Abstract
Purpose
The aim of this study was to determine whether an interchromosomal effect (ICE) occurred in embryos obtained from reciprocal translocation (rcp) and Robertsonian translocation (RT) carriers who were following a preimplantation genetic diagnosis (PGD) with whole chromosome screening with an aCGH and SNP microarray. We also analyzed the chromosomal numerical abnormalities in embryos with aneuploidy in parental chromosomes that were not involved with a translocation and balanced in involved parental translocation chromosomes.
Methods
This retrospective study included 832 embryos obtained from rcp carriers and 382 embryos from RT carriers that were biopsied in 139 PGD cycles. The control group involved embryos obtained from age-matched patient karyotypes who were undergoing preimplantation genetic screening (PGS) with non-translocation, and 579 embryos were analyzed in the control group. A single blastomere at the cleavage stage or trophectoderm from a blastocyst was biopsied, and 24-chromosomal analysis with an aCGH/SNP microarray was conducted using the PGD/PGS protocols. Statistical analyses were implemented on the incidences of cumulative aneuploidy rates between the translocation carriers and the control group.
Results
Reliable results were obtained from 138 couples, among whom only one patient was a balanced rcp or RT translocation carrier, undergoing PGD testing in our center from January 2012 to June 2014. For day 3 embryos, the aneuploidy rates were 50.7% for rcp carriers and 49.1% for RT carriers, compared with the control group, with 44.8% at a maternal age < 36 years. When the maternal age was ≥ 36 years, the aneuploidy rates were increased to 61.1% for rcp carriers, 56.7% for RT carriers, and 60.3% for the control group. There were no significant differences. In day 5 embryos, the aneuploidy rates were 24.5% for rcp carriers and 34.9% for RT carriers, compared with the control group with 53.6% at a maternal age < 36 years. When the maternal age was ≥ 36 years, the aneuploidy rates were 10.7% for rcp carriers, 26.3% for RT carriers, and 57.1% for the control group. The cumulative aneuploidy rates of chromosome translocation carriers were significantly lower than the control group. No ICE was observed in cleavage and blastocyst stage embryos obtained from these carriers. Additionally, the risk of chromosomal numerical abnormalities was observed in each of the 23 pairs of autosomes or sex chromosomes from day 3 and day 5 embryos.
Conclusion
There was not enough evidence to prove that ICE was present in embryos derived from both rcp and RT translocation carriers, regardless of the maternal age. However, chromosomal numerical abnormalities were noticed in 23 pairs of autosomes and sex chromosomes in parental structurally normal chromosomes. Thus, 24-chromosomal analysis with an aCGH/SNP microarray PGD protocol is required to decrease the risks of failure to diagnose aneuploidy in structurally normal chromosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chromosome translocation is one of the most common structural abnormalities. This translocation type generally refers to the exchange of chromosome fragments between non-homologous chromosomes, resulting in interchromosomal rearrangements. The fertilization of unbalanced gametes may cause infertility or lead to recurrent pregnancy loss or birth, such as recurrent spontaneous abortion (RSA), stillbirth, neonatal death, or birth of deformed and mentally retarded offspring [1].
Approximately 1/500 to 1/1000 of live births carry a reciprocal translocation (rcp). These carriers have normal phenotypes but can generate at least 18 different types of gametes during the meiosis process, of which only one type is normal, one type is balanced, and the rest carry unbalanced chromosomal changes. It is assumed that the reproduction of normal/balance gametes depends on the involved chromosomes, the breakpoint positions, the segregation patterns, and the gender of the translocation carrier [2]. Robertsonian translocation (RT) is the other special form of translocation that occurs in acrocentric chromosomes. During gamete formation, non-homologous Rob carriers can produce six types of gametes during meiosis, of which only one type is normal and one type is balanced.
Preimplantation genetic diagnosis (PGD) refers to an embryo biopsy in chromosomal translocation carriers, especially those with a history of RSA, to select normal or balanced embryos for intrauterine transfer, which will effectively reduce the abortion rates, improve the live birth rates, and avoid the selective termination of affected pregnancies. Multicolor fluorescence in situ hybridization (FISH) detecting the translocated chromosomes is a traditional PGD for translocation carriers. However, it cannot realize wholesome chromosome testing. One of the FISH limitations is due to the existence of interchromosomal effect (ICE).
The concept of ICE was mentioned by Lejeune in 1963. It was used for some chromosomes involved in rearrangement that affect the segregation of the structurally normal chromosomes [3,4,5,6]. Moreover, several studies have investigated the ICE in embryos derived from translocation carriers’ PGD cycles [5,6,7,8]. However, the present data on ICE are still controversial, and it is difficult to draw a conclusion about the mechanism of ICE occurrences. Some studies have suggested that an ICE exists in PGD embryos, and others have shown this effect is negligible or even not present [4, 6, 7].
With the advantage of 24-chromosomal screening, the technique of array comparative genomic hybridization (aCGH) and single nucleotide polymorphism (SNP) microarrays is extensively applied presently, which not only detects the chromosomes involved in the rearrangement but also completes aneuploidy screening to reduce the risk of aneuploidy affecting structurally normal chromosomes from ICE. It is possible that aCGH and SNP array technology could be a replacement for the traditional FISH in PGD clinical applications [9].
In early 2011, we began to apply aCGH and SNP in PGD for chromosomal translocation carriers [10]. In this paper, we retrospectively analyzed the PGD application of the two new techniques in rcp and RT carriers and explored the molecular karyotype results under the ICE of embryos from these cases.
Materials and methods
Patient information and embryo sources
In this study, 138 couples, of whom only one patient had a balanced rcp or was a RT carrier, were undergoing PGD testing in our center from January 2012 to June 2014. Translocation status was confirmed by karyotype analysis. No compound translocation cases were recruited in either group. Among these couples, the indications of PGD were 44.9% (62/138) with primary infertility, 23.9% (33/138) with RSA, and 4.3% (6/138) with recurrent implantation failure (RIF); 20.1% (29/138) had secondary infertility after once abnormal pregnancy or delivery history and 5.8% (8/138) were found to be a translocation carrier in the karyotype test before entering the IVF protocol. A total number of 1214 embryos of 95 PGD cycles from rcp carriers and 44 cycles from RT carriers were biopsied with the techniques of aCGH/SNP. Meanwhile, 76 age-matched patients with normal karyotypes in the control group were undergoing 76 PGS cycles due to RSA [82.9% (63/76)], RIF [5.3% (4/76)], and advanced maternal age (AMA) (≥ 36 y) [11] [11.8% (9/76)] (Table 1).
The Ethics Committee in the First Affiliated Hospital of Sun Yat-sen University approved this study. All patients underwent genetic and eugenics counseling and signed informed consent for PGD.
Ovarian stimulation and assessment of embryos
After controlled ovarian stimulation (COS), ova were retrieved, and intracytoplasmic sperm injection (ICSI) [12] was performed on mature oocytes. Fertilized eggs with two pronuclei and two polar bodies were considered normal fertilization. Embryonic development was observed at 3 days (day 3) after oocyte retrieval, and the number of embryonic blastomeres and the fragmentation conditions were recorded.
Embryo biopsy
Cleavage-stage biopsy and aCGH
The standards for embryos suitable for biopsy included normal fertilization, day 3 embryos containing six or more blastomeres, and embryos with less than 20% fragmentation. Mechanical biopsies were used to puncture the zona pellucida and extract a single blastomere containing a nucleus. After biopsy, the embryos were quickly rinsed with culture medium and placed into blastocyst-cultured medium to continue incubation [13]. Only blastocysts with normal/balance results were transferred to the uterus or thawed for possible later transfer.
For rcp carriers, 24 Sure+ chips (BlueGnome Ltd., Cambridge, UK) with a higher resolution were selected, while for RT carriers and patients who underwent PGS, the 24 Sure V3 chips (BlueGnome Ltd., Cambridge, UK) were chosen due to economic considerations. Every single biopsied blastomere underwent whole genome amplification (WGA) using the Sureplex kit (BlueGnome Ltd., Cambridge, UK). The amplified products were labeled with different fluorescent dyes (CY3/CY5) and hybridized to the 24 Sure V3/24 Sure+ chips (BlueGnome Ltd., Cambridge, UK). After washing, the chips were scanned using the Innoscan 710 scanner (Innopsys, Carbonne, France), and the data were analyzed using BlueFuse Multi software (BlueGnome Ltd., Cambridge, UK).
Blastocyst biopsy and SNP
All embryos were cultured to the blastocyst stage. On day 5 or day 6 after fertilization, blastocysts that were higher than the blastocyst morphology score of 3BC were chosen for biopsy [14]. Biopsy of trophectoderm (TE) from blastocysts was performed at the same-day PGD for translocation testing and aneuploidy screening. Approximately 4 to 10 TE cells were aspirated with a biopsy pipette. Whole embryos after biopsy were cryopreserved for possible later transfer.
Biopsied samples were first subjected to WGA using a RELI-g Mid Kit (Qiangen, German). WGA product was then used for SNP array screening for chromosomal abnormalities, including aneuploidy and unbalanced segment anomalies. Individual embryonic DNA samples were hybridized to a Human CytoSNP-12 panel (Illumina, USA) as Infinium HD Assay Super, Manual Protocol described, which determines approximately 300,000 SNPs with an average distance of 9.7 kb.
Microarray
According to the results of the aCGH and SNP array, each biopsied embryo PGD results were divided into five categories as following: (1) All 24 chromosomes were normal/balanced and euploidy embryos (euploid and no translocation imbalance); (2) embryos with inheritance parental translocation-related chromosome imbalance, referred to the unbalanced of the chromosomes involved with a translocation (euploid with translocation imbalance); (3) aneuploid alone without the inheritance of a parental translocation chromosome imbalance, which referred to parent non-translocation-related chromosomes with a duplicate or missing influence due to the interchromosomal effects, and normal in translocation-related chromosome structure (aneuploid and no translocation imbalance); (4) aneuploid combined with a parental translocation imbalance, which was regarded as the parental translocation-related chromosomes are unbalanced, and parts of non-translocation chromosomes were aneuploid (aneuploid with translocation imbalance); and (5) embryos detected as a failure. Embryos that were euploid and had no translocation imbalance were transferred into a uterus or frozen preserved for further transfer [15]. In the control group, embryos obtained from age-matched patients with normal karyotypes and PGS results were analyzed and sorted into three categories: (1) euploid, (2) aneuploid, and (3) embryos detected as a failure.
Statistical analyses
A combination of Student’s t test, F test for variance, and chi-squared test was performed. Student’s t test was used to verify the homogeneity of ages, and the retrieved oocyte number and the number of embryos biopsied were entered as independent variables. Differences were calculated using the chi-squared or non-parametric Kruakal-Wallis tests where appropriate, such as the chromosomal abnormality rate, translocation-non-related chromosomal aberration types, as well as the aneuploid rate. A p value less than 0.05 was considered statistically significant.
Results
In total, 832 embryos obtained from rcp carriers and 382 embryos from RT carriers were biopsied. A total of 579 embryos were analyzed in the age-matched patients with non-translocation karyotypes in the control group. Since it was known that advanced maternal age affected the presence of embryonic aneuploids in a negative and important way, the results of PGD/PGS were categorized by maternal age, < 36 and ≥ 36 years. In the maternal age-matched sub-groups, the average maternal age and paternal age; average retrieved oocytes and matured oocytes; and number of normally fertilized embryos and cleavage embryos were similar among the rcp carriers, the RT carriers, and the control group (Table 2).
There were 282 cleavage embryos from rcp carriers whose maternal age was < 36 years were biopsied in 29 cycles detached by aCGH. Overall, 19.5% (55/282) of the embryos were euploid and without a chromosomal translocation imbalance, and they were chosen for transferring or freezing, whereas 50.7% (140/282) of the embryos were aneuploid. Further, 36 cleavage embryos from rcp carriers whose maternal age was ≥ 36 years were analyzed in 4 aCGH cycles. 11.1% (4/36) of the embryos were euploid and had no translocation, which were chosen for transfer or frozen, and 61.1%(22/36) of the embryos were aneuploid.
We noticed that in the maternal age < 36 years group, the rate of euploid without a translocation imbalance from day 3 embryos from rcp carriers was significantly lower than RT carriers [19.5% (55/287) vs. 29.3% (49/167), p < 0.05], and the rate of euploids with a translocation imbalance was significantly higher in rcp carrier day 3 embryos [27.6% (78/287) vs. 18.2% (31/167), p < 0.05]. However, with advanced maternal age (≥ 36), the differences were not significant. The rate of euploids with no translocation imbalance in day 3 embryos from rcp carriers was close to being statistically significantly higher in the maternal age < 36 year group than that in the advanced maternal age group [19.5% (55/287) vs. 11.1% (4/36), p > 0.05], whereas among RT carriers, a significantly higher rate of euploids with no translocation imbalance on day 3 embryos was found between the maternal age < 36 years group and the advanced maternal age group [29.3% (49/167) vs. 10.0% (3/30), p < 0.05]. Although a higher aneuploid rate was observed in the day 3 embryos obtained from rcp carriers [50.7% (143/287) vs. 61.1% (22/36), p > 0.05] and RT carriers [49.1% (82/287) vs. 56.7% (17/30), p > 0.05] with advanced maternal age, there was no significant difference. While in the control group, 44.8% (86/192) of day 3 embryos were aneuploid derived from patients with maternal age of < 36 years and 60.3% (35/58) from patients with advanced maternal age, and the difference was significant (p < 0.05). There were no significant differences in the aneuploidy rates found in day 3 embryos from rcp carriers and RT carriers compared with the age-matched control group. Therefore, no ICE were detected in the day 3 embryos from translocation carriers.
Overall, 166 day 5 blastocysts from RT carriers whose maternal age was <36 years were biopsied in 21 cycles detected with a SNP array. Forty-four percent (73/166) of the embryos were euploid and had no translocation, which were chosen for vitrification freezing, and 34.9%(58/166) of the embryos were aneuploid. While 19 blastocysts from RT carriers whose maternal age was ≥ 36 years were biopsied in 3 cycles with a SNP array. Of these embryos, 26.3%(5/19) were euploid with no translocation and were chosen for freezing, and 26.3%(5/19) were aneuploid.
In the maternal age of < 36 year group, the rate of euploid without a translocation imbalance in day 5 embryos from rcp carriers was significantly lower than that from RT carriers [27.8% (135/486) vs. 44.0% (73/166), p < 0.05]. As maternal age advanced (≥ 36), the differences were not significant. The rate of euploids with a translocation imbalance was significantly higher in rcp carrier day 5 embryos than that in RT carriers both in the maternal age of < 36 years group [40.1% (195/486) vs. 16.7% (26/166), p < 0.05] and in the maternal age advanced group [50.0% (14/28) vs. 26.3% (5/19), p < 0.05]. The aneuploidy rates were found to be significantly lower in rcp carriers’ and RT carriers’ day 5 embryos compared with those in the control group in the maternal age of < 36 year group [24.5% (119/486) vs. 34.9% (58/140) vs. 53.6% (75/140), p < 0.05]. With advanced maternal age (≥ 36), the aneuploidy rates were found to be significantly lower in translocation carrier day 5 embryos compared with those in the control group as well [10.7% (3/28) vs. 26.3 (5/19) vs. 57.1% (108/189), p < 0.05] (Table 3). Similar to the embryos obtained from day 3, there were no apparent ICE detected in the day 5 embryos of translocation carriers.
It was noticed that the incidences of aneuploidy with or without translocation balanced in day 3 embryos in translocation-carried parent embryos had significantly increased due to maternal age in rcp and RT couples. For rcp couples, women ≥ 34 years compared to women < 30 years were 46.7% (46/98) vs. 34.6% (113/327), respectively (p < 0.05); for RT couples, it was 57.4% (54/94) vs. 33.3% (40/120), respectively (p < 0.001). However, the increase in aneuploidy with maternal age gain in day 5 embryos was not observed (Fig. 1).
We also analyzed the data of embryos from rcp and RT carriers that were aneuploid in parental chromosomes non-involved with translocation and balanced in involved parental translocation chromosomes. The risk of chromosomal numerical abnormalities was observed in each of the 23 pairs of autosomes or sex chromosomes from day 3 and day 5 embryos.
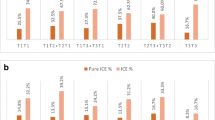
In day 3 embryos, the occurrence rates of 7.3% (11/150) in the deletion of both Chr21and Chr22, the reduplicate rate 7.3% (11/150) of Chr16, and the segmental duplication rate 6.0% (9/150) of Chr17 were significantly higher than other chromosomes in embryos derived from rcp-balanced embryos. The deletion rates of 7.7% (11/104) in Chr16 and Chr22 were significantly higher than those of other chromosomes from RT translocation balanced embryos (p < 0.05) (Fig. 2).
Comparation of the prevalence of aneuploidy without an unbalanced parental translocation chromosome under the influence of ICE rearrangement according to the abnormalities of each chromosome number in day 3 blastocsyt by aCGH. del, deletion; dup duplication; seg del, segmental deletion, seg dup, segmental duplication. *Statistical comparisons were performed using Kruskal-Wallis test and a p value of less than 0.05 was considered significant
In day 5 blastocysts, we found the segmental deletion rate 7.3% (10/126) of Chr19 and the deletion rate 8.7% (11/126) of Chr22 were significantly higher than other chromosomes in embryos derived from rcp-balanced embryos. While the segmental deletion rate 8.9% (6/68) of Chr5, 7.4% (5/68) of Chr9, and 8.9% (6/68) of Chr22 was significantly higher than those of other chromosomes from RT-balanced embryos (p < 0.05). In addition, we noticed that Chr12 and Chr23 in day 5 embryos from rcp carriers, as well as Chr14, Chr19, and Chr23 in day 5 blastocysts from RT carriers, had no copy number variants (Fig. 3).
Comparation of the prevalence of aneuploidy without an unbalanced parental translocation chromosome under the influence of ICE rearrangement according to the abnormalities of each chromosome number in day 5 embryos by SNP array. del, deletion; dup, duplication; seg del, segmental deletion; seg dup, segmental duplication. *Statistical comparisons were performed using Kruskal-Wallis test and a p value of less than 0.05 was considered significant
Discussion
In this study, the objective was to identify whether ICE presented in preimplantation embryos were obtained from rcp and RT carrier couples with whole chromosome screening of aCGH and SNP microarray. The main two reasons for these couples entering PGD cycles were primary infertility (44.9%) and RSA (23.9%). We also investigated whether selecting one or two embryos that were euploid and without translocation imbalance errors diagnosed through an aCGH and SNP array would benefit the couples.
There has been a long debate on whether ICE exists. Previous papers analyzed ICE with multiple FISH probes to detect whether the copy numbers of sperm or embryo chromosomes had changed. Several studies used multiple FISH to assess the occurrence of ICE in structural rearrangement carriers by analyzing the frequencies of numerical abnormalities in the sperm from rcp carriers [16, 17] and from RT carriers [18, 19]. Some studies suggested that ICE existed in the sperm from both rcp and RT carriers [20,21,22,23] or were only found in RT carriers [18, 19, 24,25,26,27]. A. Pujol et al. reported that the incidence of aneuploidy detected in oocytes for the first polar bodies (1PBs) biopsy for the chromosomes not involved in the RT translocations was extremely high [28]. Others proposed that ICE were not present or negligible [4, 29]. Pinar et al. investigated cleavage-stage embryos from translocation carriers undergoing PGD, which were biopsied by mFISH for the chromosomes involved in the translocation in addition to chromosomes13, 15, 16, 17, 18, 21, 22, X, and Y. They found no ICE in embryos derived from Robertsonian and reciprocal translocation carriers [30]. The mechanism for how the translocation chromosomes disrupted other non-involved chromosomes in embryogenesis was not defined clearly. Previous studies showed that the rate of ICE depended on the breakpoints and the regions of the translocated chromosomes [4, 6]. Studies have also suggested that meiotic divisions might induce ICE in the embryos at the positioning and pairing of the rearranged chromosomes with the non-translocated chromosomes may be disturbed and may alter segregation [3, 23, 25]. The segregation error of translocation chromosomes during meiosis may cause deviations during the mitosis process, which may lead to centrosome amplification, chromosome anomaly separation, and genomic instability, triggering aneuploidy in non-related chromosomes.
In our study, we analyzed the occurrence of ICE by comparing the cumulative aneuploid rates in rcp and RT carriers with those of age-matched patients without a chromosomal translocation karyotype who were undergoing PGS. Both PGD and PGS outcomes were assessed on day 3 and day 5 using an aCGH or SNP array. In our control group, the indications of PGS were 82.9% of RSA, 5.3% of RIF, and 11.8% with AMA (≥ 36 years). However, the ideal control group would be age-matched couples undergoing PGD for single gene disorders and without an infertility problem as they have no increasing risk of aneuploidy in the embryos. It was hard to congregate this patient group because there are extra costs for the patients and there is no indication of PGS. In our center, we began to try a PGD+PGS model for couples with single gene disorders and a maternal age over 36 or those that had a history of spontaneous abortion. However, the results of this new model need further study.
The cumulative aneuploidy rates of rcp carriers’ day 3 embryos were 50.7 and 61.1% whose maternal age was under 36 years and ≥ 36 years, respectively, and in Day 5 embryos from carriers whose maternal age was under 36 years and ≥ 36 years, the aneuploidy rates decreased to 24.5 and 10.7%, respectively. We assumed the decrease in aneuploidy rates between embryos from day 3 that were biopsied by aCGH and day 5 detected by SNP array, on the one hand, was caused by the two techniques of analysis, which were deeply different. It is well known that the aCGH techniques may overestimate at least 10% of embryonic aneuploidy; however, with embryonic development to the blastocyst stage and chromosome self-repair, the chromosome abnormalities decreased, so the aneuploid rate decreased. It has been reported that there were approximately 60% of day 3 “high quality embryos” that can develop into blastocysts, and approximately 60% of the top quality cleavage-stage embryos were aneuploid, while the aneuploid rate decreased to 30% of blastocyst. Array CGH analysis showed that aneuploidy is not related to the number of embryos that are generated [31, 32]. Therefore, in recent years, along with our center technology upgrade, blastocyst biopsy has been fully implemented and SNP array has been used for detection, instead of aCGH, which may improve the utilization of the embryo.
In day 3 embryos from RT carriers, 49.1% had aneuploidy with maternal age < 36 years and 56.7% with maternal age ≥ 36 years, and in day 5 embryos from RT carriers, the aneuploidy rates decreased to 34.9 and 26.3% of maternal age < 36 and ≥ 36 years, respectively. Regardless of maternal age, the differences in aneuploidy rates between rcp/RT chromosomal translocation carriers and the control group showed no significant difference. Our study suggested that ICE were not detected in cleavage- or blastocyst-stage embryos from rcp and RT carriers.
The frequency of aneuploidy was directly dependent on maternal age. We found in cleavage-stage embryos, the cumulative aneuploidy rates were increased with advanced maternal age. This finding aligned itself with the well-established trend that increased maternal age was associated with increased aneuploidy [33]. The reason for this increase could be the tendency to non-disjunction related to advanced maternal age combined with an interchromosomal effect resulting in the presence of synaptic errors in other chromosome pairs, but this phenomenon of increasing aneuploidy rates did not appear in day 5 embryos from rcp or RT carriers. This may due to a decrease in the total amount of elderly patients with good-quality embryos available for PGD biopsy, resulting in a small overall sample size for those of maternal age ≥ 36 years.
In addition, we noticed that, in day 5 embryos, the rate of aneuploidy with a translocation balance from rcp carriers was significantly lower than that from RT carriers. The results were consistent with other reports of RT carriers having a higher risk of aneuploid oocytes [28]. The synaptic defects in the trivalent formed during meiosis I in RT translocations might cause a higher aneuploid prevalence for the chromosomes not involved in the rearrangement than in rcp carriers, and this may result in heterosynapses or even the production of univalence. This may delay chromosome congression and interfere with the spindle checkpoint, producing chromosome aneuploidy [34]. Synaptic defects may also allow the maturation of MII kinetochores and result in pre-division [35].
Most of the ICE in previous studies were performed with multiple FISH as this analysis limited the analyzed number of chromosomes and there was lower accuracy with more rounds of FISH probing. In our center, 24 chromosome screening with the aCGH and SNP array technique had been applied in PGD cycles for rcps and RTs since 2011. With restrictions of biopsies in early 2011, all of the embryos were biopsied by day 3 at a cleavage stage, and the normal/balanced embryos were chosen according to the PGD results for transplanting or freezing on day 5. The progressive realization of day 5 blastocyst biopsy and SNP array technology with higher resolution were gradually achieved since 2013 in our center. In our previous study, we found that normal and/or balance rates detected by aCGH were significantly higher than those by FISH in both rcp (38.2 vs. 15.39%) and RT (67.2 vs. 30.7%) carriers [10, 13, 36]. It was demonstrated that the chromosomal translocation PGD using an aCGH and SNP array was quite effective [15]. With a wholesome 24 chromosome screening, we noticed the risk of chromosomal numerical abnormalities existed in 23 pairs of autosomes and sex chromosomes. The deletion rates of Chr21 and Chr22, the reduplicate rate of Chr16, the segmental duplication rate of Chr17 from cleavage-stage embryos obtained from rcp carrier couples, and the deletion rates of Chr16 and Chr22 from cleavage-stage embryos of RT carriers had a higher frequency than the other chromosomes tested. In day 5 blastocysts, we found the segmental deletion rate of Chr19 and Chr22, the segmental deletion rate of Chr5 and Chr9, and the deletion rate of Chr22 were significantly higher than other chromosomes from RT translocation balanced embryos. In addition, we noticed that Chr12 and Chr23 in day 5 embryos from rcp carriers as well as the Chr14, Chr19, and Chr23 in day 5 blastocysts from RT carriers had no copy number variants. Kovaleva reported that although an increased incidence of trisomy 21 is present in balanced reciprocal translocation and inversion carriers, this may not be evidence of ICE [18]. This observation agreed with the hypothesis of a relationship between an increase in non-disjunction for these chromosomes and the presence of a chromosomal rearrangement, but rather it agreed with Chr21 being more susceptible to non-disjunction than other chromosomes [37,37,39]. This implied that 24-chromosomal analysis with an aCGH/SNP microarray could supply more crucial information of aneuploidies, which is more valuable than the traditional FISH technique in the diagnosis of ICE.
However, the mechanism of ICE events has not been defined clearly yet; further larger-scale studies on structural rearrangement carriers may help clarify the still unknown mechanism of cytogenetic features that promote ICE.
Conclusion
In conclusion, according to our study, rcp and RT carriers had no significant increase in the aneuploid rate compared with age-matched RSA, RIF, and AMA patients with normal chromosomal karyotypes. There was not enough evidence to prove that ICE was present in embryos derived from both rcp and RT carriers regardless of the maternal age. However, the aneuploid changes were noticed in both day 3 and day 5 embryos. Additionally, we noticed there were risks of chromosomal numerical abnormalities in 23 pairs of autosomes and sex chromosomes from translocation carrier embryos. It is imperative for translocation carrier couples to undergo aneuploidy screening with 24-chromosomal analysis with an aCGH/SNP microarray for translocation diagnosis to reduce the abortion risk of a baby with congenital abnormalities due to misdiagnosis caused by the aneuploidy.
References
Lledo B, Ortiz JA, Morales R, Ten J, de la Fuente PE, Garcia-Ochoa C, et al. The paternal effect of chromosome translocation carriers observed from meiotic segregation in embryos. Hum Reprod. 2010;25:1843–8.
Faraut T, Mermet MA, Demongeot J, Cohen O. Cooperation of selection and meiotic mechanisms in the production of imbalances in reciprocal translocations. Cytogenet Cell Genet. 2000;88:15–21.
Lejeune J. Autosomal disorders. Pediatrics. 1963;32:326–37.
Estop AM, Cieply K, Munne S, Surti U, Wakim A, Feingold E. Is there an interchromosomal effect in reciprocal translocation carriers? Sperm fish studies. Hum Genet. 2000;106:517–24.
Munne S. Analysis of chromosome segregation during preimplantation genetic diagnosis in both male and female translocation heterozygotes. Cytogenet Genome Res. 2005;111:305–9.
Alfarawati S, Fragouli E, Colls P, Wells D. Embryos of Robertsonian translocation carriers exhibit a mitotic interchromosomal effect that enhances genetic instability during early development. PLoS Genet. 2012;8:e1003025.
Gianaroli L, Magli MC, Ferraretti AP, Munne S, Balicchia B, Escudero T, et al. Possible interchromosomal effect in embryos generated by gametes from translocation carriers. Hum Reprod. 2002;17:3201–7.
Vozdova M, Oracova E, Musilova P, Kasikova K, Prinosilova P, Gaillyova R, et al. Sperm and embryo analysis of similar t(7;10) translocations transmitted in two families. Fertil Steril. 2011;96:e66–70.
Tan YQ, Tan K, Zhang SP, Gong F, Cheng DH, Xiong B, et al. Single-nucleotide polymorphism microarray-based preimplantation genetic diagnosis is likely to improve the clinical outcome for translocation carriers. Hum Reprod. 2013;28:2581–92.
Xie Y, Xu Y, Miao B, Zeng Y, Zhou C. A preliminary study on the application of array comparative genomic hybridization for preimplantaion genetic diagnosis. Chin J Med Genet. 2013;30:283–7.
Debrock S, Melotte C, Spiessens C, Peeraer K, Vanneste E, Meeuwis L, et al. Preimplantation genetic screening for aneuploid of embryos after in vitro fertilization in women aged at least 35 years: a prospective randomized trial. Fertil Steril. 2010;93:364–73.
Harton GL, Magli MC, Lundin K, Montag M, Lemmen J, Harper JC. Eshre pgd consortium/embryology special interest group—best practice guidelines for polar body and embryo biopsy for preimplantation genetic diagnosis/screening (pgd/pgs). Hum Reprod. 2011;26:41–6.
Xu YW, Zhou CQ, Zeng YH, Liu Y, Gao L, Zhuang GL. Clinical analysis of 100 preimplantation genetic diagnosis cycles. Chin J Obstet Gynecol. 2011;46:255–9.
Gardner DK, Lane M, Schoolcraft WB. Physiology and culture of the human blastocyst. J Reprod Immunol. 2002;55:85–100.
Tobler KJ, Brezina PR, Benner AT, Du L, Xu X, Kearns WG. Two different microarray technologies for preimplantation genetic diagnosis and screening, due to reciprocal translocation imbalances, demonstrate equivalent euploidy and clinical pregnancy rates. J Assist Reprod Genet. 2014;31:843–50.
Kirkpatrick G, Ma S. Meiotic segregation and interchromosomal effects in a rare (1:2:10) complex chromosomal rearrangement. J Assist Reprod Genet. 2012;29:77–81.
Kasikova K, Vozdova M, Prinosilova P, Gaillyova R, Hanakova M, Rubes J. Sperm meiotic segregation, aneuploid and high risk of delivering an affected offspring in carriers of non-Robertsonian translocation t(13;15). J Assist Reprod Genet. 2012;29:693–8.
Kovaleva NV. Increased risk of trisomy 21 in offspring of carriers of balanced non-contributing autosomal rearrangements is not explained by interchromosomal effect. Genetika. 2013;49:259–68.
Piomboni P, Stendardi A, Gambera L. Chromosomal aberrations and aneuploid of spermatozoa. Adv Exp Med Biol. 2014;791:27–52.
Vozdova M, Oracova E, Horinova V, Rubes J. Sperm fluorescence in situ hybridization study of meiotic segregation and an interchromosomal effect in carriers of t(11;18). Hum Reprod. 2008;23:581–8.
Juchniuk DVM, Santos SA, Pereira CS, Cuzzi JF, Laureano LA, Franco JJ, et al. Meiotic segregation and interchromosomal effect in the sperm of a double translocation carrier: a case report. Mol Cytogenet. 2009;2:24.
Bonnet-Garnier A, Guardia S, Pinton A, Ducos A, Yerle M. Analysis using sperm-fish of a putative interchromosomal effect in boars carrying reciprocal translocations. Cytogenet Genome Res. 2009;126:194–201.
Anton E, Vidal F, Blanco J. Reciprocal translocations: tracing their meiotic behavior. Genet Med. 2008;10:730–8.
Anton E, Vidal F, Blanco J. Interchromosomal effect analyses by sperm fish: incidence and distribution among reorganization carriers. Syst Biol Reprod Med. 2011;57:268–78.
Anton E, Blanco J, Vidal F. Meiotic behavior of three D;G Robertsonian translocations: segregation and interchromosomal effect. J Hum Genet. 2010;55:541–5.
Alfarawati S, Fragouli E, Colls P, Wells D. First births after preimplantation genetic diagnosis of structural chromosome abnormalities using comparative genomic hybridization and microarray analysis. Hum Reprod. 2011;26:1560–74.
Fiorentino F, Spizzichino L, Bono S, Biricik A, Kokkali G, Rienzi L, et al. Pgd for reciprocal and Robertsonian translocations using array comparative genomic hybridization. Hum Reprod. 2011;26:1925–35.
Pujol A, Durban M, Benet J, Boiso I, Calafell JM, Egozcue J, et al. Multiple aneuploid in the oocytes of balanced translocation carriers: a preimplantation genetic diagnosis study using first polar body. Reproduction. 2003;126:701–11.
Munne S, Escudero T, Fischer J, Chen S, Hill J, Stelling JR, et al. Negligible interchromosomal effect in embryos of Robertsonian translocation carriers. Reprod BioMed Online. 2005;10:363–9.
Tulay P, Gultomruk M, Findikli N, Yagmur E, Bahceci M. Is the interchromosomal effect present in embryos derived from Robertsonian and reciprocal translocation carriers particularly focusing on chromosome 10 rearrangements? Zygote. 2015;23:908–15.
Majumdar G, Majumdar A, Verma IC, Upadhyaya KC. Relationship between morphology, euploidy and implantation potential of cleavage and blastocyst stage embryos. J Hum Reprod Sci. 2017;10:49–57.
Minasi MG, Fiorentino F, Ruberti A, Biricik A, Cursio E, Cotroneo E, Varricchio MT, Surdo M, Spinella F, Greco E: Genetic diseases and aneuploid can be detected with a single blastocyst biopsy: a successful clinical approach. Hum Reprod. 2017;8:1770–77.
Wapner RJ. Genetics of stillbirth. Clin Obstet Gynaecol. 2010;53:628.
Cukurcam S, Sun F, Betzendahl I, et al. Trichlorfon predisposes to aneuploidy and interferes with spindle formation in in vitro maturing mouse oocytes. Mutat Res. 2004;2:165–78.
Flatters M, Maxfield R, Dawson D. The effects of a ring chromosome on the meiotic segregation of other chromosomes in Saccharomyces cerevisiae. Mol Gen Genet. 1995;3:309–316.
Yanxin X, Yanwen X, Benyu M, Yanhong Z, Jing W, Canquan Z. Clinical investigation to compare acgh and fish in preimplantationgenetic diagnosis of chromosome translocation carriers. Chin J Obstet Gynecol. 2014;49:193–8.
Spriggs EL, Rademaker AW, Martin RH. Aneuploid in human sperm: results of two-and three-color fluorescence in situ hybridization using centromeric probes for chromosomes 1, 12, 15, 18, x, and y. Cytogenet Cell Genet. 1995;71:47–53.
Blanco J, Egozcue J, Vidal F. Incidence of chromosome 21 disomy in human spermatozoa as determined by fluorescent in-situ hybridization. Hum Reprod. 1996;11:722–6.
Pellestor F, Girardet A, Coignet L, Andreo B, Charlieu JP. Assessment of aneuploid for chromosomes 8, 9, 13, 16, and 21 in human sperm by using primed in situ labeling technique. Am J Hum Genet. 1996;58:797–802.
Acknowledgements
The Nature and Science Foundation of Guangdong Province (S2012010009176); the Guangzhou City Science and Technology Board Foundation: the establishment of genetic disease preimplantation embryos and early pregnancy screening model (201300000097).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xie, Y., Xu, Y., Wang, J. et al. Preliminary analysis of numerical chromosome abnormalities in reciprocal and Robertsonian translocation preimplantation genetic diagnosis cases with 24-chromosomal analysis with an aCGH/SNP microarray. J Assist Reprod Genet 35, 177–186 (2018). https://doi.org/10.1007/s10815-017-1045-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-017-1045-9