Abstract
Increasing evidence has shown that miRNA-451a (miR-451a) is associated with the development of systemic lupus erythematosus (SLE); however, the mechanism of this association is not fully known. The present study found an increased expression of miR-451a in the spleen and thymus of an SLE mice model. A decrease in miR-451a expression partly relieved the enlargement of the spleen and decreased the proteinuria content and immune complex deposits. The deficiency in miR-451a also decreased numbers of CD4+CD69+ and CD4+/CD8+ T cells and the levels of the serum cytokines IL-17a and IL-4. The IFN regulatory factor (IRF) 8 was highly expressed in the immune organs of wild-type mice but was suppressed in SLE-like mice. A dual-luciferase reporter assay was carried out in combination with gene silencing and overexpression to verify that IRF8 was a target of miR-451a in vitro and in vivo. The data indicate the function and a target of miR-451a in SLE, providing a new target for SLE therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by widespread organ involvement [1]. In SLE patients, many different autoantibodies are produced and deposited in the joints, liver, kidney, and skin, which result in the impairment of multi-organ functions [2]. In particular, a large number of autoantibodies and immune complexes can accumulate in the kidney, causing renal failure [3]. Epidemiologic data have shown that nearly one third of SLE patients suffer from glomerular nephritis [4]. SLE is associated with diverse features of immunologic abnormality, in which B and T lymphocytes play important roles [5, 6]. Some studies have shown that a high ratio of CD4+/CD8+ T cells [7] and an imbalance in T-helper (TH) cell subsets (TH1/TH2/TH17) [8] contribute to the pathogenesis of SLE. Although there is substantial evidence that manifold genetic, hormonal, and environmental factors are involved in the development of SLE [9], the pathogenesis of this disease is not yet fully clear. Further exploration of the etiology of SLE is thus crucial toward developing a valid therapy.
The interferon regulatory factor (IRF) family includes multiple transcription factors involved in both the development and the activation of distinct cells of the immune system by directing the expression and activities of the type I interferon system [10]. IRF [8], a member of the IRF family of transcription factors, plays a central role in the development of specific myeloid cell subsets [11]. IRF8 mutations result in immunodeficiency and dysregulated differentiation [12]. IRF8 is also expressed in lymphoid cells and is involved in the transcriptional regulation of critical elements of the germinal center reaction and B-cell lineage specification [13]. Some reports have shown that genetic variation in the IRF8 gene is relevant to SLE and multiple sclerosis in both mice and humans [14]; however, the molecular basis of such association remains unclear.
MicroRNAs (miRs), as endogenous silencers of various target genes through translational repression or mRNA degradation [15], play essential roles in regulating many biological events, including cell proliferation and differentiation [16]. Alterations in miR expression and function lead to major dysfunction of the immune system and mediate susceptibility to autoimmune diseases [17], such as SLE [18]. The abnormality of miR-451a has been reported to participate in lymphoid organ tumors [19, 20], and the overexpression of miR-451a has been found to modulate the incidence of lymphoma [21]. Furthermore, the ectopic expression of miR-451a is involved in various autoimmune diseases, such as multiple sclerosis and rheumatoid arthritis [22]. Although the relationship between miR-451a and SLE has been partly reported, its molecular mechanism is not fully known.
In the present study, we constructed SLE mouse models and knocked out miR-451a in SLE mice. The deficiency of miR-451a in SLE mice relieved the immune complex deposits in the kidney, decreased the ratio of CD4+CD69+ and CD4+/CD8+ T cells, and suppressed the expression of inflammatory cytokines. Furthermore, miR-451 negatively regulated the expression of IRF8 in vitro and in vivo. Taken together, we concluded that miR-451a promoted the progression of SLE and that IRF8 was a target of miR-451a in SLE. These findings provide new insights into a potential biomarker for SLE and represent a valuable therapeutic strategy for SLE treatment.
MATERIALS AND METHODS
Animal culture
Faslpr/lpr and miR-451a−/− (miR-451a deletion) of C57B/L6 mice were purchased from Cyagen Biosciences (USA). The mice were kept under specific pathogen-free conditions at a temperature of 23 ± 2 °C. Standard rodent feed and water were given ad libitum. Animal care was provided according to the procedures outlined in the “Guide for the Care and Use of Laboratory Animals.” A heterozygous F1 generation was obtained from the hybridization between B6.MRL/lpr and miR-451a−/−. After a cascade of backcrossing and intercrossing, littermates were produced with the genotypes miR-451a−/−Faslpr/lpr and Faslpr/lpr. We chose wild-type (WT, n = 10), Faslpr/lpr (n = 10), and miR-451a−/−Faslpr/lpr (n = 10) female mice for the phenotypic assay. The animals were killed, and their physiologic indexes were recorded.
Coomassie brilliant blue (CBB) detection of urine protein
Mice urine collected from different groups over 24 h was used for the proteinuria assay. Urine (0.1 mL) was added into 5 mL of CBB reagent solution as the test tube, and 0.1 mL normal saline was added into 5 mL of CBB solution as the blank control tube. The mixture was kept at room temperature for 5 min, after which a colorimetric assay was carried out at 595 nm absorbance by using a spectrophotometer (Vita Easyshade). The numerical values were recorded, and the standard curve was constructed for the measurement of the protein concentration. One milliliter of 5% stock protein solution was added into 49 mL normal saline to dilute the standard protein solution and obtain a concentration of 1 mg/mL. The diluted solution (0.1 mL) was drawn from each tube and mixed with 5 mL CBB for the colorimetric assay. After each test, the colorimetric tube was washed with 95% ethyl and then with ddH2O. The concentration of proteinuria was obtained as (ODtest − ODblank)/(ODstandard − ODblank) × 750 mg/L.
Flow cytometry
Cells were incubated for 30 min at 4 °C with fluorochrome-labeled monoclonal antibodies (mAbs): FITC-conjugated anti-mouse CD4, FITC-conjugated anti-mouse CD8, and FITC-conjugated anti-mouse CD69 (Santa Cruz, California). The control was incubated with isotype-matched FITC-conjugated mouse antibody (Santa Cruz, California). Erythrocyte lysis and cell fixation were carried out by using OptiLyse C lysing solution (cat. no. 1401; Beckman). The treated blood samples were passed through a Coulter Epics XL flow cytometer (Beckman) and a Coulter FC 500 analyzer (Beckman Coulter) for various subset analyses. The relevant data were obtained and analyzed by using the FlowJo software version 7.6 (Tree Star, Inc., Ashland, OR, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Spleen and thymus tissues were isolated for total RNA extraction by using Trizol buffer (Sigma, USA). The quality of RNA was determined by agarose electrophoresis, and the concentration was measured by using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA) at absorbances of 260 and 280 nm. One microgram of RNA was reverse transcribed into complementary DNA (cDNA) by using the One-Step cDNA synthesis kit (TaKaRa, Dalian, China) according to the manufacturer’s instruction. The relative expressions of the interest genes were analyzed with the One-Step real-time PCR system (Invitrogen, USA). The real-time PCR reaction mixture (10 μL) consisted of 1 μL of cDNA product, 5 μL of 2× SYBR green mix (TaKaRa, Dalian, China), 0.5 μL of primers, and 3 μL of ddH2O. The reactions were carried out at 95 °C for 10 min, followed by 40 cycles at 95 °C for 5 s and then at 60 °C for 10 s. The 2−ΔΔCT method was used to determine the relative individual gene quantities. For the qRT-PCR, we used the miR-451a forward primer 5′-AAACCGTTACCATTACTGAGTT-3′ and the reverse primer provided by the miScript SYBR Green PCR kit, the U6 forward primer 5′-CTCGCTTCGGCAGCACA-3′ and the reverse primer 5′-ACGCTTCACGAATTTGCGT-3′, the IRF8 forward primer 5′-CCAGCAGTTCTTCCGA-3′ and the reverse primer 5′-CCTCTTCTGCCAGTTGCC-3′, and the GAPDH forward primer 5′-GCCAAAAGGGTCATCATCTC-3′ and the reverse primer 5′-GTAGAGGCAGGGATGATGTTC-3′; GAPDH was used as an internal standard for IRF8 detection.
Western blot
Proteins were extracted from spleen and thymus tissues by using RIPA buffer (Beyotime, Shanghai, China). The protein concentration was determined with the use of a bicinchoninic acid kit (Pierce, Germany) according to the protocol. An equal amount of protein was subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis. When the indicator bromophenol blue reached the end, the proteins were electro-transferred to a polyvinylidene fluoride membrane (Millipore, Boston, MA, USA). The membrane was blocked by using 5% nonfat milk in Tris-buffered saline with 0.05% Tween 20 (TBST) for 1 h at room temperature and then was incubated with rabbit monoclonal anti-IRF8 antibody overnight at 4 °C. After being washed thrice with TBST, the membrane was incubated with the corresponding horseradish peroxidase-conjugated secondary antibody (Bio-Rad, Hercules, CA, USA) for 1 h at room temperature. The membrane was developed with an electrochemiluminescence solution (Pierce, Germany). The IRF8 protein bands were quantitatively analyzed by using the Quantity One software, normalized to GAPDH.
Enzyme-linked immunosorbent assay (ELISA)
Before the experiments were carried out, the interest antibody was diluted to 10 μg/mL with the use of carbonate buffer. An equal volume (0.1 mL/well) of antibody dilution was added into a polystyrene plate (Corning Co., USA), which was kept at 4 °C overnight. The next morning, the polystyrene plate was incubated at 37 °C for 1 h after 0.1 mL/well diluted sample was added. The plate was incubated with fresh enzyme-labeled antibody for 1 h at 37 °C. After being washed, the plate was incubated with tetramethylbenzidine at 37 °C for 20 min to develop color changes. The OD at 450 nm was measured with a NanoDrop ND-2000 spectrophotometer, with the blank control having an OD of zero. If the ODtest > 2 × ODnegative test, the OD was defined as a positive value.
Immunofluorescence assay
The kidney was fixed in 4% fresh ice-cold paraformaldehyde buffer for 15 min, embedded in paraffin wax, and then cut into 6-μm-thick sections. For the morphology study, hematoxylin and eosin staining was done, and the signal was observed under a light microscope (Olympus, Japan). For the immunohistochemistry, the sections were cultured with 3% hydrogen peroxide in methanol for 15 min to block the activity of endoperoxidase. After being permeabilized with 0.5% Triton X-100 for 15 min, the slides were blocked with 5% bovine serum albumin (Sigma, USA) for 30 min and then probed with rabbit monoclonal primary antibody at 4 °C overnight. The next morning, the slides were incubated with biotin-labeled secondary antibody and stained with streptavidin-biotin complex–fluorescein isothiocyanate (SABC-FITC; Boster Co., Wuhan, China) and streptavidin-biotin complex–Cy3 (SABC-Cy3; Boster Co., Wuhan, China). For each sample, five non-overlapped fields in the slides were randomly captured by using a laser confocal microscope (Olympus, Japan).
Statistical analysis
All experiments were done at least thrice by using independent samples. The measurement data were expressed as means ± standard deviation. Statistical analysis was carried out by using the SPSS 20.0 software (SPSS, Chicago, IL, USA). The homogeneity of variance was subjected to least significant difference (LSD) multiple comparisons among groups. Statistical significance was set at P < 0.05.
RESULTS
MiR-451a was highly expressed in the spleen and thymus of the Faslpr/lpr mice
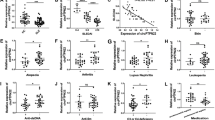
To explore the relationship between miR-451a and SLE, the levels of miR-451a in the spleen and thymus tissues were compared between the WT and Faslpr/lpr mice through qRT-PCR analysis. As shown in Fig. 1a, b, the miR-451a levels were significantly higher in the spleen and thymus tissues from the Faslpr/lpr mice than in those from the WT mice, suggesting that miR-451a might be related to SLE. Thus, miR-451a knockout SLE mice (miR-451a−/−Faslpr/lpr) were constructed to verify this hypothesis. To examine the efficiency of gene knockout, we analyzed the expressions of miR-451a and miR-144, the coding sequences of which are very close to that of miR-451a. As shown in Fig. 3c, the expression of miR-451a was suppressed in the miR-451a−/−Faslpr/lpr mice compared with the Faslpr/lpr mice; however, the knockout of miR-451a had no significant effect on the miR-144 expression. These results indicated that miR-451a was successfully knocked out without off-target effects.
MiR-451a was highly expressed in the spleen and thymus in the Faslpr/lpr mice. The relative expression of miR-451a in spleen (a) and thymus (b) was analyzed by qRT-PCR analysis. c miR-451a was significantly suppressed in the miR-451a−/−Faslpr/lpr mice. Total RNA of spleen was isolated for qRT-PCR. The expression of miR-144 was analyzed for off-target examination. U6 was used as the internal control. d The knockout of miR-451a suppressed the enlargement of spleen. e The ratio of spleen weight/body weight in the miR-451a−/−Faslpr/lpr mice was lesser than that in the Faslpr/lpr mice. f CBB detection was used to examine the content of proteinuria of 24 h at the age of 30 weeks and 40–50 weeks. **P < 0.01.
The effect of miR-451a on the spleen weight, spleen weight/body weight ratio, and 24-h proteinuria was then analyzed. Compared with the WT mice, the Faslpr/lpr mice showed a progressive enlargement of the spleen; however, the spleen enlargement in the miR-451a−/−Faslpr/lpr mice was effectively relieved compared with that in the Faslpr/lpr mice (Fig. 1d). A similar change trend in the spleen weight/body weight ratio was found in the WT, Faslpr/lpr, and miR-451a−/−Faslpr/lpr mice (Fig. 1e). Further, the proteinuria content over 24 h in Faslpr/lpr and miR-451a−/−Faslpr/lpr mice was analyzed. As shown in Fig. 1f, there was no significant difference between Faslpr/lpr and miR-451a−/−Faslpr/lpr mice at the age of 30 weeks (P > 0.05), whereas at the age of 40–50 weeks, the proteinuria content in Faslpr/lpr mice was obviously higher than that in miR-451a−/−Faslpr/lpr mice (P < 0.01). These results indicated that miR-451a was highly expressed in SLE mice and affected the spleen weight and 24-h proteinuria content.
miR-451a deficiency suppressed the levels of autoantibodies
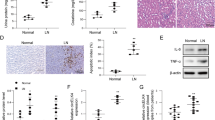
To examine the effect of the knockout of miR-451a on the kidney, histopathology by HE staining was carried out (Fig. 2a). The Faslpr/lpr mice showed increased mononuclear cell infiltration, enlarged glomeruli, and tissue structure disorder. However, the deletion of miR-451a significantly relieved the glomerular enlargement and decreased the infiltration of mononuclear cells.
Deficiency of miR-451a suppressed the immune complex deposit and serum autoantibodies levels. a The knockout of miR-451a inhibited the immune complex deposit in the kidney. The kidney tissues from the Faslpr/lpr and miR-451a−/−Faslpr/lpr mice were extracted for HE staining and immunofluorescence assay. Scale bar (yellow) = 200 μm; scale bar (red) = 100 μm. The signals of IgM, C1q, IgA, and IgG were stained in green and quantitatively analyzed in (b). Serum IgM (c), IgG (d), and anti-dsDNA antibody (e) levels were suppressed in the miR-451a−/−Faslpr/lpr mice analyzed by ELISA. **P < 0.01.
SLE is a chronic systemic autoimmune disease characterized by the generation of high-affinity pathogenic autoantibodies. To investigate the immune complex deposit in the kidney, the fluorescence intensities were compared between the Faslpr/lpr and miR-451a−/−Faslpr/lpr mice. As shown in Fig. 2a, b, the signals of IgM, C1q, IgA, and IgG in the miR-451a−/−Faslpr/lpr mice were significantly suppressed compared with those in the Faslpr/lpr mice (P < 0.01), suggesting that the knockout of miR-451a mitigated the immune complex deposit in the kidney in the SLE mice. The levels of serum autoantibodies, as analyzed by ELISA, showed similar levels of IgM, IgG, and anti-double-stranded DNA (dsDNA) antibody, as shown in Fig. 2c–e. These results indicated that miR-451a modulated the levels of autoantibodies in organs and serum induced by SLE.
miR-451a knockout decreased T-follicular helper (Tfh) cells and Tfh-related cytokines
T-follicular helper (Tfh) cells, a kind of specialized CD4+ T cell subset, have emerged as central players in the production of autoantibodies associated with SLE. This study analyzed the number of Tfh cells and the level of the Tfh-related cytokine IL-21. The number of Tfh cells in the Faslpr/lpr mice lymph node was higher than that in the WT mice (P < 0.01); however, the number of Tfh cells was lower in the miR-451a−/−Faslpr/lpr mice than in the Faslpr/lpr mice (P < 0.05; Fig. 3a). A similar trend was observed in the serum IL-21 levels of the WT, Faslpr/lpr, and miR-451a−/−Faslpr/lpr groups, as shown in Fig. 3b. Meanwhile, the mRNA levels of Bcl-6 and IL-21 in the spleen were further analyzed by qRT-PCR (Fig. 3c, d). The levels of Bcl-6 and IL-21 were higher in the Faslpr/lpr mice than in the WT mice; however, compared with the Faslpr/lpr mice, the knockout of the miR-451a mice decreased the levels of Bcl-6 and IL-21 (P < 0.05). The results of PNA staining in the spleen showed that the number of germinal centers was large in the Faslpr/lpr mice but was decreased in miR-451a−/−Faslpr/lpr mice (Fig. 3e). These data indicated that the deficiency of miR-451a suppressed the numbers of Tfh cells and germinal centers.
Knockout of miR-451a reduced Tfh cells and Tfh-related cytokine. a Deficiency of miR-451a reduced the number of Tfh cells in lymph node detected by flow cytometry. b Serum IL-21 cytokine level was decreased after the deletion of miR-451a in SLE mice analyzed by ELISA. The mRNA levels of Bcl-6 (c) and IL-21 (d) in miR-451a−/−Faslpr/lpr spleen were suppressed compared with Faslpr/lpr. GAPDH was used as the internal control. e The deficiency of miR-451a inhibited the number of germinal center in spleen examined by PNA stain. Germinal center was stained in red. Scale bar (blue) = 100 μm. *P < 0.05, **P < 0.01.
miR-451a knockout decreased the ratio of CD4+CD69+ and CD4+/CD8+ in the spleen and lymph node
The subsets of cells in the spleen and lymph node were studied by using flow cytometry. As shown in Fig. 4, the ratio of CD4+CD69+ T cells in the spleen and lymph node from the Faslpr/lpr group was higher compared with the WT mice (P < 0.05). However, the silencing of miR-451a suppressed an increase of the ratio in the Faslpr/lpr mice (Fig. 4a, b). Similar results on the ratio of CD4+/CD8+ were found in the WT, Faslpr/lpr, and miR-451a−/−Faslpr/lpr mice (Fig. 4c, d). These data indicated that miR-451a affected the ratio of inflammatory T subsets in SLE.
Deficiency of miR-451a reduced the ratio of CD4+CD69+ and CD4+/CD8+ T cells in the spleen and lymph node. The ratio of CD4+CD69+ T cells in the spleen (a) and lymph node (b) from the miR-451a−/−Faslpr/lpr mice was inhibited compared to that in the Faslpr/lpr mice. The ratio of CD4+/CD8+ in the spleen (c) and lymph node (d) from the miR-451a−/−Faslpr/lpr mice was lesser than that from the Faslpr/lpr mice. The levels of serum IL-17a (e), IL-4 (f), and IFN-γ (g) were detected by ELISA. **P < 0.01.
The levels of serum inflammatory cytokines were also further investigated. Compared with the WT mice, the Faslpr/lpr group showed increased levels of IL-17a and IL-4; however, the knockout of miR-451a suppressed the increase (P < 0.05; Fig. 4e, f). The level of INF-γ was lower in the Faslpr/lpr than in the WT mice, whereas no obvious difference was found between the Faslpr/lpr and miR-451a−/−Faslpr/lpr mice (Fig. 4g). These data indicated that miR-451a modulated T subsets and the levels of inflammatory cytokines in the SLE mice.
IRF8 was suppressed in the Faslpr/lpr mice
Some studies have reported that IRF8 was related to SLE. To confirm the function of IRF8 in the SLE mice, the mRNA levels of IRF8 were first detected in various tissues of WT or SLE-like mice. In the WT mice, IRF8 was highly expressed in the spleen, blood, and lymph node; in the Faslpr/lpr mice, the expression of IRF8 in various tissues was suppressed (Fig. 5a). Compared with the WT mice, the Faslpr/lpr group showed markedly inhibited mRNA and protein levels of IRF8 (P < 0.01; Fig. 5b–d). However, the deficiency of miR-451a relieved the decrease in IRF expression. These results indicated that IRF8 was involved in the progression of SLE and that some relationship between miR-451a and IRF8 might exist.
IRF8 was associated with the progression of SLE. a The mRNA level of IRF8 in various tissues from the WT and Faslpr/lpr mice was detected by qRT-PCR. Total RNA was extracted from various tissues from the WT and Faslpr/lpr mice. GAPDH was used as the internal control. The mRNA (b) and protein (c) levels of IRF8 in the WT, Faslpr/lpr, and miR-451a−/−Faslpr/lpr spleen were analyzed by qRT-PCR and western blot. d IRF8 protein bands were quantitatively analyzed by Quantity One software normalized to GAPDH. **P < 0.01.
miR-451a targeted IRF8 in vitro and in vivo
The miR-451a target gene predicted by miRecords and the Venny 2.0 software indicated that IRF8 might be a miR-451a target gene in humans and mice. To characterize the interaction between miR-451a and IRF8, a dual-luciferase reporter vector was constructed in HEK293 cells. As shown in Fig. 6a, compared with the negative control miRNA (WT + Neg), the miR-451a mimic significantly suppressed the activity of the reporter luciferase that was fused with the WT IRF8 3′-untranslated region (UTR). Compared with the co-transfection of the WT IRF8 3′-UTR and miR-451a mimic (WT + mimic), the co-transfection of the mutated IRF8 3′-UTR and miR-451a mimic (MT + mimic) increased the activity of the reporter luciferase (P < 0.01). There was no difference between WT + Neg and MT + mimic (P > 0.05). In addition, compared with WT + Neg, the co-transfection of the WT IRF8 3′-UTR and miR-451a inhibitor (WT + inhibitor) obviously increased the activity (P < 0.01). These results indicated a direct interaction between miR-451a and IRF8. The miR-451a mimic or inhibitor was further used to identify the interaction. As shown in Fig. 6b, the miR-451a inhibitor upregulated the protein level of IRF8, whereas the miR-451a mimic suppressed the expression of IRF8. These data indicated that miR-451a negatively modulated the expression of IRF8 in vitro and in vivo.
MiR-451a targeted IRF8 in vitro and in vivo. a miR-451a interacted with IRF8 3′-UTR in HEK293 cells. HEK293 cells were transfected with WT IRF8 3′-UTR+miR-451a negative control (WT + Neg), WT IRF8 3′-UTR+miR-451a mimic (WT + mimic), mutated IRF8 3′-UTR+miR-451a mimic (MT + mimic), and WT IRF8 3′-UTR+miR-451a inhibitor (WT + inhibitor), respectively. A dual-luciferase reporter assay was performed. b miR-451 mimic and inhibitor regulated the protein level of IRF8 in HEK293 cells. GAPDH was used as the internal control. **P < 0.01.
DISCUSSION
In the present study, the level of miR-451a was found to be increased in SLE mice. The deficiency of miR-451a in SLE mice relieved the enlargement of the spleen, reduced the proteinuria content and immune complex deposits, and decreased the ratios of CD4+CD69+ and CD4+/CD8+ T cells and the expression of inflammatory cytokines. The expression of IRF8 was highly enhanced in immune organs of WT mice but was suppressed in SLE mice. A dual-luciferase reporter assay was done to verify whether IRF8 was a target of miR-451a. Taken together, the data indicated the function and a direct target of miR-451a in SLE, providing a new target for SLE therapy.
The deficiency of miR-451a mitigated immune complex deposits in the kidney
SLE is a chronic autoimmune disease characterized by immunologic abnormalities and multi-organ injury involving the kidneys and central nervous system [2]. In our study, miR-451a was found to be highly expressed in immune organs, including the spleen and thymus, suggesting that miR-451a might be associated with immune diseases. Proteinuria is not only one of the most common clinical manifestations of renal injury but is also an important factor related to the continuous progression of renal diseases [23]. The 24-h proteinuria contents in Faslpr/lpr and miR-451a−/−Faslpr/lpr mice were compared in the current work. The knockout of miR-451a was found to decrease the content of proteinuria, relieve the glomerular enlargement, and reduce the infiltration of mononuclear cells compared with the SLE mice. These results suggested that the knockout of miR-451a partly mitigated the kidney damage in SLE. Abnormalities in B and T cells contribute to loss of self-tolerance and the production of autoantibodies, which are hallmarks of SLE [2]. In our study, the signals of IgM, C1q, IgA, and IgG in the kidney and the titers of serum IgM, IgG, and SLE-specific autoantibody (anti-dsDNA antibody) were stronger in SLE than in WT mice; this finding was in agreement with previous results. When miR-451a was suppressed, the improvement of the above-mentioned autoantibodies in SLE mice was inhibited, suggesting that miR-451a enhanced the immune complex deposit in SLE. In SLE, anti-dsDNA antibodies bind to nucleosomes trapped in the glomerular basement membrane, with subsequent renal inflammation [24]. Combined with the above results, we concluded that miR-451a enhanced the immune complex deposit in the kidney, causing kidney damage and proteinuria.
miR-451a knockout modulated inflammatory cell subsets and related cytokines
Tfh cells are characterized by increasing expressions of the important surface markers CXCR5, CD40 ligand (CD40L), and ICOS [25]. In this work, the number of Tfh cells in mice was determined by using the surface marker CXCR5+CD40L+ICOS+. The number of Tfh cells was found to be increased in SLE mice, which provided new evidence in support of the previous result that Tfh cells were associated with SLE. Tfh cells are the main source of IL-21 cytokine, and the emergence of IL-21 has been implicated in SLE [26]. Bcl-6 is a transcriptional repressor expressed in Tfh cells and is involved in the development and differentiation of Tfh cells [27]. In SLE, pathologic germinal centers develop due to the increased numbers of Tfh cells [27]. In the present study, the upregulation of Tfh cell surface markers, Bcl-6, IL-21, and germinal centers in SLE mice was suppressed after the silencing of miR-451, suggesting that miR-451a was involved in SLE by regulating the numbers of inflammatory cell subsets and related cytokines.
Many studies have indicated that defective regulation of the Th1, Th2, and Th17 cell function is implicated in the pathophysiology of SLE [28]. The balance of cytokines plays an important role in the etiology and pathogenesis of SLE [29]. Inflammatory cytokines, such as Th1-derived IFN-γ, Th2-derived IL-4, cytokines IL-10 and IL-13, and Th17-derived IL-17a [30], have been identified as important players in SLE [31]. In SLE patients, the Th1 function has been found to be suppressed by the downregulation of IFN-γ, whereas the Th2 function was enhanced [31]. Similarly to this previous study, the current work found that the level of IFN-γ was decreased in Faslpr/lpr SLE mice. The plasma levels of IL-17a [32] and IL-4 [33] have been found to be increased in SLE patients compared with normal controls. Our results are similar to these previous results; the upregulation of IL-17a and IL-4 was suppressed in miR-451a−/−Faslpr/lpr mice, suggesting that miR-451a affected the abnormality of Th2 and Th17. Although IFN-γ is strongly associated with Th1 response, no significant difference in the level of IFN-γ between Faslpr/lpr and miR-451a−/−Faslpr/lpr was observed in our study. Thus, further evidence is needed to verify the effect of miR-451a on Th1.
IRF8 is a target of miR-451a in SLE
In the past decade, another important step toward understanding the pathogenesis of SLE was achieved, that is, the discovery of the role of the type I interferon (IFN) pathway [34]. The transcription of IFN is monitored by members of the IRF family, which in humans consists of nine distinct proteins [35]. IRF genes, including IRF5, IRF7, and IRF8, are important in the SLE pathogenesis [36]. IRF8 contributes to innate and adaptive immune responses, and common variants in the IRF8 gene are associated with susceptibility to SLE [37]. The association between IRF8 and SLE was corroborated in European populations [38] and in the Han Chinese population [39]. In our experiments, IRF8 was highly expressed in immune organs but was inhibited in SLE mice. IRF8 functions as a strong repressor of BAFF expression in B cells [40], and increased BAFF levels have been suggested to play a role in SLE [41]. miR-451a is located on chromosome 17q11.2 and exerts a tumor suppressive role by targeting MIF, c-MYC, and AKT1 [42]. However, the target of miR-451a in SLE development is unclear. In our study, miR-451a directly negatively regulated the expression of IRF8 in SLE, thus providing a new miR-451a target for SLE therapy. These data provide new evidence that IRF8 is associated with SLE. However, more experiments are needed to determine the molecular mechanism by which such association occurs.
CONCLUSION
The knockout of miR-451a affected the enlargement of the spleen, reduced the urine protein content and immune complex deposits, and decreased the ratio of CD4+CD69+ and CD4+/CD8+ T cells. IRF8 was highly expressed in immune organs but was suppressed in SLE mice. A dual-luciferase reporter assay showed that IRF8 was a target of miR-451a. The data indicated the function of miR-451a in SLE and showed a new direct target of miR-451a, thus providing a new target in SLE therapy.
Abbreviations
- CBB:
-
Coomassie brilliant blue
- ELISA:
-
Enzyme-linked immunosorbent assay
- IRF8:
-
IFN regulatory factor 8
- miR-451a:
-
miRNA-451a
- SLE:
-
Systemic lupus erythematosus
- Tfh:
-
T-follicular helper
References
Wahren-Herlenius, M., and T. Dorner. 2013. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet 382(9894): 819–831.
Bengtsson, A.A. and Ronnblom, L. 2016. Systemic lupus erythematosus: still a challenge for physicians. J Intern Med
Mok, C.C., R.C. Kwok, and P.S. Yip. 2013. Effect of renal disease on the standardized mortality ratio and life expectancy of patients with systemic lupus erythematosus. Arthritis Rheum 65(8): 2154–2160.
Anaya, J.M., C. Canas, R.D. Mantilla, R. Pineda-Tamayo, G.J. Tobon, C. Herrera-Diaz, D.M. Rendon, A. Rojas-Villarraga, and M. Uribe. 2011. Lupus nephritis in Colombians: contrasts and comparisons with other populations. Clin Rev Allergy Immunol 40(3): 199–207.
Taher, T.E., H.A. Muhammad, E. Bariller, F. Flores-Borja, Y. Renaudineau, D.A. Isenberg, and R.A. Mageed. 2013. B-lymphocyte signalling abnormalities and lupus immunopathology. Int Rev Immunol 32(4): 428–444.
Moulton, V.R., and G.C. Tsokos. 2015. T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. J Clin Invest 125(6): 2220–2227.
Asano, S., K. Mori, K. Yamazaki, T. Sata, A. Kurata, Y. Sato, H. Odajima, Y. Akaike, H. Wakasa, and M. Kojima. 2014. Necrotizing lymphadenitis (NEL) is a systemic disease characterized by blastic transformation of CD8+ cells and apoptosis of CD4+ cells. Virchows Arch 464(1): 95–103.
Talaat, R.M., S.F. Mohamed, I.H. Bassyouni, and A.A. Raouf. 2015. Th1/Th2/Th17/Treg cytokine imbalance in systemic lupus erythematosus (SLE) patients: correlation with disease activity. Cytokine 72(2): 146–153.
Schoenfeld, S.R., S. Kasturi, and K.H. Costenbader. 2013. The epidemiology of atherosclerotic cardiovascular disease among patients with SLE: a systematic review. Semin Arthritis Rheum 43(1): 77–95.
Mancino, A., and G. Natoli. 2016. Specificity and Function of IRF Family Transcription Factors: Insights from Genomics. J Interferon Cytokine Res 36(7): 462–469.
Yanez, A., and H.S. Goodridge. 2016. Interferon regulatory factor 8 and the regulation of neutrophil, monocyte, and dendritic cell production. Curr Opin Hematol 23(1): 11–17.
Abrams, S.I., C.S. Netherby, D.Y. Twum, and M.N. Messmer. 2016. Relevance of Interferon Regulatory Factor-8 Expression in Myeloid-Tumor Interactions. J Interferon Cytokine Res 36(7): 442–453.
Wang, H., and H.C. Morse 3rd. 2009. IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol Res 43(1-3): 109–117.
Salloum, R., and T.B. Niewold. 2011. Interferon regulatory factors in human lupus pathogenesis. Transl Res 157(6): 326–331.
Ambros, V. 2004. The functions of animal microRNAs. Nature 431(7006): 350–355.
Kloosterman, W.P., and R.H. Plasterk. 2006. The diverse functions of microRNAs in animal development and disease. Dev Cell 11(4): 441–450.
Singh, R.P., I. Massachi, S. Manickavel, S. Singh, N.P. Rao, S. Hasan, D.K. Mc Curdy, S. Sharma, D. Wong, B.H. Hahn, and H. Rehimi. 2013. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev 12(12): 1160–1165.
Qu, B., and N. Shen. 2015. miRNAs in the Pathogenesis of Systemic Lupus Erythematosus. Int J Mol Sci 16(5): 9557–9572.
Su, Z., L. Ni, W. Yu, Z. Yu, D. Chen, E. Zhang, Y. Li, Y. Wang, X. Li, S. Yang, Y. Gui, Y. Lai, and J. Ye. 2015. MicroRNA-451a is associated with cell proliferation, migration and apoptosis in renal cell carcinoma. Mol Med Rep 11(3): 2248–2254.
Chen, Q., H. Hu, D. Jiao, J. Yan, W. Xu, X. Tang, J. Chen, and J. Wang. 2016. miR-126-3p and miR-451a correlate with clinicopathological features of lung adenocarcinoma: The underlying molecular mechanisms. Oncol Rep 36(2): 909–917.
Gorur, A., S. Balci Fidanci, N. Dogruer Unal, L. Ayaz, S. Akbayir, H. Yildirim Yaroglu, M. Dirlik, M.S. Serin, and L. Tamer. 2013. Determination of plasma microRNA for early detection of gastric cancer. Mol Biol Rep 40(3): 2091–2096.
Baulina, N.M., O.G. Kulakova, and O.O. Favorova. 2016. MicroRNAs: The Role in Autoimmune Inflammation. Acta Naturae 8(1): 21–33.
Xu, L.B., N. Chi, and W. Shi. 2015. Amiloride, a urokinase-type plasminogen activator receptor (uTPA) inhibitor, reduces proteinurea in podocytes. Gen Mol Res 14(3): 9518–9529.
Olin, A.I., M. Morgelin, L. Truedsson, G. Sturfelt, and A.A. Bengtsson. 2014. Pathogenic mechanisms in lupus nephritis: Nucleosomes bind aberrant laminin beta1 with high affinity and colocalize in the electron-dense deposits. Arthritis Rheumatol 66(2): 397–406.
Deenick, E.K., A. Chan, C.S. Ma, D. Gatto, P.L. Schwartzberg, R. Brink, and S.G. Tangye. 2010. Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity 33(2): 241–253.
Bubier, J.A., T.J. Sproule, O. Foreman, R. Spolski, D.J. Shaffer, H.C. Morse 3rd, W.J. Leonard, and D.C. Roopenian. 2009. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. Proc Natl Acad Sci U S A 106(5): 1518–1523.
Simpson, N., P.A. Gatenby, A. Wilson, S. Malik, D.A. Fulcher, S.G. Tangye, H. Manku, T.J. Vyse, G. Roncador, G.A. Huttley, C.C. Goodnow, C.G. Vinuesa, and M.C. Cook. 2010. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum 62(1): 234–244.
Mak, A., and N.Y. Kow. 2014. The pathology of T cells in systemic lupus erythematosus. J Immunol Res 2014: 419029.
Martin, J.C., D.L. Baeten, and R. Josien. 2014. Emerging role of IL-17 and Th17 cells in systemic lupus erythematosus. Clin Immunol 154(1): 1–12.
Panda, A.K. and Das, B.K. 2016. Diminished IL-17A levels may protect filarial-infected individuals from development of rheumatoid arthritis and systemic lupus erythematosus. Lupus
Ohl, K., and K. Tenbrock. 2011. Inflammatory cytokines in systemic lupus erythematosus. J Biomed Biotechnol 2011: 432595.
Li, D., B. Guo, H. Wu, L. Tan, C. Chang, and Q. Lu. 2015. Interleukin-17 in systemic lupus erythematosus: a comprehensive review. Autoimmunity 48(6): 353–361.
Wong, C.K., C.Y. Ho, E.K. Li, and C.W. Lam. 2000. Elevation of proinflammatory cytokine (IL-18, IL-17, IL-12) and Th2 cytokine (IL-4) concentrations in patients with systemic lupus erythematosus. Lupus 9(8): 589–593.
Luo, S., Y. Wang, M. Zhao, and Q. Lu. 2016. The important roles of type I interferon and interferon-inducible genes in systemic lupus erythematosus. Int Immunopharmacol 40: 542–549.
Lazzari, E., and C.A. Jefferies. 2014. IRF5-mediated signaling and implications for SLE. Clin Immunol 153(2): 343–352.
Graham, R.R., S.V. Kozyrev, E.C. Baechler, M.V. Reddy, R.M. Plenge, J.W. Bauer, W.A. Ortmann, T. Koeuth, M.F. Gonzalez Escribano, B. Pons-Estel, M. Petri, M. Daly, P.K. Gregersen, J. Martin, D. Altshuler, T.W. Behrens, and M.E. Alarcon-Riquelme. 2006. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet 38(5): 550–555.
Lessard, C.J., I. Adrianto, J.A. Ice, G.B. Wiley, J.A. Kelly, S.B. Glenn, A.J. Adler, H. Li, A. Rasmussen, A.H. Williams, J. Ziegler, M.E. Comeau, M. Marion, B.E. Wakeland, C. Liang, P.S. Ramos, K.M. Grundahl, C.J. Gallant, M.E. Alarcon-Riquelme, G.S. Alarcon, J.M. Anaya, S.C. Bae, S.A. Boackle, E.E. Brown, D.M. Chang, S.K. Cho, L.A. Criswell, J.C. Edberg, B.I. Freedman, G.S. Gilkeson, C.O. Jacob, J.A. James, D.L. Kamen, R.P. Kimberly, J.H. Kim, J. Martin, J.T. Merrill, T.B. Niewold, S.Y. Park, M.A. Petri, B.A. Pons-Estel, R. Ramsey-Goldman, J.D. Reveille, R.H. Scofield, Y.W. Song, A.M. Stevens, B.P. Tsao, L.M. Vila, T.J. Vyse, C.Y. Yu, J.M. Guthridge, K.M. Kaufman, J.B. Harley, E.K. Wakeland, C.D. Langefeld, P.M. Gaffney, C.G. Montgomery, and K.L. Moser. 2012. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am J Hum Genet 90(4): 648–660.
Ruiz-Larranaga, O., P. Migliorini, M. Uribarri, L. Czirjak, M.C. Alcaro, J. Del Amo, M. Iriondo, C. Manzano, S. Escorza-Trevino, and A. Estonba. 2016. Genetic association study of systemic lupus erythematosus and disease subphenotypes in European populations. Clin Rheumatol 35(5): 1161–1168.
Li, S.W., Y. He, Z.H. Zheng, D.W. Liu, and Z.S. Liu. 2014. Single-nucleotide polymorphisms of IRF8 gene are associated with systemic lupus erythematosus in Chinese Han population. Int J Immunogenet 41(2): 112–118.
Shin, D.M., C.H. Lee, and H.C. Morse 3rd. 2011. IRF8 governs expression of genes involved in innate and adaptive immunity in human and mouse germinal center B cells. PLoS ONE 6(11), e27384.
Sjostrand, M., A. Johansson, L. Aqrawi, T. Olsson, M. Wahren-Herlenius, and A. Espinosa. 2016. The Expression of BAFF Is Controlled by IRF Transcription Factors. J Immunol 196(1): 91–96.
Pan, X., R. Wang, and Z.X. Wang. 2013. The potential role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol Cancer Ther 12(7): 1153–1162.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Cheng, J., Wu, R., Long, L. et al. miRNA-451a Targets IFN Regulatory Factor 8 for the Progression of Systemic Lupus Erythematosus. Inflammation 40, 676–687 (2017). https://doi.org/10.1007/s10753-017-0514-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-017-0514-8