Abstract
Feeding upon the scales of other fish—lepidophagy—is a highly specialized foraging strategy in fish. Scale-eating is rare in teleosts, yet has evolved several times in East African cichlids, the most famous case being the Perissodini clade in Lake Tanganyika. Here, we examined the prey spectrum of the scale-eater Perissodus microlepis (Boulenger, 1898) via morphological assessment and targeted sequencing (barcoding) of ingested scales. We found that the size of the ingested scales, but not their number, correlates with the body size of scale-eaters. Sequencing of a segment of the mitochondrial ND2 gene in more than 300 scales revealed that P. microlepis feed upon a broad spectrum of prey species. In total, we detected 39 different prey species, reflecting the cichlid community in the rocky littoral zone of Lake Tanganyika. The most common prey were the algae-eaters Petrochromis polyodon, Pe. ephippium, Eretmodus cyanostictus, Tropheus moorii, and Simochromis diagramma, which make up more than half of the diet. The diversity of scales found within scale-eaters and the overall broad prey spectrum suggest that P. microlepis is an opportunistic feeder. Mouth-handedness and body color hue of the scale-eaters do not seem to have an influence on prey choice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lake Tanganyika, the oldest of the African Great Lakes, is home to a particularly diverse freshwater fauna with a great degree of endemism, including the morphologically, ecologically and genetically most diverse assemblage of cichlid fishes in Africa (Fryer & Iles, 1972; Salzburger et al., 2002, 2014). Within a period of about 10 million years, more than 200 cichlid species have evolved in Lake Tanganyika, occupying a broad range of ecological niches (Koblmüller et al., 2008; Muschick et al., 2012; Meyer et al., 2017). Taxonomically, the Tanganyikan cichlid species have been grouped into 12–16 tribes, i.e., the taxonomic rank between the subfamily and genus level (Poll, 1986; Takahashi, 2003). The Tanganyikan cichlid tribes differ with respect to species numbers (one species in, e.g., Boulengerochromini to roughly 100 species in Lamprologini), breeding mode (mouthbrooding vs. substrate spawning), as well as the range of foraging strategies (see e.g., Koblmüller et al., 2008). One of the most peculiar feeding modes in cichlids from Lake Tanganyika is the scale-eating behavior of several members of the Perissodini tribe (Marlier & Leloup, 1954; Takahashi et al., 2007).
The strategy to forage on scales of other fishes (‘lepidophagy’) is known from a few species in a few fish families only (Sazima, 1983; Martin & Wainwright, 2013; Kolman et al., 2018). In Lake Tanganyika, however, there are six Perissodini species that more or less exclusively feed upon scales of other fish (Takahashi et al., 2007, 2016). To this end, scale-eaters ambush their prey from the rear, suddenly attack and bite out a single or a few scales together with epidermis from the flanks of their victims. Scale-eaters are a common component of the littoral fish communities in Lake Tanganyika (Hori et al., 1993) and show a number of adaptations with regard to their specific way of feeding including hook-like teeth, asymmetry of the mouth opening as well as aggressive mimicry (Liem & Stewart, 1976; Hori, 1993; Hori & Watanabe, 2000; Takahashi et al., 2007; Boileau et al., 2015). In particular the mouth dimorphism of scale-eaters has received considerable scientific attention, following Hori’s initial report of negative frequency-dependent selection in Perissodus microlepis (Boulenger, 1898) (Hori, 1993; Indermaur et al., 2018; see below).
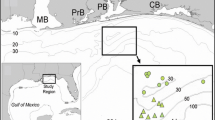
Perissodus microlepis (Fig. 1) is the most common of the Tanganyikan scale-eaters; it has a lake-wide distribution and occurs across all habitat types to a depth of up to 70 m (Takahashi et al., 2007; Konings, 2015). Two discrete morphs with respect to mouth morphology were recognized in P. microlepis, one with a mouth opening to the right side (‘right morph’), and one to the left side (‘left morph’) (Hori, 1993). The mouth polymorphism in P. microlepis has long been implicated with a lateralized feeding behavior, whereby the right morph attacks the left flank of prey fish and vice versa (Hori, 1993). This has later been confirmed experimentally (Van Dooren et al., 2010; Lee et al., 2012; Takeuchi et al., 2012; Takeuchi and Oda, 2017). More recently, it has been suggested that the angle of mouth opening is continuously distributed in P. microlepis instead of having two discrete modes (Kusche et al., 2012), whereas an analysis of lower-jaw bones by Takeuchi et al. (2016) revealed a bimodal distribution of mouth orientation. In addition to the polymorphism in mouth orientation, there is also a difference in body coloration in P. microlepis. Nshombo (1994) reported the existence of four different color morphs in P. microlepis, and found that these color morphs exhibit different attack strategies. Hori & Watanabe (2000), on the other hand, suggested that a local morph of P. microlepis found in the South of Lake Tanganyika characterized by a yellow anal fin has adopted an aggressive mimicry strategy to feed upon Cyprichromini.
In this study, we examined the prey spectrum of the Tanganyikan scale-eater Perissodus microlepis using a DNA-barcoding approach. It has previously been suggested, based on direct observations in the field, that P. microlepis is an opportunistic feeder attacking a broad range of fish species (Nshombo et al., 1985; Hori et al., 1993); it has further been found that juvenile scale-eaters feed on copepods and become more and more specialized with age (Takeuchi et al., 2016). Similarly, in the scale-eater Plecodus straeleni, a diverse range of prey species has been found, in this case using a molecular approach to taxonomically assign ingested scales (Boileau et al., 2015). Here, we adopted the strategy of Boileau et al. (2015) and sequenced a fragment of the mitochondrial NADH Dehydrogenase Subunit II (ND2) gene in hundreds of scales extracted from the intestinal tracts of more than 100 specimens of P. microlepis collected in the South of Lake Tanganyika, following scale counts and the morphological classification of all scales. We then tested if the prey species spectrum correlated with mouth orientation (left vs. right) or body color hue (dark vs. light) of the scale-eaters.
Methods
Sampling
In total, we collected 203 adult specimens of Perissodus microlepis in the southern part of Lake Tanganyika in February and March 2010 under permits issued by the Lake Tanganyika Research Unit, the Department of Fisheries, Food and Agriculture, Republic of Zambia; and the Department of Immigration, Republic of Zambia. Fish were collected at eight sampling sites by means of gill-nets on SCUBA in a depth between 5 and 20 m (see Fig. 1A, Table 1, and Supplementary Table 1 for details). Specimens were photographed, weighed, sexed via inspecting their gonads, and measured for standard (SL) and total length (TL); we also determined, whenever possible, the mouth orientation (left vs. right) as well as body color hue (dark vs. light) on fresh specimens. The intestinal tract of each specimen was removed in the field and preserved in 96% ethanol; specimens were then individually labeled and preserved in 96% ethanol. Samples were transported to the Zoological Institute of the University of Basel for further analyses.
Scale morphological analysis and counting
The intestinal tracts of all collected specimens of P. microlepis were inspected in detail in the laboratory, using a Leica MZ75 stereomicroscope. Eighty-four intestinal tracts were empty or their stomach and gut contents were too digested to allow further assignment, leaving us with a total of 119 intestinal tracts for further inspections. Of these, 117 intestinal tracts contained exclusively fish scales, whereas in one stomach we found, in addition to scales, a fish eye and in another one a fish embryo. The scales recovered from these 119 intestinal tracts were photographed with a Leica DFC310 FX digital camera mounted on to a Leica M205FA stereomicroscope; scales were counted and sorted according to size (into four different quartiles, XS, S, M, and L), general morphology (ctenoid vs. cycloid), and pigmentation, following the criteria described in Kuusipalo (1998). Scales were then rinsed with ethanol and collected in separate 2 ml microcentrifuge tubes filled with 96% ethanol for molecular analyses via DNA barcoding.
DNA barcoding of scales
For DNA barcoding of scales recovered from the intestinal tracts of P. microlepis, we followed a modified version of the protocol described in Boileau et al. (2015). In short, we extracted DNA from individual scales applying a phenol–chloroform-isoamyl alcohol precipitation following digestion in 180 μl 2× CTAB Buffer and 7.5 μl Proteinase K (10 mg/ml) overnight at 37°C and 300 rpm on an Eppendorf® Thermomixer compact. DNA quality was determined with a NanoDrop ND-1000 spectrophotometer (ThermoFisher Scientific).
For PCR amplification of a 407-bp-long section of the mitochondrial NADH Dehydrogenase Subunit II (ND2) gene, a standard marker in East African cichlid fish (e.g., Kocher et al., 1995; Salzburger et al., 2002, 2005), we used the primers ND2-327 (5′-CCC TCT TCA TGC TTG ACT CC-3′) and ND2-733 (5′-GGG GTG TGA GAG CTG TTA GG-3′) (Boileau et al., 2015). To avoid amplification of P. microlepis endogenous DNA, we added blocking oligonucleotides Perplex-488-blocH (5′-ctg GCC CTT GTT GGG GGC TGA ttt-3′) and Perplex-617-blocL (5′-aaa CAT AAT GAA GTA GGT AAG AAG GGT ctc-3′). These blocking oligonucleotides were designed to specifically anneal within the ND2 genes of several Tanganyikan scale-eaters of the tribe Perissodini including P. microlepis and Plecodus straeleni (Boileau et al., 2015) to inhibit the amplification of ND2 from those species. PCR was performed with the enzyme AmpliTaq (Applied Biosystems) on a Veriti® 96-well Thermal Cycler (Applied Biosystems) following the protocol described in Boileau et al. (2015); PCR products were then purified with ExoSAP-IT® (Affymetrix) and Sanger-sequenced on an ABI 3130xl genetic analyzer using the BigDye® 3.1 kit (Applied Biosystems). The sequences have been deposited on GenBank under the accession numbers MH275093–MH275427.
Sequence analysis and taxonomic assignments
Due to the close relatedness of the potential prey taxa, we used a combination of BLAST searches and phylogenetic analyses to assign the ND2 sequences derived from individual scales recovered from the intestinal tracts of P. microlepis to particular prey species (see Boileau et al., 2015). In a first step, individual sequences were inspected by eye with the software CodonCode Aligner v.3.7.1.1 (CodonCode Corporation). BLAST searches (BLASTN) were performed with BLAST + version 2.2.31 against GenBank’s nucleotide collection (nr/nt) database (version December 2017) on the sciCORE computer cluster of the University of Basel. For phylogenetic analyses, the obtained sequences were assembled to a reference set containing sequence information of 180 species of cichlid fishes from Lake Tanganyika (Boileau et al., 2015; Meyer et al., 2015) using CodonCode Aligner. Maximum likelihood based phylogenetic analyses were performed with a heuristic search in Paup* 4.0a, build 159 (Swofford, 2002), applying the GTR+G+I model of molecular evolution and using the model parameters estimated from the data. The sequences obtained from ingested scales were assigned to a particular cichlid species whenever the best BLAST hit (according to BLAST score and E-value) was identical to the taxon with which the unknown sequence clustered in the phylogenetic tree. In cases where the best BLAST hit was shared between several sequences belonging to more than one species (usually a consequence of the short length of the query sequence), we used the phylogenetic clustering for taxonomic assignment. Sequences that could not be unequivocally assigned to a species according to this strategy were excluded from further analyses.
Statistical analyses
All statistical analyses were performed in R version 3.4.0. El Capitan build (7338) (R Development Core Team, 2008). In a first step, we used the scale-count information to test, via an ANCOVA in the R package Vegan version 2.4-5 (Oksanen et al., 2017), whether the number of scales in the intestinal tracts of scale-eaters is dependent on body size (analyzing SL and TL separately) and sex. In a second step, we used the size-class information of the ingested scales (XS, S, M, L) to test whether scale size correlates with the body size of the scale-eaters, again applying an ANCOVA and analyzing SL and TL separately. We then used the taxonomic assignment of the ingested scales based on DNA barcoding together with the information on mouth orientation (left vs. right) and body color hue (dark vs. light) of the scale-eaters, as well as on sampling location, to test whether there are differences in the prey spectrum with respect to mouth orientation, body color hue or sampling location. The latter test was performed as there is evidence for genetic structuring within P. microlepis in the South of Lake Tanganyika (Koblmüller et al., 2009). We used permutational multivariate analysis of variance on distance matrices, as implemented in the adonis function of the R-package Vegan. Distance matrices were generated from binary presence/absence data using the function vegdist with Raup-Crick dissimilarity index.
Results
In total, we recovered 10,749 scales in the intestinal tracts of 119 P. microlepis specimens (SL distribution of P. microlepis 53–115 mm; mean = 75.07 ± 9.65 mm). The number of scales per intestinal tract varied from 1 to 342 (median = 81; mean = 88.1 ± 70.1), which is in the range of what has been reported previously (Takeuchi et al., 2016). Based on morphological grounds (according to Kuusipalo, 1998), we classified the recovered scaled into 53 different scale types, without knowing their taxonomic identity at this stage. The number of scale types per intestinal tract varied from 1 to 16 (median = 5; mean = 5.9 ± 3.2).
On the basis of the scale counts per intestinal tract, we found that the number of scales ingested by a scale-eater did not depend on its body size (ANCOVA; P-value = 0.2378). However, we found that the size of the ingested scales correlated with the body size of the scale-eaters (P-value < 0.001). Note that no difference in standard or total length was found between male and female scale-eaters in our dataset (Wilcoxon test, SL: P-value = 0.6208, TL: P-value = 0.9416), so that we did not treat the sexes differentially in this analysis.
The DNA of 384 scales could successfully be amplified, of which 333 samples could successfully be sequenced. Despite the use of blocking primers, 107 sequences obtained from ingested scales matched Perissodus reference sequences, rendering it impossible to distinguish between endogenous contamination and potential prey on the basis of the data at hand. These sequences were consequently excluded from further analyses. Another twelve sequences were too short to allow the unambiguous taxonomic assignment to a single reference and were also excluded from further analyses. Thus, a total of 214 scales could be assigned to a particular prey species (in 194 cases based on both BLAST searches and phylogenetic analyses; see Supplementary Table 2).
Among the 214 scales from 88 intestinal tracts of P. microlepis, we identified a total of 39 prey species belonging to 8 different cichlid tribes (Fig. 2). According to our analyses, the five most common prey species of P. microlepis were Petrochromis polyodon (N = 37), Pe. ephippium (26), Eretmodus cyanostictus (20), Tropheus moorii (20) and Simochromis diagramma (15). These five species, which accounted for more than 50% of the scales found in the intestinal tracts of P. microlepis, are all herbivorous. The most commonly found prey species, Pe. polyodon, was also the only one detected at all of the eight sampling sites. At the tribal level, the by far most common prey of P. microlepis belonged to Tropheini/Haplochromini (N = 136; 12 different prey species) followed by the most species-rich tribe in Lake Tanganyika, Lamprologini (N = 25, 14 species), the Ectodini (N = 20; 6 species) and Eretmodini (N = 20; 1 species). Members of the Cyprichromini (N = 6; 2 species), Boulengerochromini (N = 2; 1 species), Limnochromini (N = 2; 1 species), and other Perissodini (N = 2; 2 species) were apparently infrequently attacked by P. microlepis.
The prey spectrum of the scale-eater Perissodus microlepis in the southern part of Lake Tanganyika. The bars indicate the number of scales extracted from the intestinal tracts of scale-eaters that were assigned to one of 39 different prey species by means of a molecular barcoding approach. The bars are color coded according to the tribe to which the respective species belongs
The prey choice of individuals appeared to not be significantly correlated with body color (partial R2: 0.006, P = 0.83), mouth orientation (partial R2: 0.013, P = 0.50), or sampling location (partial R2: 0.112, P = 0.14).
Discussion
In this study, we examined the prey spectrum of the Tanganyikan scale-eater Perissodus microlepis. To this end, we first counted and characterized the scales retrieved from the intestinal tracts of more than one hundred individuals of P. microlepis collected at eight sites in the southern part of Lake Tanganyika and then determined the sequence of a fragment of the mitochondrial ND2 gene of hundreds of individual scales to permit their taxonomic assignment to specific prey species.
The morphological examination of ingested scales as well as the molecular characterization of a subset of those revealed that the Tanganyikan scale-eater P. microlepis feeds upon a broad spectrum of prey species, as already reported by Nshombo et al. (1985) based on field observations. When applying the criteria established for Lake Malawi cichlids (Kuusipalo, 1998), we identified 53 different scale types among the 10,749 scales examined based on morphological grounds. The grouping of scales into scale types is not expected to directly correspond to prey species, since several types of scales differing in size and morphology can be found on a single Tanganyikan cichlid (Lippitsch, 1990, 1993). However, this high number already suggests a relatively broad prey spectrum. This was confirmed by the molecular characterization of ingested scales, which led to the identification of 39 different prey species belonging to 8 different cichlid tribes (Fig. 2).
The by far most common prey species belonged to the Tropheini, followed by Lamprologini, Ectodini and Eretmodini, whereas members of other tribes were only infrequently attacked. The prey spectrum of P. microlepis, thus, more or less reflects the community composition of cichlids in the rocky littoral zone of Lake Tanganyika (Hori et al., 1993; Muschick et al., 2012; Konings, 2015), where P. microlepis is most common. Interestingly, the prey spectrum of P. microlepis is rather similar to the one reported previously for another Tanganyikan scale-eater, Plecodus straeleni (Boileau et al., 2015). (Note, however, that in the present study, we did not perform additional PCR experiments that would permit detecting mastacembelid eels, which account for 23% of the scales in the intestinal tracts of Pl. straeleni.) An apparent difference between the prey spectra of P. microlepis and Pl. straeleni is that P. microlepis is more frequently feeding on Eretmodus cyanostictus, which usually occurs in relatively shallow waters (< 10 m). This somewhat reflects the distribution of the two scale-eating species in question: While overlapping in large parts of their depth distribution, P. microlepis is also common in shallow waters, where Pl. straeleni is rarely observed (Takeuchi et al., 2010).
Our analyses characterize the Tanganyikan scale-eater P. microlepis as a rather opportunistic feeder—both at the level of species and the level of individuals—with a broad spectrum of potential prey species. Furthermore, the fact that up to 16 different scale types were present in a single intestinal tract and that neither the mouth orientation (left vs. right) nor the body color hue (light vs. dark) or the sampling location correlated with prey species, indicate that individual scale-eaters do not seem to be specialized toward a particular prey species. However, we found that the intestinal tracts of larger scale-eaters contain, on average, larger scales, which indicates that adult P. microlepis preferentially attack different size classes of prey species according to their own body size. No correlation was found between body size and the number of ingested scales. This is different to the findings of Takeuchi et al. (2016) who showed that the intestinal tracts of larger and more lateralized scale-eaters contained more scales. Note, however, that they inspected scales over a much larger range of body lengths including juveniles that feed on copepods, whereas we here focused on adult specimen only.
Both traits, mouth orientation and body color, could potentially influence the prey choice of P. microlepis individuals. Body color correlates with the preference for microhabitats (Nshombo, 1994), which presumably differ in their composition of potential prey. Such habitat preferences of color morphs also occur in other predatory cichlid species in Lake Tanganyika (Kohda and Hori, 1993). Mouth-handedness of P. microlepis has been shown to be under negative frequency-dependent selection (Hori, 1993). In a population of scale-eaters with an unbalanced frequency of the two morphs, the more frequent morph will soon be at a disadvantage as prey fish will guard more on the side they are attacked on more often, decreasing the success rate for that morph.
Overall, our study once more demonstrates that molecular barcoding of ingested material is a powerful tool to obtain information on prey composition in general (Symondson, 2002) and for scale-eating cichlids in particular (Boileau et al., 2015). It also shows that this approach is highly sensitive to local faunal differences. For example, we only detected scales of Lepidiolamprologus kendalli in the intestinal tracts of P. microlepis collected from the western shoreline of Lake Tanganyika, corresponding to the localities where L. kendalli occurs (see Konings, 2015). Likewise, we detected scales of the haplochromine Astatotilapia burtoni at a location for which we had previous evidence of its occurrence (Pauquet et al., 2018).
References
Boileau, N., F. Cortesi, B. Egger, M. Muschick, A. Indermaur, A. Theis, H. H. Büscher & W. Salzburger, 2015. A complex mode of aggressive mimicry in a scale-eating cichlid fish. Biology Letters 11: 20150521.
Fryer, G. & T. D. Iles, 1972. The cichlid fishes of the Great Lakes of Africa: their biology and evolution. Oliver & Boyd, Edinburgh.
Hori, M., 1993. Frequency-dependent natural selection in the handedness of scale-eating cichlid fish. Science 260: 216–219.
Hori, M. & K. Watanabe, 2000. Aggressive mimicry in the intra-populational color variation of the Tanganyikan scale-eater Perissodus microlepis (Cichlidae). Environmental Biology of Fishes 59: 111–115.
Hori, M., M. M. Gashagaza, M. Nshombo & H. Kawanabe, 1993. Littoral fish communities in Lake Tanganyika: irreplaceable diversity supported by intricate interactions among species. Conservation Biology 7: 657–666.
Indermaur, M., A. Theis, B. Egger & W. Salzburger, 2018. Mouth dimorphism in scale-eating cichlid fish from lake Tanganyika advances individual fitness. Evolution. https://doi.org/10.1111/evo.13552.
Koblmüller, S., K. M. Sefc & C. Sturmbauer, 2008. The Lake Tanganyika cichlid species assemblage: recent advances in molecular phylogenetics. Hydrobiologia 615: 5–20.
Koblmüller, S., N. Duftner, K. M. Sefc, U. Aigner, M. Rogetzer & C. Sturmbauer, 2009. Phylogeographic structure and gene flow in the scale-eating cichlid Perissodus microlepis (Teleostei, Perciformes, Cichlidae) in southern Lake Tanganyika. Zoologica Scripta 38: 257–268.
Kocher, T. D., J. A. Conroy, K. R. McKaye, J. R. Stauffer & S. F. Lockwood, 1995. Evolution of NADH dehydrogenase subunit 2 in east African cichlid fish. Molecular Phylogenetics and Evolution 4: 420–432.
Kohda, M. & M. Hori, 1993. Dichromatism in relation to the trophic biology of predatory cichlid fishes in Lake Tanganyika, East Africa. Journal of Zoology 229: 447–455.
Kolmann, M. A., J. M. Huie, K. Evans & A. P. Summers, 2018. Specialized specialists and the narrow niche fallacy: a tale of scale-feeding fishes. Royal Society Open Science 5: 171581.
Konings, A., 2015. Tanganyika Cichlids in Their Natural Habitat, 3rd ed. Cichlid Press, El Paso.
Kusche, H., H. J. Lee & A. Meyer, 2012. Mouth asymmetry in the textbook example of scale-eating cichlid fish is not a discrete dimorphism after all. Proceedings of the Royal Society B 279: 4715–4723.
Kuusipalo, L., 1998. Scale morphology in Malawian cichlids. Journal of Fish Biology 52: 771–781.
Lee, H. J., H. Kusche & A. Meyer, 2012. Handed foraging behavior in scale-eating cichlid fish: its potential role in shaping morphological asymmetry. PLoS ONE 7: e44670.
Liem, K. F. & D. E. Stewart, 1976. Evolution of scale-eating cichlid fishes of Lake Tanganyika: a generic revision with a description of a new species. Bulletin of the Museum of Comparative Zoology at Harvard College 147: 319–350.
Lippitsch, E., 1990. Scale morphology and squamation patterns in cichlids (Teleostei, Perciformes): a comparative study. Journal of Fish Biology 37: 265–291.
Lippitsch, E., 1993. A phyletic study on lacustrine haplochromine fishes (Perciformes, Cichlidae) of East Africa, based on scale and squamation characters. Journal of Fish Biology 42: 903–946.
Marlier, G. & N. Leleup, 1954. A curious ecological niche among the fishes of Lake Tanganyika. Nature 174: 935–936.
Martin, C. H. & P. C. Wainwright, 2013. On the measurement of ecological novelty: scale-eating pupfish are separated by 168 My from other scale-eating fishes. PLoS ONE 8: e71164.
Meyer, B. S., M. Matschiner & W. Salzburger, 2015. A tribal level phylogeny of Lake Tanganyika cichlid fishes based on a genomic multi-marker approach. Molecular Phylogenetics and Evolution 83: 56–71.
Meyer, B. S., M. Matschiner & W. Salzburger, 2017. Disentangling incomplete lineage sorting and introgression to refine species-tree estimates for Lake Tanganyika cichlid fishes. Systematic Biology 66: 531–550.
Muschick, M., A. Indermaur & W. Salzburger, 2012. Convergent evolution within an adaptive radiation of cichlid fishes. Current Biology 22: 2362–2368.
Nshombo, M., 1994. Polychromatism of the scale-eater Perissodus microlepis (Cichlidae, Teleostei) in relation to foraging behavior. Journal of Ethology 12: 141–161.
Nshombo, M., Y. Yanagisawa & M. Nagoshi, 1985. Scale-eating in Perissodus microlepis (Cichlidae) and change of its food-habits with growth. Japanese Journal of Ichthyology 32: 66–73.
Oksanen, J., F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs & H. Wagner, 2017. vegan: community ecology package. R package version 2.4-5. https://CRAN.R-project.org/package=vegan.
Pauquet, G., W. Salzburger & B. Egger, 2018. The puzzling phylogeography of the haplochromine cichild fish Astatotilapia burtoni. Ecology and Evolution 8(11): 5637–5648.
Poll, M., 1986. Classification des Cichlidae du lac Tanganika: tribus, genres et especes. Memoires de la classe des sciences. T. XLV. Fascicule, 2nd ed. Academie Royale de Belgique, Brussels.
R Development Core Team, 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. http://www.R-project.org.
Salzburger, W., A. Meyer, S. Baric, E. Verheyen & C. Sturmbauer, 2002. Phylogeny of the Lake Tanganyika cichlid species flock and its relationship to the Central and East African haplochromine cichlid fish faunas. Systematic Biology 51: 113–135.
Salzburger, W., T. Mack, E. Verheyen & A. Meyer, 2005. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evolutionary Biology 5: 17.
Salzburger, W., B. Van Bocxlaer & A. S. Cohen, 2014. Ecology and evolution of the African Great Lakes and their faunas. Annual Review of Ecology, Evolution, and Systematics 45: 519–545.
Sazima, I., 1983. Scale-eating in characoids and other fishes. Environmental Biology of Fishes 9: 87–101.
Swofford, D. L., 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland.
Symondson, W. O. C., 2002. Molecular identification of prey in predator diets. Molecular Ecology 11: 627–641.
Takahashi, T., 2003. Systematics of Tanganyikan cichlid fishes (Teleostei: Perciformes). Ichthyological Research 50: 367–382.
Takahashi, R., K. Watanabe, M. Nishida & M. Hori, 2007. Evolution of feeding specialization in Tanganyikan scale- eating cichlids: a molecular phylogenetic approach. BMC Evolutionary Biology 7: 195.
Takeuchi, Y. & Y. Oda, 2017. Lateralized scale-eating behaviour of cichlid is acquired by learning to use the naturally stronger side. Scientific Reports 7: 8984.
Takeuchi, Y., H. Ochi, M. Kohda & M. Hori, 2010. A 20-year census of a rocky littoral fish community in Lake Tanganyika. Ecology of Freshwater Fish 19: 239–248.
Takeuchi, Y., M. Hori & Y. Oda, 2012. Lateralized kinematics of predation behavior in a Lake Tanganyika scale-eating cichlid fish. PLoS ONE 7: e29272.
Takeuchi, Y., M. Hori, S. Tada & Y. Oda, 2016. Acquisition of lateralized predation behaviour associated with development of mouth asymmetry in a Lake Tanganyika scale-eating cichlid fish. PLoS ONE 11: e0147476.
Van Dooren, T. J. M., H. A. Van Goor & M. Van Putten, 2010. Handedness and asymmetry in scale-eating cichlids: antisymmetries of different strength. Evolution 64: 2159–2165.
Acknowledgements
We would like to thank our helpers in the field M. Colombo, M. T. Dittmann, A. Indermaur, and M. Roesti; T. Veall for logistic support; L. Makasa and the Department of Fisheries, Republic of Zambia, for research permits; J. Meunier for help with the statistical analyses; R. Isenschmid and A. Schett for assistance with imaging of scales; and three anonymous reviewers and the Guest Editors of this species issue for valuable comments. This study was supported by grants from the European Research Council (ERC, Starting Grant ‘INTERGENADAPT’ and Consolidator Grant ‘CICHLID~X’), the University of Basel, and the Swiss National Science Foundation to W. S.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: S. Koblmüller, R. C. Albertson, M. J. Genner, K. M. Sefc & T. Takahashi / Advances in Cichlid Research III: Behavior, Ecology and Evolutionary Biology
Electronic supplementary material
Below is the link to the electronic supplementary material.
10750_2018_3714_MOESM1_ESM.xlsx
Supplementary material 1 Supplementary Table 1. Information about the specimens of Perissodus microlepis examined in this study (including sampling information). (XLSX 73 kb)
10750_2018_3714_MOESM2_ESM.xlsx
Supplementary material 2 Supplementary Table 2. Information about the scales examined in this study. Sample_ID… identifier of an individual scale, Fish_ID… identifier of the specimen of Perissodus microlepis from which the scale was extracted, Species (BLAST)… Assignment of the mtDNA sequence obtained from the scale via BLAST, Species (phylogeny)… Assignment of the mtDNA sequence obtained from the scale based on a phylogenetic analysis, Scale type… Assignment of scales according to morphology into one of 53 scale types, arbitrarily and consecutively named A to AZ. In cases of a discrepancy in the assignment between BLAST and the phylogenetic analyses, we used the assignment identified in bold. (XLSX 19 kb)
Rights and permissions
About this article
Cite this article
Kovac, R., Boileau, N., Muschick, M. et al. The diverse prey spectrum of the Tanganyikan scale-eater Perissodus microlepis (Boulenger, 1898). Hydrobiologia 832, 85–92 (2019). https://doi.org/10.1007/s10750-018-3714-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-018-3714-9