Abstract
Multiprotein bridging factor 1 (MBF1) is a highly conserved transcriptional co-activator involved in the regulation of diverse processes, such as environmental stress responses. We recently identified a novel MBF1 gene, PyMBF1, from the marine red alga Pyropia yezoensis. In this study, quantitative real-time PCR analysis revealed that PyMBF1 transcripts were upregulated in P. yezoensis cells during exposure to oxidative and heat stresses. We also examined heat signaling in P. yezoensis cells by monitoring the accumulation of PyMBF1 transcripts. Heat activation of PyMBF1 was inhibited by the membrane rigidifier dimethylsulfoxide, whereas it was induced without heat stress by the membrane fluidizer benzyl alcohol (BA). Induction of PyMBF1 transcripts by heat and BA was inhibited by 1-butanol, an inhibitor of phospholipase D (PLD). The results suggest that the heat activation of PyMBF1 requires membrane fluidization and activation of PLD. These findings provide an initial step toward understanding heat signaling in marine red algae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The growth of plants is greatly affected by a variety of environmental factors, such as dehydration, temperature, and salinity. Understanding the mechanisms by which plants perceive environmental signals, transmit signals to the cellular machinery, and regulate the expression of stress-responsive genes is important in fundamental and applied biology. The cellular and molecular responses to environmental stress have been studied extensively in higher plants (Miller et al. 2010; Huang et al. 2012; Mizoi et al. 2012). For example, it has been reported that the extracellular stress signal is perceived at the membrane level and then activates a complex signaling cascade of secondary signal molecules, such as Ca2+, inositol phosphates, and abscisic acid. The stress signal also regulates transcription factors to induce multiple stress-responsive genes, which leads to the adaptation of the plant, thus, providing stress tolerance (Mahajan and Tuteja 2005). However, the mechanisms by which macroalgae, in contrast to higher plants, respond to environmental stresses are largely unknown.
The red macroalga Pyropia yezoensis (formerly Porphyra yezoensis; Sutherland et al. 2011) has received attention as a model organism for studies on the physiology and molecular biology of seaweeds because, in addition to its economic importance, it has several advantages for biological research (Saga and Kitade 2002). In P. yezoensis, large-scale expressed sequence tag (EST) analyses have been carried out (Nikaido et al. 2000; Asamizu et al. 2003), and these are being exploited to identify genes involved in environmental stress in this species (Uji et al. 2012a, b; Li et al. 2012). In addition, transient gene expression systems have been developed in our laboratory (Fukuda et al. 2008; Mikami et al. 2009; Uji et al. 2010). Therefore, P. yezoensis is a suitable species for the elucidation of molecular mechanisms regulating environmental stress responses in marine red algae.
Transcriptional coactivators play an important role in eukaryotic gene regulation by interacting with transcription factors (Näär et al. 2001). Multiprotein bridging factor 1 (MBF1) is a highly conserved transcriptional coactivator involved in the regulation of diverse processes, such as endothelial cell differentiation, histidine metabolism, hormone-regulated lipid metabolism, and central nervous system development (Takemaru et al. 1997, 1998; Brendel et al. 2002; Liu et al. 2003). In higher plants, it has been shown that AtMBF1c from Arabidopsis thaliana is upregulated by salinity, drought, hydrogen peroxide (H2O2), and heat (Tsuda and Yamazaki 2004). Constitutive expression of AtMBF1c in transgenic plants enhances the tolerance to bacterial infection, heat, and osmotic stress (Suzuki et al. 2005). Moreover, the accumulation of MBF1 mRNA increases through the combined effect of drought stress and heat shock in Retama raetam (Pnueli et al. 2002) and Nicotiana tabacum (Rizhsky et al. 2002). Thus, MBF1 may have a role that is of general importance in plant stress responses; however, the role of MBF1 in marine macroalgae remains obscure.
We have recently reported the molecular characterization and nuclear localization of PyMBF1, an MBF1 from P. yezoensis (Uji et al. 2010). In this study, we showed that PyMBF1 transcripts were upregulated by heat and oxidative stresses. In addition, we investigated heat stress signaling in P. yezoensis by monitoring the accumulation of PyMBF1 transcripts.
Materials and methods
Algal material and stress treatment
Leafy gametophytes of the P. yezoensis strain TU-1 were cultured at 15 °C in enriched sealife (ESL) medium adjusted to pH 8.0, as described previously by Uji et al. (2012a). The cultured algae were used for expression analysis of PyMBF1. Heat stress was induced by increasing culture medium temperature from 15 to 25 °C. Cold stress was induced by decreasing the temperature from 15 to 5 °C. To induce oxidative stress, cultured algae were treated with ESL medium containing 1 mM H2O2. Algal materials were placed directly into liquid nitrogen and stored at −80 °C until use for transcription analysis.
Chemical treatments
Benzyl alcohol (BA) and dimethylsulfoxide (DMSO) were used to fluidize and rigidify the plasma membrane, respectively. Lanthanum chloride (LaCl3) was used as a calcium channel blocker and 1-butanol as an inhibitor of phospholipase D (PLD). Chemicals were freshly prepared by dissolving these in ESL medium. O′-bis(2-aminoethyl)ethyleneglycol-N,N,N′,N′-tetraacetic acid (EGTA), a calcium chelator, was dissolved in ESL medium and the solution was adjusted to pH 8.0 with NaOH. Concentrations of the above chemicals used in experiments are provided in the corresponding figure legends. Algal materials were frozen immediately in liquid nitrogen and stored at −80 °C until use for transcription analysis.
Transcription analysis of PyMBF1
Total RNA extraction from gametophytes and sporophytes was carried out with an RNeasy Plant Mini Kit (Qiagen), and the resulting total RNA was further purified with a TURBO DNA-free Kit (Applied Biosystems/Life Technologies). Total RNA (0.5 μg/reaction) was used for first-strand cDNA synthesis with a PrimeScript II First Strand cDNA Synthesis Kit (TaKaRa Bio). Real-time PCR was performed with an ABI Prism 7300 Sequence Detection System and software (Applied Biosystems/Life Technologies). All real-time PCR was performed under the following conditions: 30 s at 95 °C followed by 40 cycles of 5 s at 95 °C and 31 s at 60 °C. Gene-specific primers for PyMBF1 (forward, 5′-TCGCCGAGAAGAAGCACGG-3′; reverse, 5′-CCCGTCTACACCTCCTTCC-3′) and Py18SrRNA (forward, 5′-TGATAGTCCTGGGTCGGAAG-3′; reverse, 5′-TGATGACCTGCGCCTACAAG-3′) were used in real-time PCR. Specificity of the PCR products was confirmed by analyzing the dissociation curve at the end of each reaction (15 s at 95 °C, 1 min at 60 °C, and 15 s at 95 °C). The reaction mixture (20 μL) contained 10 μL SYBR Premix Ex Taq GC (TaKaRa Bio), 0.8 μL of each forward and reverse primer (5 μM), 0.4 μL of ROX Reference Dye, and 4 μL of cDNA template (100-fold dilution). The Py18SrRNA gene, whose transcriptional activity does not fluctuate significantly under heat, cold, and oxidative stresses, was used as an internal control to normalize the amount of mRNA in each reaction. The amounts of PyMBF1 mRNA were calculated on the basis of a standard curve. The standard curve for each primer set was prepared by serial cDNA dilution (1:10–1:105) and plotted against the threshold cycle. All experiments were conducted in triplicate.
Phylogenetic analysis of MBF1
A phylogenetic tree was constructed using the neighbor-joining method with MEGA version 5.0 (http://www.megasoftware.net). The amino acid sequences of MBF1s were retrieved from the GenBank and Genome databases.
Results
Phylogenetic analysis of MBF1
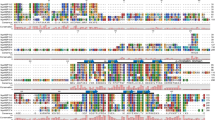
To identify the group with which PyMBF1 clusters, we constructed a phylogenetic tree based on the amino acid sequence alignment of PyMBF1 and other MBF1s. Figure 1 shows that PyMBF1 is not classifiable in either of plant group I or II, and closest to Cyanidioschyzon merolae, a unicellular red alga.
Phylogenetic tree of MBF1. The phylogenetic tree is based on the amino acid sequences of the ORF of MBF1 using the neighbor-joining method. Bootstrap values are indicated only for nodes with greater than 50 %. Branch lengths indicate evolutionary distance with a scale of 0.1. Species and accession numbers (in alphabetical order): A. thaliana (AtMBF1a), NP_565981; A. thaliana (AtMBF1b), NP_191427; A. thaliana (AtMBF1c), NP_189093; Bombyx mori, NP_001036824; C. merolae, CMJ111C; Chlamydomonas reinhardtii, XM_001699673; Danio rerio, BC059541; Drosophila melanogaster, AB031273; Ectocarpus siliculosus Esi0061_0121; Homo sapiens (hMBF1alpha), AB002282; H. sapiens (hMBF1beta), AB002283; Lycopersicon esculentum, AF096246; Neurospora crassa, XP_960690; N. tabacum, AB072698; Phaeodactylum tricornutum, XP_002182087; P. yezoensis, AB480828; R. raetam, AF439278; Saccharomyces cerevisiae, AB017593; Solanum tuberosum, AF232062; and Thalassiosira pseudonana, XP_002293430. The amino acid sequences of MBF1 from C. merolae and E. siliculosus were derived from the C. merolae Genome Project (http://merolae.biol.s.u-tokyo.ac.jp/) and the E. siliculosus Genome Project (http://bioinformatics.psb.ugent.be/webtools/bogas/overview/Ectsi), respectively. Other sequences were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/)
Effects of heat, cold, and oxidative stresses on PyMBF1 transcripts
We tested whether mRNA levels of PyMBF1 were affected by heat and H2O2 stresses as well as cold stress (Fig. 2). Figure 2b shows that cold stress had little effect on the expression patterns of PyMBF1. However, expression levels of PyMBF1 after 60 min of exposure to heat (Fig. 2a) and H2O2 treatments (Fig. 2c) were 4.3- and 2.5-fold higher than those at baseline. We also tested the time-dependent change in mRNA accumulation of PyMBF1 under heat treatment (Fig. 3). The transcript level of PyMBF1 slightly increased within 5 min of initiating the heat shock and continued to increase during the incubation period.
Expression of PyMBF1 transcripts in P. yezoensis gametophytes under various stresses. a Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of PyMBF1 expression in response to heat stress. RNA samples were prepared from gametophytes 60 min after a shift from 15 to 25 °C. Py18SrRNA was used as an internal control. Results are presented as relative expression compared with that in nonstressed material (0 h). Data are presented as mean ± standard deviation (n = 3). b qRT-PCR analysis of PyMBF1 expression in response to cold stress. RNA samples were prepared from gametophytes 60 min after a shift from 15 to 5 °C. c qRT-PCR analysis of PyMBF1 expression in response to oxidative stress. RNA samples were prepared from gametophytes 60 min after treatment with 1 mM H2O2
Expression analysis of time-dependent change of PyMBF1 transcripts in P. yezoensis gametophytes by heat stress. qRT-PCR analysis of PyMBF1 expression in response to heat stress at different time points. RNA samples were prepared from gametophytes 0, 5, 15, 30, 45, or 60 min after a shift from 15 to 25 °C. The Py18SrRNA gene was used as an internal control. Results are presented as relative expression compared with that in nonstressed material (0 h). Data are presented as mean ± standard deviation (n = 3)
Change in membrane fluidity is required for heat activation of PyMBF1
To test whether changes in membrane fluidity play a fundamental role in heat signal transduction in P. yezoensis, transcript accumulation of PyMBF1 in the gametophytes was used as an end-point marker. Treatment with BA at 15 °C for 1 h increased the transcript level of PyMBF1, but the level was lower than that observed with heat treatment at 25 °C (Fig. 4a). When thalli were incubated at 25 °C for 1 h in the presence of DMSO at concentrations of 4–8 %, heat activation of PyMBF1 was inhibited at all concentrations tested (Fig. 4b).
Changes in membrane fluidity mediate heat activation of PyMBF1. a Membrane fluidization induces PyMBF1 transcript accumulation in P. yezoensis gametophytic cells. Gametophytic cells were incubated at 15 °C for 1 h with the indicated concentrations of the membrane fluidizer benzyl alcohol (BA) or at 25 °C for 1 h without BA (25 °C) before RNA extraction. Treatment with 10 m BA did not affect the viability of intact cells. Py18SrRNA was used as an internal control. Results are presented as relative expression compared with that in nontreated material (0 mM). Data are presented as mean ± standard deviation (n = 3). b Membrane rigidification inhibits heat-triggered PyMBF1 transcript accumulation in P. yezoensis gametophytic cells. Gametophytic cells were pretreated for 1 h at 15 °C with the indicated concentrations of the membrane rigidifier dimethylsulfoxide (DMSO) and subsequently incubated at 25 °C for 1 h or were incubated at 15 °C for 1 h without DMSO (15 °C) before RNA extraction. Treatment with 8 % DMSO did not affect the viability of intact cells
Ca2+ and PLD are involved in heat activation of PyMBF1
We examined whether modulators of Ca2+ influx and PLD activity affect the heat-induced PyMBF1. Thalli of P. yezoensis were treated with a Ca2+ chelator EGTA, a Ca2+ channel blocker LaCl3, or a PLD inhibitor 1-butanol. Figure 5 shows that levels of PyMBF1 activation by heat or BA were partially reduced by EGTA and LaCl3. In addition, treatment with 1-butanol reduced the level of activation to that seen without heat stress or BA treatment (Fig. 5).
Effects of addition of chemicals on heat- and BA-triggered PyMBF1 transcript accumulation in P. yezoensis gametophytic cells. Gametophytic cells were pretreated for 1 h at 15 °C with the Ca2+ chelator O′-bis(2-aminoethyl)ethyleneglycol-N,N,N′,N′-tetraacetic acid (EGTA; 1 mM), the Ca2+ channel blocker lanthanum chloride (LaCl 3 ; 100 μM), or the phospholipase D inhibitor 1-butanol (0.4 %), and then either heat shocked at 25 °C (a) or treated with 10 mM BA at 15 °C (b) for 1 h before RNA extraction
Discussion
Plant MBF1 is definitively classified into two groups. Plant group I includes AtMBF1a and AtMBF1b. Their expression is tissue-specific and is not changed by stresses (Tsuda and Yamazaki 2004). Plant group II contains AtMBF1c which is inducible under various stresses such as heat and oxidative stresses (Tsuda and Yamazaki 2004). In addition, N. tabacum MBF1 and R. raetam MBF1 (ERTCA) which are upregulated by combination of drought and heat (Pnueli et al. 2002; Rizhsky et al. 2002) are categorized to plant groups I and II, respectively. As shown in Fig. 2, the mRNA transcripts of PyMBF1 were accumulated by environmental stresses like MBF1 genes from higher plants, but PyMBF1 is not classifiable in either of the two plant groups (Fig. 1). The expression of PyMBF1 was increased within 60 min after H2O2 or heat treatment (Figs. 2 and 3). These results suggest that PyMBF1 plays a role in the oxidative and heat stress response pathways in P. yezoensis, although it is no clear yet whether MBF1s from red algae show the diversity like in higher plants. The level of AtMBF1c mRNA increases as a result of dehydration or high salinity as well as H2O2 or heat treatment (Tsuda and Yamazaki 2004). In addition, dehydration and high salinity lead to increased production of reactive oxygen species, such as H2O2 in marine macroalgae, including Pyropia (Lu et al. 2006; Contreras-Porcia et al. 2011). Thus, the level of PyMBF1 transcripts probably also fluctuates in response to dehydration and high salinity stresses.
Membrane fluidity is rapidly and reversibly affected by temperature change, and changes in membrane fluidity play a crucial role in temperature sensing events in plants (Los and Murata 2004). For example, Synechocystis cells deficient in fatty acid desaturase with altered membrane fluidity have defects in temperature-regulated gene expression (Inaba et al. 2003). It has been shown that changes in membrane fluidity mediate the temperature-induced activation of genes in land plants by using the membrane fluidizer BA and the membrane rigidifier DMSO (Örvar et al. 2000; Sangwan et al. 2001, 2002). For example, the effects of heat shock on tobacco cells were countered by treating the cells with DMSO and were mimicked by BA without heat stress (Sangwan et al. 2002). Our results showed that the accumulation of PyMBF1 transcripts in response to heat is prevented by membrane rigidification; whereas, it is increased by membrane fluidization without heat shock, demonstrating that temperature-induced activation requires alteration of membrane fluidity also in red algae.
The accumulation of PyMBF1 transcripts with 10 mM BA treatment was lower than that with heat treatment (Fig. 4a). We therefore examined the transcription level of PyMBF1 by treating the cells with 20 mM BA. However, the expression level did not increase in comparison with 10 mM BA treatment (data not shown). DNA microarray analysis of the gene expression profile of Synechocystis cells showed that there were genes whose expression was markedly induced by heat but not by BA treatment and vice versa (Inaba et al. 2003). These findings suggest different heat-sensing mechanisms from those involved in membrane fluidization.
Phospholipid-based signaling plays an important role in the responses to a variety of abiotic and biotic stresses including cold shock, drought, and pathogen attack (Zonia and Munnik 2006). PLD hydrolyses phospholipids to form phosphatidic acid (PA), which is an important phospholipid signal (Testerink and Munnik 2005). It has been suggested that PLD is involved in the heat stress responses of higher plants. For example, heat stress led to rapid PA accumulation in the cell membranes of tobacco and Arabidopsis through activation of a PLD pathway (Mishkind et al. 2009). In addition, PLD from the grape berry and Jatropha curcas were increasingly expressed in response to heat stress (Wan et al. 2007; Liu et al. 2010). In the present study, heat- and BA-induced activation of PyMBF1 was inhibited by a PLD inhibitor (Fig. 5), suggesting that PLD activation is required for the heat stress response at a more downstream position than membrane fluidization in heat signaling of P. yezoensis.
In response to high temperature, an elevation in cytosolic-free Ca2+ concentration has been found in several plants (Knight and Knight 2000) and the induction of Ca2+ influx by membrane fluidization upon heat stress is required for the heat activation of MAP kinase in alfalfa cells (Sangwan et al. 2002). In tobacco cells, heat-triggered accumulation of HSP70 depends on the activation of Ca2+ channels by increasing membrane fluidity (Suri and Dhindsa 2008). EGTA, a Ca2+ chelator, and LaCl3, a Ca2+ channel blocker, have previously been shown to prevent Ca2+ influx into the cytosol in plant cells (Monroy and Dhindsa 1995; Knight et al. 1996). 1-butanol is a useful PLD inhibitor in plant cells (Munnik et al. 1995; Gardiner et al. 2003). We used 1 mM EGTA, 100 μM LaCl3, and 0.4 % 1-butanol, because it has been reported that these reagent concentrations inhibit the establishment of cell polarity in monospores from P. yezoensis (Li et al. 2009). In the present study, levels of heat- or BA-activation of PyMBF1 were partially inhibited by the addition of the Ca2+ chelator or the Ca2+ channel blocker, though these activations were completely inhibited by the PLD inhibitor (Fig. 5). Plant PLDs have groups whose activity may or may not require Ca2+ (Qin and Wang 2002). Our findings suggest that the heat response of P. yezoensis is activated by both Ca2+-dependent and Ca2+-independent PLDs. We found at least two genes homologous to plant PLD in the P. yezoensis EST database (EST accession numbers AU193295 and AV430741). These EST clones do not contain a C2 domain, which is a Ca2+-binding motif identified in Ca2+-dependent PLDs near the N terminus. Since they however are not full-length cDNAs, cloning of the full-length PLD genes from P. yezoensis and analyses of their function and regulation are needed for our understanding of the roles of Ca2+ and PLD in the heat stress responses of red marine algae.
In conclusion, we showed that PyMBF1 was increasingly expressed in P. yezoensis cells undergoing oxidative or heat stresses. We also revealed that the heat activation of PyMBF1 requires membrane fluidization and PLD activation. To our knowledge, this is the first report demonstrating that changes in membrane fluidity and PLD activity regulate stress-inducible gene expression in marine macroalgae.
References
Asamizu E, Nagajima M, Kitade Y, Saga N, Nakamura Y, Tabata S (2003) Comparison of RNA expression profiles between the two generations of Porphyra yezoensis (Rhodophyta), based on expressed sequence tag frequency analysis. J Phycol 39:923–930
Brendel C, Gelman L, Auwerx J (2002) Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism. Mol Endocrinol 16:1367–1377
Contreras-Porcia L, Thomas D, Flores V, Correa JA (2011) Tolerance to oxidative stress induced by desiccation in Porphyra columbina (Bangiales, Rhodophyta). J Exp Bot 62:1815–1829
Fukuda S, Mikami K, Uji T, Park EJ, Ohba T, Asada K, Kitade Y, Endo H, Kato I, Saga N (2008) Factors influencing efficiency of transient gene expression in the red macrophyte Porphyra yezoensis. Plant Sci 174:329–339
Gardiner J, Collings DA, Harper JDI, Marc J (2003) The effects of the phospholipase D-antagonist 1-butanol on seedling development and microtubule organisation in Arabidopsis. Plant Cell Physiol 44:687–696
Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo ZF (2012) Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep 39:969–987
Inaba M, Suzuki I, Szalontai B, Kanesaki Y, Los DA, Hayashi H, Murata N (2003) Gene-engineered rigidification of membrane lipids enhances the cold inducibility of gene expression in Synechocystis. J Biol Chem 278:12191–12198
Knight H, Knight MR (2000) Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J Exp Bot 51:1679–1686
Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8:489–503
Li L, Saga N, Mikami K (2009) Ca2+ influx and phosphoinositide signalling are essential for the establishment and maintenance of cell polarity in monospores from the red alga Porphyra yezoensis. J Exp Bot 60:3477–3489
Li XC, Xing YZ, Jiang X, Qiao J, Tan HL, Tian Y, Zhou B (2012) Identification and characterization of the catalase gene PyCAT from the red alga Pyropia yezoensis (Bangiales, Rhodophyta). J Phycol 48:664–669
Liu B, Yao L, Wang WG, Gao JH, Chen F, Wang SH, Xu Y, Tang L, Jia YJ (2010) Molecular cloning and characterization of phospholipase D from Jatropha curcas. Mol Biol Rep 37:939–946
Liu QX, Jindra M, Ueda H, Hiromi Y, Hirose S (2003) Drosophila MBF1 is a co-activator for tracheae defective and contributes to the formation of tracheal and nervous systems. Development 130:719–728
Los DA, Murata N (2004) Membrane fluidity and its roles in the perception of environmental signals. Biochim Biophys Acta 1666:142–157
Lu IF, Sung MS, Lee TM (2006) Salinity stress and hydrogen peroxide regulation of antioxidant defense system in Ulva fasciata. Mar Biol 150:1–15
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158
Mikami K, Uji T, Li L, Takahashi M, Yasui H, Saga N (2009) Visualization of phosphoinositides via the development of the transient expression system of a cyan fluorescent protein in the red alga Porphyra yezoensis. Mar Biotechnol 11:563–569
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33:453–467
Mishkind M, Vermeer JEM, Darwish E, Munnik T (2009) Heat stress activates phospholipase D and triggers PIP2 accumulation at the plasma membrane and nucleus. Plant J 60:10–21
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:86–96
Monroy AF, Dhindsa RS (1995) Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25 °C. Plant Cell 7:321–331
Munnik T, Arisz SA, deVrije T, Musgrave A (1995) G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7:2197–2210
Näär AM, Lemon BD, Tjian R (2001) Transcriptional coactivator complexes. Annu Rev Biochem 70:475–501
Nikaido I, Asamizu E, Nakajima M, Nakamura Y, Saga N, Tabata S (2000) Generation of 10,154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Res 7:223–227
Örvar BL, Sangwan V, Omann F, Dhindsa RS (2000) Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J 23:785–794
Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R (2002) Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J 31:319–330
Qin C, Wang X (2002) The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol 128:1057–1068
Rizhsky L, Liang HJ, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130:1143–1151
Saga N, Kitade Y (2002) Porphyra: a model plant in marine sciences. Fish Sci 68(Suppl):1075–1078
Sangwan V, Foulds I, Singh J, Dhindsa RS (2001) Cold-activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant J 27:1–12
Sangwan V, Orvar BL, Beyerly J, Hirt H, Dhindsa RS (2002) Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J 31:629–638
Suri SS, Dhindsa RS (2008) A heat-activated MAP kinase (HAMK) as a mediator of heat shock response in tobacco cells. Plant Cell Environ 31:218–226
Sutherland JE, Lindstrom SC, Nelson WA, Brodie J, Lynch MDJ, Hwang MS, Choi HG, Miyata M, Kikuchi N, Oliveira MC, Farr T, Neefus C, Mols-Mortensen A, Milstein D, Müller KM (2011) A new look at an ancient order: generic revision of the Bangiales (Rhodophyta). J Phycol 47:1131–1151
Suzuki N, Rizhsky L, Liang HJ, Shuman J, Shulaev V, Mittler R (2005) Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol 139:1313–1322
Takemaru K, Harashima S, Ueda H, Hirose S (1998) Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol Cell Biol 18:4971–4976
Takemaru K, Li FQ, Ueda H, Hirose S (1997) Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc Natl Acad Sci U S A 94:7251–7256
Testerink C, Munnik T (2005) Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 10:368–375
Tsuda K, Yamazaki K (2004) Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim Biophys Acta 1680:1–10
Uji T, Hirata R, Mikami K, Mizuta H, Saga N (2012a) Molecular characterization and expression analysis of sodium pump genes in the marine red alga Porphyra yezoensis. Mol Biol Rep 39:7973–7980
Uji T, Monma R, Mizuta H, Saga N (2012b) Molecular characterization and expression analysis of two Na+/H+ antiporter genes in the marine red alga Porphyra yezoensis. Fish Sci 78:985–991
Uji T, Takahashi M, Saga N, Mikami K (2010) Visualization of nuclear localization of transcription factors with cyan and green fluorescent proteins in the red alga Porphyra yezoensis. Mar Biotechnol 12:150–159
Wan SB, Wang W, Wen PF, Chen JY, Kong WF, Pan QH, Zhan JC, Tian L, Liu HT, Huang WD (2007) Cloning of phospholipase D from grape berry and its expression under heat acclimation. J Biochem Mol Biol 40:595–603
Zonia L, Munnik T (2006) Cracking the green paradigm: functional coding of phosphoinositide signals in plant stress responses. Subcell Biochem 39:207–237
Acknowledgments
This study was supported in part by a grant from the Regional Innovation Cluster Program (Global Type) of the Ministry of Education, Culture, Sports, Science and Technology of Japan to N.S.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Uji, T., Sato, R., Mizuta, H. et al. Changes in membrane fluidity and phospholipase D activity are required for heat activation of PyMBF1 in Pyropia yezoensis (Rhodophyta). J Appl Phycol 25, 1887–1893 (2013). https://doi.org/10.1007/s10811-013-0006-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-013-0006-7