate how incorporating different aspects of modeling protein–ligand interactions impact the accuracy of the predictions, including protein dielectric models, charged versus neutral ligands, ΔΔGs solvation energies, and induced conformational stress. The core method is based on annealing of chemical potential in a Grand Canonical Monte Carlo (GC/MC) simulation. By imposing an initially very high chemical potential and then automatically running a sequence of simulations at successively decreasing chemical potentials, the GC/MC simulation efficiently discovers statistical distributions of bound fragment locations and orientations not found reliably without the annealing. This method accounts for configurational entropy, the role of bound water molecules, and results in a prediction of all the locations on the protein that have any affinity for the fragment. Disregarding any of these factors in affinity-rank prediction leads to significantly worse correlation with experimentally-determined free energies of binding. We relate three important conclusions from this challenge as applied to GC/MC: (1) modeling neutral ligands—regardless of the charged state in the active site—produced better affinity ranking than using charged ligands, although, in both cases, the poses were almost exactly overlaid; (2) simulating explicit water molecules in the GC/MC gave better affinity and pose predictions; and (3) applying a ΔΔGs solvation correction further improved the ranking of the neutral ligands. Using the GC/MC method under a variety of parameters in the blinded SAMPL3 Challenge provided important insights to the relevant parameters and boundaries in predicting binding affinities using simulated annealing of chemical potential calculations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The third Statistical Assessment of the Modeling of Proteins and Ligands (SAMPL3) Challenge [1] was, in part, a blinded test of how well various computational technologies predict binding affinities and binding poses of 34 ligands to a protein. Forecasting such properties for charged compounds, ligands with rotatable bonds, as well as dealing with protein flexibility and solvation were all part of the challenge. To approach SAMPL3, we used our fragment-based molecular design platform [2, 3] to (1) rank the component fragments of SAMPL3 ligands by binding affinities on the test protein, which reveals the lowest free energy poses, (2) re-construct the SAMPL3 ligands from these fragments, and (3) evaluate the assembled ligand poses and binding affinities. Within the dataset, different models were tested in the blinded challenge: charged fragments, neutral fragments, and ΔΔGs solvation corrected neutral fragments. Further studies examined two protein dielectric models, neutral ligands simulated against a protein with a neutral Asp189 in the binding site, and explicit versus implicit water models. These tests allowed us to probe the effectiveness of our method and examine the role of water, electrostatics, and dielectric parameters.
The key to predictive rank ordering by affinity is adequately modeling the binding free energy of fragments or ligands to proteins (or other macromolecules), which critically depends upon two factors: complete sampling of the relevant degrees of freedom and accurate potential functions. The degree of difficulty of this problem may be appreciated by realizing that even the relatively simple problem of predicting the free energy difference between d and l alanine binding to small synthetic host molecules of ~300 atoms and 2–5 torsional degrees of freedom [4] has proven to be an enormous challenge. Accomplishing this required creating a new simulation technique that simultaneously samples configuration space with Monte Carlo and stochastic dynamics [5, 6] within a free energy perturbation formalism. Numerical convergence of the free energy to within 0.3 kcal/mol required generating tens of millions of configurations. Thus, this work was characterized as going to “extraordinary lengths” [4] to obtain the free energy of interchanging a proton with a methyl group in the context of a simplistic synthetic “receptor”. Because such free energy simulations are impractical for realistic systems such as ligands binding to proteins, “their main utility has been to obtain additional insights concerning the origin of free energy differences, in synergy with experiment” [7]. Simonson and Karplus give a good perspective on the increasing use of empirical methods such as free energy component analysis and Poisson-Boltzmann free energy simulations [8]. For example, linear interaction energy component analysis was used to create two novel potent benzimidazole analogs from the binding analysis of 20 benzimidazole derivatives to HIV reverse transcriptase [9], while a similar analysis on nine ligands binding to avidin was performed with a Poisson-Boltzmann Surface Area (PBSA) method [10]. Interestingly, Kuhn and coworkers obtained a correlation coefficient of 0.92 with PBSA and only 0.55 with LIE. Yet when Pearlman [11] performed a PBSA analysis on a set of ligands that bind to p38 he concluded that PBSA, “yielded results much inferior to Dock Energy Score, … but at appreciably larger computational costs.” Such divergent results from different investigators using the same methods may be because these empirical methods require significant knowledge of the protein, the structure–activity relationships of the active molecule training set and special expertise in the computational methods, and thus the relative outcomes may have hinged on the skill of the practitioners. Other computational methods such as docking and scoring functions are fast models for (1) sampling ligand binding mode, which is fairly straightforward; and (2) predicting affinity, but do not account for entropy, and thus are not adequately predictive of affinity ranking in practice [12, 13].
Our computational fragment-based approach, which is based on annealing of the chemical potential, in GC/MC simulations (ACPS) [2, 3], only requires input of the protein structure and a set of organic fragments and thus the results are objective and not dependent on user expertise. The method starts with a protein bathed in a solvent of a particular fragment at high chemical potential. Tens of millions of trial insertions and deletions of the fragment into protein simulation cell are rapidly carried out at a given chemical potential until the protein–ligand system comes to equilibrium. Although this might appear computationally expensive, currently available multi-core computer platforms enable this to be practical. This process is repeated with stepwise lowering of the chemical potential, until the system goes through an abrupt phase transition, resulting in evacuation of almost all fragments from the protein simulation cell with a small group of fragments left tightly bound to collection of sites distributed over the surface of the protein. The result is a “fragment map” that depicts the distribution of location, orientation and binding affinity of each of the fragments on the surface of the target protein. A site on a protein where a chemically diverse group of fragments clusters but where water molecules do not bind has been shown to be a “hot spot”, likely the location of ligand binding or protein–protein interaction [3]. Unlike other computational fragment-based schemes, this method rigorously accounts for the configurational free energy, makes reliable predictions, samples more efficiently and comprehensively, has fewer limiting assumptions, and produces statistically-principled distributions of fragment locations and orientations—all critical for molecule design. With design software that can effectively mine and search large sets of data, the fragments can be visualized and linked together into larger ligands in literally millions of ways. The relative binding affinities of these putative ligands are predicted by combining the free energy metric (excess chemical potential) of its component fragments. We used this fragment map to construct ligands and then evaluated the full ligands by resubmitting them into the simulation.
One of the most important and key unsolved problems in fragment-based drug discovery is the linker problem [14, 15]. When is it found that two distinct fragments bind with high affinity in adjacent pockets of the protein, either by NMR, X-ray, or computation, the key question is whether or not they can be linked to create a high affinity compound. Linking two proximal fragments covalently is fraught with challenges, because bond length or bond angle strain may be introduced, a high energy conformation may be created, desolvation penalties may change, or the act of forming a chemical bond could change the electronic properties such that the linked fragments do not bind in the same way or to the same degree as the sum of the two individual fragments [16]. The simulated annealing of chemical potential algorithm applied to rigid fragments does not address these issues, so we have augmented it with a new algorithm called constrained-fragment annealing (CFA). This algorithm, as described in more detail below, co-anneals two or more fragments in the GC/MC simulation with an importance sampling technique that biases the search such that appropriate bond angles and bond lengths between the two distinct fragments are preferentially sampled.
In the blinded part of the study we tested three variations on our method (charged ligands, neutral ligands, and applied a solvation correction to the neutral ligands). In the post-submission phase, we used the conformation of six ligands from X-ray co-crystal structures (6 had alternate occupancies in the file so a total of seven compounds were simulated), and the corresponding protein structure, to test two different electrostatic states: (1) charged ligands and charged Asp189 residue and (2) neutralized ligands and neutralized Asp189. Asp189 is located at the bottom of the binding pocket, and we calculated its pKa to be 6.29 [17–20]. In addition to electrostatics, two other factors that influence calculations of ligand binding are waters (i.e., discrete and continuum models) and the protein dielectric constant. Accordingly, we tested the role of these two aspects of the model by splitting the electrostatic categories into four additional sub-categories: (a) dielectric constant of 1, (b) dielectric constant of 1 with calculated water molecules, (c) dielectric constant of 4, and (d) dielectric constant of 4 with water molecules. This gave 12 different tests of our method on the seven ligand co-crystal structures for a total of 84 additional simulations. Finally, we reran all the active ligands in the challenge data set with the optimal parameters determined above.
Methods
Protein structure preparation
To begin our analysis we selected the first protein structure provided in the SAMPL3 Challenge data (tryp1). To account for missing residues or atoms, we checked the tryp1 structure with the Profix program in the JACKAL molecular modeling package [21] and found no corrections needed to be made. Next, we added hydrogen atoms to the tryp1 structure using the Reduce program [22], which also flipped the imidazole ring of His57 into the correct conformation. Applying the PROPKA method [17–20], we found that the pKa[Asp189] = 6.29. For the blinded part of the study, we kept the residue charged. After the SAMPL3 Challenge was unblinded, we learned that the assays were run at pH 7.2 and the compounds were crystalized at pH 6.4 [23]. This led us to try a neutral form of Asp189 in a post-submission study. Since the calcium ion in the structure appeared sufficiently remote from the binding site, we did not use density functional theory to calculate the electron distribution around the calcium and the partial charges of residue atoms in close proximity to it, a process that we normally perform for ions near binding sites. After the protein structure was prepared, we applied ACPS to compute fragment distributions.
Annealing of chemical potential simulations (ACPS)
The algorithm requires the input of a fragment structure, a protein structure, and atomic force field parameters (Amber). The process consists of a sequence of grand canonical ensemble Monte Carlo (GC/MC) simulations where a chemical potential is imposed between an ideal gas reservoir of fragments and a simulation cell of sufficient size to enclose three “solvent” layers of fragments around the target protein. The simulations start with a very high excess chemical potential where the probability of inserting a fragment (conceptually, moving from the reservoir to the system) is dramatically higher than the probability of deleting the fragment from the system. The system will adapt to this chemical potential until an equilibrium is attained where the average number of fragments becomes stable [24, 25]. In addition to initially causing the simulation cell to be filled and packed with fragments, the high chemical potential allows difficult to find fragment configurations to be efficiently discovered that require passing through energetically unfavorable configurations before reaching optimal positions. The imposed chemical potential is lowered, or “annealed”, in discrete steps from high positive values to low negative values [2]. For each change in chemical potential, GC/MC is run until equilibrium is reached. This process is repeated automatically until the system goes through an abrupt transition, whereby all of the bulk solvent fragment molecules leave the protein cell, because the probabilities of deletion are greater than the probabilities of fragment insertion (a state designated as a phase transition). In the post-phase-transition regime of the simulation, a small number of fragment probes remain tightly bound to diverse pockets spread all over the protein and the fragments have higher affinity for the protein than they do for each other. We continually lower the imposed chemical potential until no fragments remain in the protein cell.
The Monte Carlo method used to compute the fragment distributions employed in this study follows the GC/MC scheme formulated by Adams [26]. In this method, small rigid molecules (fragments) are inserted, deleted, rotated, and translated. In each Monte Carlo step, a type of move is chosen at random, and energy associated with the new configuration is calculated. The step is accepted or rejected based on a criterion designed to cause the Markov chain of Monte Carlo steps to converge to a fragment population characterized by a Boltzmann probability distribution, \( e^{{\left( { - E/k_{B} T} \right)}} /Q, \) where Q is the partition function that normalizes the probabilities. The insertion acceptance probability is
The deletion acceptance probability is
The acceptance for moves (translations or rotations) is
where μ is the chemical potential, V is the volume of the system, N is the number of fragments in the system, ΔE is the change in non-bonded protein-fragment interaction energies (Coulomb plus Lennard-Jones).
Following Adams [26], it is more convenient to express the acceptance probabilities involving chemical potential in terms of a parameter B defined as
Hot-spot identification
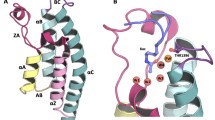
Taking the Challenge protein structure, tryp1, as an unknown, we simulated a series of fragments, including water, to determine the protein hot spot (location of high affinity for a diversity of fragments but lower affinity for water). Acetate, ammonia, aniline, benzene, ethanol, ethylamine, furan, methanol, pyrrole, thiazole, and water were all simulated on a dehydrated form of the protein. After following a hot-spot identification method described elsewhere [3], the method produced a single site (Fig. 1). Thus, the binding site does not need to be known a priori, although it was in this case. For inclusion of water in the next simulations, we scanned increasingly negative values of free energy from the water simulation until the binding pocket was void of water molecules, and kept all other calculated water molecules for a total of 108 water molecules. All figures were rendered with the PyMOL program [37].
Building SAMPL3 Challenge ligands
Each Challenge ligand contained multiple functional groups and multiple rotatable bonds, so we deconstructed the ligands into fragments that have no rotatable bonds (Table S5). These rigid fragments were simulated and then joined to reconstruct the Challenge ligands. For these linked fragments, new atom types were determined using Antechamber [27, 28] and partial charges were calculated with AM1-BCC [29]. The binding energies of the assembled ligands are then evaluated using additional annealing techniques (see below).
The systematic assembly process begins by selecting the pose of an “anchor” fragment, typically the amine or acid, which binds with the highest affinity. In some cases, there are multiple such poses, and all need to be evaluated. Fragments that had overlapped bonding locations with the anchor fragment, such as 11, where the ethylamine anchor overlapped the correct bonding position of the 2,3-dihydrobenzofuran, were directly bonded. In situations where the secondary fragments did not overlap, for instance 6, we aligned the second fragment to the highest affinity anchor fragment at the bottom of the pocket, amine or alcohol. For the acidic moieties, we determined, from the excess chemical potential, that the acidic ligands would still bind in the pocket with the acid facing up, so the acidic fragment was aligned and bonded to the aromatic fragment in the binding pocket whose pose was determined from the fragment map. For ligands with isolated ring systems, such as 29 (Fig. 5), we bonded the first two fragments as mentioned above, and then aligned and bonded the third fragment at both a 45° angle and planar to the other aromatic fragment (up to six positions for ligands with two heterocycles, for instance 7). The conformation that showed the lowest energy determined from energy minimization of the ligand with a rigid protein was selected for evaluation. The constructed ligands were energy minimized with local protein flexibility using 100 steepest descent steps to minimize any bond strain. At this point, the constructed ligands were resubmitted to the GC/MC simulator to calculate the final pose and energy, which included the new minimized structure for the protein.
Solvation correction
The full solvation free energy of each neutral ligand was initially determined using the SM8 model [30] within GAMESS [31] followed by a correction that accounted for the solvent accessible surface area of the ligand as modified by the protein. While incorrect treatment of the disulfide bridge at the periphery of the binding pocket led to solvation corrections of limited accuracy in the blinded part of the study, we found in the unblinded studies that the solvation correction depends strongly on the orientation of the ligands. For instance, for five ligand orientations of equal excess chemical potential that deviate by less than 0.1 Å, solvation values vary by ±0.5 kcal/mol. The solvation model is a linear empirical correlation model following that of Junmei Wang et al. [32] that implements
where \( \Updelta Gs\left( {P|L} \right) \) is the desolvation of the protein occluded by the ligand and \( \Updelta Gs\left( {L|P} \right) \) is the desolvation of the ligand occluded by the protein. Each term is computed by using the solvent-accessible surface of each atom (SASA), as restricted by other atoms,
where SASA is the atom solvent accessible surface, Q is the partial charge of the atom, and \( c^{vdw} \) and \( c^{q} \) are Van der Waals and electrostatic coefficients, respectively, trained on an experimental data set of solvation energies from ca. 600 molecules [33].
Constrained-fragment annealing (CFA)
CFA evaluates the binding of assembled ligands by applying ACPS to the component fragments of the ligand, subject to bond constraints between the fragments. The energy potential includes bonded energy terms, and non-bonded energy terms, for both intra-molecular interactions between fragments and fragment-protein interactions. CFA starts with two fragments in near-bonding configurations. These can be positions calculated directly in the GC/MC simulation, or they can be produced using an align-fragment function where the aligned fragment does not correspond exactly to a fragment position calculated in the GC/MC. Bond constraints are applied as harmonic energy penalties to the fragments, which limit the distance and angle between fragments. Such a constraint is usually a direct bond. However, for ligands such as 30 (methylene group between the benzene and morpholino groups), a CH2 constraint would be added, or we would utilize fragments with methyl groups off the bonding positions and directly add a bond constraint to the methyl group. CFA evaluates the binding of linked fragments that are mutually constrained by the geometry of bonds implementing the links. Rather than simply summing the free energies of unconstrained component fragments, CFA is designed to derive component fragment free energies that account for entropy reductions due to the restricted range of motion of a bonded fragment. Further, it allows the overall binding pose of the ligand assembled from the linked fragments to be adjusted as the component fragments are subject to a multiplicity of non-bonded protein interactions, non-bonded fragment–fragment interactions, and bonded fragment–fragment forces. CFA proceeds by designating one of the component fragments at a time to be subject to chemical potential annealing (delete/insert steps) in a GC/MC simulation, while all the fragments are allowed to rotate and translate in Monte Carlo steps. In addition to rotation steps that rotate around a fragment’s center of mass, rotations around the constrained bonds are implemented. The energy computed for the acceptance probabilities in the GC/MC now has extra terms that implement the bonded energies (length stretching, bond angle bending),
where the non-bonded terms include both Coulomb and van der Waals (Lennard-Jones model) interactions. This allows interplay between various the bonded and non-bond energies to achieve an energetically-favorable configuration of ligand pose and conformation. The final free energy scoring is the sum of the constrained free energies of the component fragments. This methodology allows for bond rotations at the joints, accounts for the change in entropy due to the bond constraint limiting motion, and includes intra-molecular non-bonded interactions.
Results and discussion
Hot-spot identification
Hot spots, or local regions of high interaction energy, are important for protein recognition and ligand binding, and identifying these sites is a critical step to finding catalytic active sites or allosteric binding sites. Here, stepwise lowering of the imposed chemical potential isolates fragment-associated sites on the protein surface, such that each site is ranked by the free energy per molecule. Using diverse fragment clustering, affinity ranking, and water exclusion, we identified a single hot-spot site on the Challenge protein structure tryp1 (Fig. 1). To validate this site, the Protein Data Bank was searched for co-crystal structures that had high protein sequence homology to tryp1 and that contained a small Challenge-like ligand. Structure 2bza contains bound benzylamine bound to a bovine pancreas β-trypsin. When this structure is compared to the structure of tryp1, the benzylamine ligand bound in the site determined by our hot-spot identification method.
Water
Accounting for the effects of water on ligand binding is necessary to predict the relative binding affinity for a series of ligands. Tightly-bound water molecules may block access to sites, link fragments to sites, or shield critical residues electrostatically. Chemical potential annealing of water identifies high affinity water sites, ranks water molecules present in crystal structures by affinity, and reveals multi-body water configurations resulting from hydrogen-bond networks, which are difficult or impossible to find with other methods. Since the tryp1 Challenge protein structure does not contain any water molecules, we used ACPS to calculate the position of water molecules prior to running the fragments. For our fragment simulations, we kept 108 water molecules—these were the predicted tightly bound water molecules as identified by post phase transition occupancy of a protein site. Four of the water molecules were proximal to the binding pocket (Fig. 2). Three were buried behind Asp189. One water molecule, near the carbonyl group of Gly219 and with its hydrogen pointing into the pocket, could influence ligand binding.
Keeping these water molecules, we simulated fragments corresponding to motifs in the ligands (Table S5). To ensure that the water molecules did not cause problems with binding—such as steric clashes with fragment binding—the tryp1 structure, with calculated ethylamine and benzene fragments at their lowest binding energy, was aligned to 2bza (Fig. 3). The fragment data overlaid well with the benzylamine in the X-ray crystal structure. The fragments displayed the same pose in a simulation run with no water molecules, but the energies were different. This suggests that the water molecules do not change the pose of the amine containing compounds, but only their relative rankings.
Calculated fragments benzene and ethylamine (green carbons) overlaid with benzylamine from 2bza structure. Structures were aligned with PyMOL [37], first with a sequence alignment and then an alignment that minimized the root mean square deviation
Building SAMPL3 ligands from fragments
The Challenge ligands could be constructed from 2 to 3 fragments either from the fragment map or from bonding positions (ligand 3 was the only single fragment ligand). Examining the fragment-map data exposed several trends including (1) all amines—aniline, ammonia, ammonium, methylamine, ethylamine, dimethylamine, and methylethaneamine—were bound at the bottom of the pocket near Asp189; and that (2) all aromatics and heterocycles—benzene, benzofuran, benzofuran-2,3-dihydro, benzothiophene, morpholine, piperazine, piperidine, pyrazole, pyridine, pyridine-1,2α-imidazole, pyrrole, pyrrolidine, thiadiazole, thiazole, and thiophene—were bound just above the amines, in the main body of the binding pocket. While our fragment map often displayed overlapping binding modes, in certain cases, as with isolated ring systems, one ring would not be found in a binding position. Ligand 26 can be constructed from a benzene, ethylamine, and pyrrole fragments, but the benzene and pyrrole have overlapping positions in the fragment map because the location of highest affinity for each fragment is within the binding cavity. Locating binding positions for the second ring by admitting fragments of lesser affinity produces other binding modes around the periphery of the binding pocket but not in bonding positions to the other, more negative affinity ring fragment. These second rings are in non-optimal positions for ligand affinity. To overcome this hurdle, we added the second ring in the correct bonding position and energy minimized. Because local energy minima might bias certain rotamers of the second fragment, multiple orientations were built and minimized. The lowest energy conformation was selected and reevaluated using the GC/MC simulation. This process is summarized in Scheme 1 and Fig. 4. Ideally, the protein would dictate the highest affinity fragments and linking would occur only with those fragments found in the fragment map.
Single-fragment annealing (SFA) uses a rigid compound in the ACPS. To assemble the compound, we would select an anchor fragment, ethylamine in this example, and align the second fragment to the anchor fragment. Multiple different starting conformations were generated. Next, the fragments were linked and energy minimized. The lowest energy conformation(s) was evaluated with SFA. All parts of this procedure are completed within the context of the protein
Analysis of pose prediction
Single-fragment annealing (SFA)
When analyzing a complex fragment that can be decomposed into two or three simpler fragments, it is often observed that each simpler fragment is pulled somewhat away from its optimal binding position on the protein. The overall molecule binds in a compromised position, so that all groups can bind into their respective sub-pockets simultaneously. SFA performs annealing of the chemical potential with an additional biased sampling protocol. This oversamples the larger complex fragment in the binding site in order to discover the optimal total complex fragment binding mode. Thus, the degree of deviation from the individual simpler fragment optimal binding modes may be analyzed. Of course, the more the complex fragment binding mode mirrors the collection of individual simple fragment binding modes, the more likely the complex linked fragment is a better binding candidate. We used SFA to determine the binding poses and affinities of the Challenge ligands.
The co-crystal structures of six (of seventeen) active compounds (6, 7, 11, 12, 26, 29) were determined. Ligand 6 had two occupancies in the X-ray crystal structure and these alternative structures (6A and 6B) differed by the placement of the amine in the bottom of the pocket. Our calculated pose for the ethylamine fragment, whether charged or neutral, overlaid well with the amine group of 6A (Fig. 5, left panel for 6). In both the 6A and 6B co-crystal structures, the benzothiophene was in the same position, but in our fragment map, neither the benzothiophene nor 3-methyl-benzothiophene fragments were in ideal bonding positions. After aligning the 3-methyl-benzothiophene to the ethylamine, energy minimizing, and resubmitting to the GC/MC simulations, the calculated ligand almost perfectly overlaid 6A (Fig. 5, middle panel for 6).
Predicted poses (green line representation) overlaid with the six experimental X-ray crystal structures (magenta stick representation); Numbers in the top left of each row correspond to the ligand Challenge numbers. Ligand 6 has two occupancies in the crystal structure; they mainly differ by the placement of the amine group (6A has the amine coming out of the plane and matches our fragment data). Each column presents a different result from our simulation; left is raw fragment map with fragments corresponding to ligand functionalities; middle is our constructed ligands used in the affinity study; right is our constrained-fragment annealing done after the release of the results
For 7, the calculated ethylamine overlaid well with the primary amine of the ligand in the crystal structure (Fig. 5, left panel for 7). The predicted thiazole fragments (2,4-dimethylthiazole, 2-methylthiazole, 4-methylthiazole) all had the nitrogen and sulfur in the relatively correct positions in the binding pocket. After linking these fragments, the bond angle between the primary amine and thiazole differed by about 75° compared to the ligand in the crystal structure, which caused the predicted ligand to orient toward Trp215 whereas the crystal ligand bound to the other side of the pocket near Gln192/Cys191. We predicted the thiophene to be planar to the thiazole, but there was a slight twist in the crystal structure. However, the sulfur of the thiophene was oriented correctly with respect to the thiazole.
The fragment map for 11 had the ethylamine perfectly overlaid with the crystal structure amine. The benzofuran fragment was in the correct relative position in the pocket with the same orientation of the oxygen. The constructed fragment had the primary amine in the correct position but the benzofuran ring was twisted by about 40° with respect to the crystal structure. The fragment map for 12 had fragments that perfectly overlaid bonding positions. The predicted ligand and the X-ray crystal structure overlaid perfectly on the sulfur atom and only deviated a small amount in the position of the bromine. The ethylamine in our prediction pointed toward Asp189/Cys220 whereas the crystal ligand pointed toward Asp189/Tyr228. For 26 and 29, the ethylamine and benzene fragments showed good agreement with the X-ray structures. The pyrrole of 26 was displaced by about 1Å, whereas the piperazine of 29 exhibited accurate overlay with the crystal.
The pose predictability of the ligands from fragment data tended to depend on the number of rotatable bonds and thus the number of fragments used to construct the ligands. For instance, 6, 11, and 12 have one rotatable bond and were assembled from two fragments. In this case our pose prediction was in good agreement with experimental data. 7, 26, and 29 have two rotatable bonds and were built from three fragments. Here our predictability decreased slightly but we still identified the correct binding pocket and relative position of the ligands within the binding pocket.
Constrained-fragment annealing (CFA)
CFA addresses one of the core problems of fragment-based design—binding evaluation of linked fragments. Binding poses of fragments provide a strong hypothesis for where and with what affinities ligands built by linking fragment will bind. However, when fragments are linked, charges in the resulting ligand are often redistributed, and the binding pose can shift to accommodate a trade-off in the binding of each component. Further, the entropy of the fragments is changed by the restriction on motion imposed by the bonds. The CFA algorithm calculates the free energy, including configurational entropy, of linked fragments with rotatable bonds (Fig. 6). There are several variations of this technique, but in essence it co-anneals two or more distinct fragments using simulated annealing of chemical potential with restraints on the bond angles and bond lengths; unlike SFA which using a rigid ligand assembled from fragments. In CFA, one or more fragments are constrained to move subject to bonded forces to other fragments or ligand. One fragment is annealed while the other fragments, if any, are allowed to move and rotate as well. This provides a characterization of the free energy that includes changes in configurational entropy due to the limited range of motion under the bonded constraint. Further, the interconnected fragments can now explore somewhat different poses. The simulation monitors the chemical potential required for the two fragments to populate optimal linkage geometry. It is often the case that observing such a population requires the algorithm to go to high chemical potentials (poor free energies), which indicates that those two fragments are not good candidates for linkage. When two fragments can be co-annealed with the CFA method and they show populations of proper geometry without substantial degradation of the interaction free energy as measured by the chemical potential, then these are predicted to be good candidates to link.
By using CFA, we were able to correct some of the erroneous angles in our constructed ligands (right-hand column of Fig. 5). CFA constrains the bond angles and distances of fragments, yet samples sufficiently to analyze all possible bond angles between the fragments—thus yielding the angles corresponding to the lowest energies. After running CFA, the relative positions of the heavy atoms did not change suggesting that, in this case, assembling fragments, applying energy minimization, and running SFAs gave the same result as our more rigorous CFA analysis. The calculated CFA poses for the fragments did overlay the experimental structures with less bond and spatial difference (lower RMSD values, Table 1), yet compound 7 still lay to the wrong side of the pocket. Examining this structure in greater detail, the experimental structure had a water molecule (number 136 in pdb file) that fit between the thiazole and thiophene of 7 and would push the ligand into a more upright position. We decided to run a simulation keeping all water molecules that we calculated within 4 Å of the protein surface and did not sterically interfere with the ligand. This starting point did not have a calculated water molecule near the crystallographic water #136. After GC/MC, where the water molecules have freedom to move during sampling, we calculated water molecules that did match with the crystallographic water molecules and kept the orientation of 7 similar to the X-ray structure (Figure S5). This water molecule forms a bridging interaction between the ligand and the protein and interacts with another water molecule. Our GC/MC simulations allowed for the discovery of this complex interaction (Figure S5), which is difficult to detect using other methods. The co-annealing of water and ligand 7 (renamed ligand 7 water ) significantly improved the RMSD value (Table 1). This led to the conclusion that keeping buried or structural water molecules was not sufficient to correctly calculate the pose of 7.
Analysis of affinity prediction
In the blinded part of this study, we tested three datasets on the SAMPL3 Challenge ligands. We chose to look at charged ligands, neutral ligands, and neutral ligands with a solvation correction. Without properly accounting for desolvation and electrostatics, limited by the inaccuracy of the forcefield models for charge–charge interactions, comparison of predicted ligand affinities across functional groups showed little correlation due to poor characterization of charge–charge interactions (but see subsequent results below when these are neutralized). For our results, we broke the dataset into three categories: amines, alcohols, and acids.
Amines
As mentioned above in the pose prediction, our best results derived from ligands with a minimal number of rotatable bonds and minimal number of constructed fragments. Ligands with only one rotatable bond and two constructed fragments were 6, 11, 12, and 20. Our prediction for the active compounds showed a linear trend with the experimental data for neutral ligand simulations (Figure S1). The least-squares fitting value became slightly better with a solvation correction, although with only three active compounds and only three data points, it was hard to say if this was significant. For charged ligands, the data did not correlate to any trend. 20 and 6 are isomers, where the ethylamine is located at the 5 and 3 position, respectively, on the benzothiophene. 20 was inactive. Comparing isomers 20 and 6, we predicted 6 to have higher affinity in all three cases: charged, neutral, and a solvation corrected neutral ligand.
The secondary amines (19 and 23) were inactive, and our predicted energies were two of the three most positive energies out of the predicted set (both neutral and solvation-corrected predictions), so we correctly ranked the secondary amines. The ligands containing aniline functionalities (3, 27, 34) were predicted to have the following ranking: 34 > 27 > 3 (both neutral and solvation corrected data); 34, with a p-substituted thiadiazole, was predicted to have significantly higher affinity than the other two ligands. The experimental data had 27, with a m-substituted pyrrole, active and the other two ligands inactive. We are still investigating the overestimation of 34. For the rest of the amine dataset, amines with isolated ring systems, we could not find any interesting correlations from our predictions to the experimental data. As mentioned above in the pose prediction, this could be due to the inadequacy of only keeping structural, buried, and tightly bound water molecules or incorrect poses.
Alcohols
From our calculations, we found the pose of the alcohols to be in the binding pocket with the hydroxyl functionalities pointing down in the pocket and interacting with Asp189. The energy for the alcohol pointing up in the pocket was slightly less favorable and could be an alternate orientation. We determined this from our fragment data and simulating constructed ligands oriented with the alcohol up or down in the binding pocket.
Out of the 34 challenge ligands, seven contained hydroxyl functionalities and only one of the alcohols was active (compound 16). Within the error bars of ca. ±0.5 kcal/mol, we ranked 16 tied for third with compound 5 and compounds 14 and 16 to be of higher affinity. The two ligands with over-predicted energies both contained a methylene group between the two rings, adding additional degrees of freedom. Using a conformational memories algorithm [34–36], which predicts the conformational energy penalty of ligands going from their unbound to bound confirmation and includes solvation, we found the energies for the over-predicted ligands to have between 3 and 5 times higher energy penalty going from an unbound confirmation to the predicted bound confirmation. Correcting for the unbound to bound stress energy cost of these molecules allowed us to correctly predict the active alcohol ligand.
Carboxylic acids
The pose of the carboxylic acids was the most difficult to determine. Initially, the data suggested that the acids could be interacting with Lys60, which resides close to the catalytic triad. This interaction was prioritized lower because the fragment data for the acids bound to the amine of Lys60 were not optimally oriented toward the triad, and the fragments representing the ring systems of the ligands were not high affinity in the area between Lys60 and the catalytic triad. Our calculated pose for the carboxylic acid ligands oriented the aromatic ring systems into the shaft of the binding pocket with acid group out of the pocket toward the catalytic triad and Gln192. For all of the other pose predictions (alcohols and amines), we did not see significant conformational changes within the protein, but the acid ligands did cause a conformational change in the protein. The acid pose predication had Gln192 swing in toward the pocket so the hydrogens on the amide nitrogen interact with the acid.
Three of the 34 ligands had acid functionalities (1, 15, and 25). 15 was the only active ligand of the acid group, and we correctly predicted it to be the highest affinity in the charged and neutral states. The solvation correction normally dampened the calculated free energy, but for 25 the energy became more negative, which was possibly due to the handling of the disulfide bridge as mentioned in the materials and methods.
X-ray crystal ligands
After submission of the results, the Challenge organizers unblinded six X-ray crystal structures for ligands 6, 7, 11, 12, 26, and 29, and we used these compounds to further test our method using different parameters that focused on the role of electrostatics, water, and dielectric. 6 offered an interesting test case since the X-ray crystal showed two occupancies at 0.4 (orientation A pointing toward Asp189/Cys220) and 0.5 (orientation B pointing toward Asp189/Tyr228, down into the other side of the pocket). Previously we tested our method using neutral ligands, but we did note that the calculated pKa of Asp189 was 6.29, so in this round of testing we simulated neutral ligands to a neutral form of Asp189 (designated Ash189). In addition to neutral simulations, we also performed charged ligand–charged protein simulations (charged Asp189). The neutral and charged simulations were run using a dielectric of 1 and 4, and at each of these values, we simulated explicit and implicit water for a total of eight categories and seven ligands for an overall count of 56 additional simulations. For the neutral simulations, 28 out of the 56 simulations, a solvation correction was applied.
Constrained-fragment annealing (CFA) co-anneals fragments with restraints in the context of the protein, which evaluates the binding of linked fragments with rotatable bonds. In CFA, the fragments are allowed to minimize their free energy only in a specific, bonded position relative to other fragments
Looking at the affinity changes due to the two different confirmations of 6 reveals a large difference in the energy based on the conformational changes (Tables S2, S3, and S4), which stems from the amine pointing in two different directions separated by about 160° rotation compared to the benzothiophene ring. Confirmation B continually ranked better within all of the datasets. Simulating neutral ligands and Ash189 gave significant improvement over charged ligands and Asp189 (Fig. 7, Figures S2 and S3). In the presence of explicit water molecules, a dielectric of 1 resulted in a higher correlation with experimental values. In the absence of explicit water molecules, dielectric 4 improved the correlation (Figures S3 and S4). Finally, the solvation correction further increased the predictions (Figs. 5 and S4).
Summary of results for testing different applications of our GC/MC method reveals that simulating charged ligands against a charged Asp189 gives lowest correlation (blue), simulating neutral ligands against a neutral form of Asp189 (ASH189) yields significant improvement in correlation (red), correcting for solvation (green) results in an higher R 2, and using explicit waters gave the maximum correlation. These results were produced at a dielectric of 4, for full results see Figures S2, S3, and S4 and corresponding Tables S2, S3, and S4
Conclusions
Anthony Nicholls designed the SAMPL challenges to create a forum for objectively testing and reporting what techniques work, what does not work, and paths to improvement so that all techniques have the opportunity to learn and evolve in an objective manner. In the case of simulated annealing of chemical potential, a method designed to effectively and completely sample the entire ensemble of fragment-protein interactions with no human bias, we clearly learned important lessons on the impact of neutralizing ligands and neutralizing binding residues, solvation, and the role of water.
In the blinded study, 34 challenge ligands were run under three different conditions—charged ligands, neutral ligands, and neutral ligands with a solvation correction. We found that the predictability of this method delivered the highest correlation to experimental data when the ligands were categorized into main function group—alcohol, carboxylic acid, and amine—and thus treated as a congeneric series. The rank-order affinity predictions for the neutral fragments were accurate in most cases, but there was little correlation between the predicted and actual affinity for the charged fragments. The pose prediction gave very good agreement with experimental data when we used the fragment map, bonded the fragments, energy minimized the constructed ligands, and resubmitted them to the GC/MC simulation. Angle and spatial deviations of the predicted poses improved when we applied our constrained-fragment annealing.
After the experimental data was made publicly available, the so-called unblinded part of the study, we reran the simulations under separate conditions, (1) protein dielectric 1 and 4, (2) explicit and implicit water molecules, (3) charged amines with a charged Asp189 in the binding site, (4) deprotonated the amines thus neutralizing them and protonated the aspartate in the protein binding site so it was also neutralized, and (5) we included the water solvation energy penalty of the neutral ligands. The dielectric of 4 damped the electrostatic part of the binding energies and gave better correlation to experimental data in the absence of water, while dielectric 1 delivered better correlation with explicit waters. Explicit waters improved the predictability of our method. While binding poses were accurately predicted in all cases, we could not predict the rank ordering of charged ligands. Rerunning the amine fragments in the neutralized form with the aspartate in the binding site of the protein also neutralized resulted in quite accurate rank-order binding predictions and including the ligand solvation correction increased the accuracy of the predictions even further. Challenged with predicting pose and affinity prediction, we found both the boundaries of what works well and the cause of outliers, which should help improve our technique for the next challenge.
References
Geballe M, Skillman G, Nicholls A (2011) Statistical assessment of the modeling of proteins and Ligands (SAMPL3) Challenge. http://sampl.eyesopen.com/
Guarnieri F, Mezei M (1996) J Am Chem Soc 118(35):8493
Kulp JL III, Kulp JL Jr, Pompliano DL, Guarnieri F (2011) J Am Chem Soc 133(28):10740
Burger MT, Armstrong A, Guarnieri F, McDonald DQ, Still WC (1994) J Am Chem Soc 116:3593
Guarnieri F, Still WC (1994) J Comput Chem 15:1302
Guarnieri F (1995) J Math Chem 18:25
Brandsdal BO, Österberg F, Almlöf M, Feierberg I, Luzhkov VB, Åqvist J (2003) Free energy calculations and ligand binding. In: Valerie D (ed) Advances in protein chemistry, vol 66. Academic Press, p 123
Simonson T, Archontis G, Karplus M (2002) Acc Chem Res 35:430
Kroeger Smith MB, Hose BM, Hawkins A, Lipchock J, Farnsworth DW, Rizzo RC, Tirado-Rives J, Arnold E, Zhang W, Hughes SH, Jorgensen WL, Michejda CJ, Smith RH Jr (2003) J Med Chem 46(10):1940
Kuhn B, Kollman PA (2000) J Med Chem 43(20):3786
Pearlman DA (2005) J Med Chem 48(24):7796
Warren GL, Andrews CW, Capelli A-M, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS (2006) J Med Chem 49(20):5912
Leach AR, Shoichet BK, Peishoff CE (2006) J Med Chem 49(20):5851
Ichihara O, Barker J, Law RJ, Whittaker M (2011) Mol Inform 30(4):298
Chung S, Parker JB, Bianchet M, Amzel LM, Stivers JT (2009) Nat Chem Biol 5(6):407
Moumne R, Larue V, Seijo B, Lecourt T, Micouin L, Tisne C (2010) Org Biomol Chem 8(5):1154
Bas DC, Rogers DM, Jensen JH (2008) Proteins Struct Function Bioinform 73(3):765
Li H, Robertson AD, Jensen JH (2005) Proteins Struct Function Bioinform 61(4):704
Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH (2011) J Chem Theory Comput 7(2):525
Søndergaard CR, Olsson MHM, Rostkowski M, Jensen JH (2011) J Chem Theory Comput 7(7):2284
Xiang J (2002) JACKAL: a protein structure modeling package. Columbia University, New York
Word JM, Lovell SC, Richardson JS, Richardson DC (1999) J Mol Biol 285(4):1735
Newman J, Fazio VJ, Caradoc-Davies TT, Branson K, Peat TS (2009) J Biomol Screen 14(10):1245
Frenkel D, Smit B (2001) Understanding molecular simulation: from algorithms to applications, 2nd edn, vol 1, Academic Press, New York
Allen MP, Tildesley DJ (1989) Computer simulation of liquids. Oxford University Press, New York
Adams DJ (1975) Mol Phys 29:307
Wang J, Wang W, Kollman PA, Case DA (2006) J Mol Graph Model 25(2):247
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25(9):1157
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) J Am Chem Soc 117(19):5179
Cramer CJ, Truhlar DG (2008) Acc Chem Res 41(6):760
Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA (1993) J Comput Chem 14(11):1347
Wang J, Wang W, Huo S, Lee M, Kollman PA (2001) J Phys Chem B 105(21):5055
Boyer R, Bryan RL (2012) J Phys Chem B submitted for publication
Guarnieri F, Weinstein H (1996) J Am Chem Soc 118(24):5580
Guarnieri F, Wilson SR (1995) J Comput Chem 16(5):648
Whitnell RM, Hurst DP, Reggio PH, Guarnieri F (2008) J Comput Chem 29(5):741
The PyMOL Molecular Graphics System, Version 1.4, Schrödinger, LLC
Acknowledgments
We thank Dr. William Chiang, Dr. John L. Kulp Jr., and Dr. David L. Pompliano for helpful discussions and commentary.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kulp, J.L., Blumenthal, S.N., Wang, Q. et al. A fragment-based approach to the SAMPL3 Challenge. J Comput Aided Mol Des 26, 583–594 (2012). https://doi.org/10.1007/s10822-012-9546-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-012-9546-1