Abstract
The cyanobacterial population in the Cajati waste stabilization pond system (WSP) from São Paulo State, Brazil was assessed by cell isolation and direct microscope counting techniques. Ten strains, belonging to five genera (Synechococcus, Merismopedia, Leptolyngbya, Limnothrix, and Nostoc), were isolated and identified by morphological and molecular analyses. Morphological identification of the isolated strains was congruent with their phylogenetic analyses based on 16S rDNA gene sequences. Six cyanobacterial genera (Synechocystis, Aphanocapsa, Merismopedia, Lyngbya, Phormidium, and Pseudanabaena) were identified by direct microscope inspection. Both techniques were complementary, since, of the six genera identified by direct microscopic inspection, only Merismopedia was isolated, and the four other isolated genera were not detected by direct inspection. Direct microscope counting of preserved cells showed that cyanobacteria were the dominant members (>90%) of the phytoplankton community during both periods evaluated (summer and autumn). ELISA tests specific for hepatotoxic microcystins gave positive results for six strains (Synechococcus CENA108, Merismopedia CENA106, Leptolyngbya CENA103, Leptolyngbya CENA112, Limnothrix CENA109, and Limnothrix CENA110), and for wastewater samples collected from raw influent (3.70 μg microcystins/l) and treated effluent (3.74 μg microcystins/l) in summer. Our findings indicate that toxic cyanobacteria in WSP systems are of concern, since the treated effluent containing cyanotoxins will be discharged into rivers, irrigation channels, estuaries, or reservoirs, and can affect human and animal health.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Waste stabilization pond (WSP) systems constitute the most commonly used method of domestic and municipal wastewater treatment in tropical countries, where the climate favors their operation. They use solar energy alone and do not occupy much more land than conventional electromechanical treatment systems, such as activated sludge plants. Thus, WSP systems are low-cost and low maintenance, highly efficient, and sustainable for low to moderate treatment volumes. This treatment technology produces quality effluent that satisfies the requirements of several guidelines and recommendations (e.g., ANZECC & ARMCANZ, 2000; CONAMA, 2005).
A typical WSP system consists of three pond types: anaerobic, facultative (anaerobic–aerobic), and maturation. Following chlorination or other forms of disinfection, the effluent is usually sufficiently clean to be discharged into receiving waters. Anaerobic and facultative ponds are used for the removal of organic matter, and their effectiveness can be evaluated using the 5-day biochemical oxygen demand (BOD5). In facultative ponds, the organic matter is decomposed by the action of aerobic bacteria, which receive oxygen via algal and cyanobacterial photosynthesis. The presence of these organisms is very important for the maintenance of the aerobic conditions of the system. However, several cyanobacterial strains produce toxins that can affect the aquatic biota and also cause health problems in human and animal populations. Several planktonic toxic cyanobacterial strains can proliferate intensively on nutrient-enriched water surfaces, forming what is known as a "bloom." Blooms may contain one or more cyanobacterial genera/populations that synthesize toxins such as hepatotoxin, neurotoxin, or dermatotoxin (Sivonen & Jones, 1999). These cyanotoxins can be classified into three broad groups based on their chemical structure: cyclic peptides, alkaloids, and lipopolysaccharides. Globally, the most frequent cyanobacterial toxins found in freshwater blooms are cyclic peptides of the microcystin family (Sivonen & Jones, 1999). Microcystin-synthesizing strains have been found in all cyanobacterial orders, although the principal species belong to the planktonic genera Microcystis, Planktothrix, and Anabaena (Sivonen & Jones, 1999).
Few studies have focused on natural cyanobacteria diversity in WSP systems (Nandini, 1999; Vasconcelos & Pereira, 2001; Oudra et al., 2002). In Brazil, the only study available is a floristic investigation of cyanobacterial strains that was conducted in WSP systems from Minas Gerais State (Von Sperling, 1996). However, there have been no investigations involving molecular and phylogenetic analyses of the cyanobacterial populations found in WSP systems. Furthermore, information concerning cyanobacterial toxins in this environment is also scarce (Vasconcelos & Pereira, 2001; Oudra et al., 2002). The presence of toxin-producing cyanobacteria in WSP systems constitutes a potential health hazard, since the treated effluent may still contain cyanotoxins and will be discharged into rivers, irrigation channels, estuaries, or reservoirs. The objectives of this study were therefore to characterize cyanobacterial isolates from a WSP system from São Paulo State, Brazil, using morphological and molecular analyses, and to evaluate the production of the hepatotoxin microcystins by cyanobacterial isolates as well as their presence in wastewater samples. A related aim was to estimate the cyanobacterial population in the WSP system using direct microscope counting for comparative purposes with the isolation technique.
Materials and methods
Site description and sample collection
Samples were collected from a facultative WSP located in the city of Cajati (24°43′23″S, 48°05′39″W), São Paulo State, Brazil. This pond is operated by SABESP (São Paulo State Sanitation Company) and belongs to the Ribeira de Iguape Valley Treatment System. The system consists of a sand tank, an anaerobic pond followed by a facultative pond, and a chlorination baffle tank at the exit area. The morphometric and operational parameters of the facultative WSP are shown in Table 1. The Cajati WSP receives a maximal influent flow of 53.4 l/s and a maximum organic load of 1,753 kg BOD5/day. The removal efficiency of Cajati WSP was evaluated in raw influent- and effluent-treated samples through analyses of pH, temperature, and dissolved oxygen (DO) at the site using a YSI 556 multiparameter probe (Yellow Springs Instruments, Inc., Yellow Springs, OH, USA), BOD5, chemical oxygen demand (COD), total suspended solids (TSS), total nitrogen, total phosphorus, thermotolerant coliforms, and phytoplankton density according to APHA (1998).
The Cajati WSP was sampled twice in summer (December 16, 2004 and January 25, 2005) and once in autumn (April 13, 2005) at the entrance, central area and outflow of the facultative pond, and in the baffle tank during a no chlorination operation. Samples were collected from the pond surface water using phytoplankton flasks and were transported to the laboratory in a cooling box containing ice packs. Five milliliters of each wastewater sample was stored at −20°C for microcystin analysis, 1 ml was used for cyanobacterial isolation, and 100 ml was preserved in 4% formaldehyde (final concentration) for microscopic inspection.
Cyanobacterial isolation, identification and densities
In order to obtain a monoculture of cyanobacterial strains, 1 ml of each wastewater sample collected from the Cajati facultative pond was dispensed into sterile test tubes containing 9 ml of liquid BG-11 medium (Allen, 1968), with or without a nitrogen source, and cycloheximide (70 mg/l) to inhibit eukaryotic cell growth. After mixing, 10-fold serial dilutions (to 10−7) were used to inoculate test tubes containing the same medium. The tubes were incubated in a growth chamber for 30 days at 24 ± 1°C under constant, white fluorescent illumination (30 μmol photon/m2/s). After this period, cells were repeatedly streaked onto solid BG-11 medium until isolated cultures were established. After isolation of uni-cyanobacterial cultures, strains were examined by a microscope (Zeiss Axioskop 40, Carl Zeiss, Jena, Germany). Morphological descriptions were made according to the classification systems devised by Komárek & Anagnostidis (1989, 1999, 2005). Where possible, the bacteriological classification (Castenholz, 2001) is provided following botanical designations (order/genera).
Cyanobacterial and algal cells of WSP samples preserved with formaldehyde (4%) were counted by direct microscopic inspection using the Utermöhl counting technique (APHA, 1998) and an inverted light microscope (Olympus CK2, Olympus, Tokyo, Japan). A few drops of Lugol’s iodine were added to the samples to induce cell sedimentation onto the glass bottom of the chamber. At least 400 cells were counted with a ±10% error, calculated considering 95% confidence interval, and a random distribution was assumed for the samples onto the bottom of the counting camera (APHA, 1998). Morphological identifications were performed as mentioned above.
DNA extraction, amplification, sequencing, and phylogenetic analysis
An aliquot (3 ml) of each cultured cyanobacterial isolates was harvested in mid to late exponential phase (10–25 days) by centrifugation (12,000×g for 5 min at 25°C) in a sterile 1.5-ml microcentrifuge tube. Total genomic DNA was extracted using a modified cetyl-trimethyl-ammonium bromide (CTAB)-based extraction method adapted for cyanobacteria (Fiore et al., 2000). The 16S rDNA sequence was amplified from genomic DNA using the cyanobacteria-specific primer-set 27F1 and 1494Rc (Neilan et al., 1997). Amplification was performed in a 25-μl volume reaction containing 10 ng genomic DNA, 5 pmol/μl of each oligonucleotide primer, 0.2 mM of each dNTP, 3.0 mM MgCl2, 1× PCR buffer, and 1.5 U Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA). A Gene Amp PCR System 2400 (Applied Biosystems, Foster City, CA, USA) was used. Thermal cycling was performed with an initial denaturation step at 94°C for 4 min, followed by 30 cycles of DNA denaturation at 94°C for 20 s, primer annealing at 50°C for 30 s, strand extension at 72°C for 2 min, and a final extension step at 72°C for 7 min. On completion of the 16S rDNA gene amplification, PCR products were immediately cloned using a pGEM®-T Easy Vector System (Promega, Madison, WI, USA) according to the manufacturer’s instructions, and transformed into E. coli DH5α. Plasmid DNA containing the clone was then extracted by an alkaline lysis method (Birnboim & Doly, 1979). DNA sequencing was performed using the cloned PCR products and a DYEnamic ET Terminator Cycle Sequencing kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK). Primers used for the cycle sequencing were the vector’s T7 and M13 primer sites and the internal primer sets 341-357F, 357-341R, 685-704F, 704-685R, 1099-1114F, and 1114-1099R (Lane, 1991). The cycle sequencing reaction was performed using a Gene Amp PCR System 2400 (Applied Biosystems), and the reaction conditions were 25 cycles of the following: 20 s at 95°C, 15 s at 50°C, and 1 min at 60°C. After the sequencing reaction was completed, residual dye terminators were removed by ethanol precipitation using sodium acetate/EDTA buffer provided with the DYEnamic ET Terminator Cycle Sequencing Kit, following the manufacturer’s instructions. The purified reaction was then resuspended in HiDi formamide (Applied Biosystems) and the samples read in an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Cloned PCR products were bidirectionally sequenced and each set of sequencing data was obtained from at least two independent experiments. The sequenced fragments were assembled into one contig using the software Phred/Phrap/Consed program (Philip Green, University of Washington, Seattle, USA) and only bases with a quality >20 were considered. The 16S rDNA sequences obtained in this study were aligned with related sequences retrieved from GenBank using the Clustal X program (Thompson et al., 1997). Phylogenetic trees were constructed by neighbor-joining (NJ) and maximum-parsimony (MP) algorithms using the MEGA version 3.1 package program (Kumar et al., 2004). The NJ and MP stability of the relationships were assessed by bootstrapping (1,000 replicates). When the best hit came from uncultured clones using the BLAST comparative analysis (Altschul et al., 1990), the closest cultured strain was chosen. All sequence data were deposited in GenBank under the following accession numbers: Leptolyngbya sp. CENA103—EF088339, Leptolyngbya sp. CENA104—EF088333, Nostoc sp. CENA105—EF088340, Merismopedia sp. CENA106—EF088332, Nostoc sp. CENA107—EF088341, Synechococcus sp. CENA108—EF088334, Limnothrix sp. CENA109—EF088335, Limnothrix sp. CENA110—EF088338, Limnothrix sp. CENA111—EF088336 and Leptolyngbya sp. CENA112—EF088337.
Microcystin analyses
Microcystin analyses on both cyanobacterial isolates and wastewater samples were performed by enzyme-linked immunosorbent assay (ELISA) using a commercially available diagnostic kit (Beacon Analytical Systems Inc., Portland, ME, USA) following the manufacturer’s recommendations, with at least three replicates per sample. Aliquots of 2 ml of wastewater samples previously frozen (−20°C) were used for total microcystin analysis. Lyophilized cyanobacterial cells (0.02 g) of cultured isolates grown on BG-11 liquid medium were resuspended in 2 ml of water (Milli Q, Millipore) and used for intracellular microcystin analyses. The isolates and environmental samples were microwaved for 1 min. The extracts were centrifuged (10,000×g for 15 min), and the collected supernatants were used for the ELISA assay. The detection limit of this method is 0.1 μg/l.
Results
Characterization of Cajati WSP
The physical and chemical parameters of the Cajati WSP are showed in Table 2. Although some of the parameters (BOD5, COD (of autumn sample), TSS, total nitrogen, total phosphorus, and thermotolerant coliforms) analyzed are below the maximum values expected for WSP systems according to the PROSAB (Researches Program on Basic Sanitation—Brazil) (Chernicharo et al., 2006), most of them showed no removal efficiency or efficiency values lower than the guideline recommendations. The Cajati WSP pond was projected to have a hydraulic retention time (HRT) of 5 days in the anaerobic pond and 24 days in the facultative pond. However, we observed HRT values that were five times higher (25.4 and 49.5 days in anaerobic and facultative ponds, respectively). These environmental conditions can be expected to favor algae and cyanobacteria growth.
Cyanobacterial isolation, identification, and densities
The isolation technique used allowed for cultivation of 10 cyanobacterial strains belonging to five genera (Synechococcus, Merismopedia, Limnothrix, Leptolyngbya, and Nostoc) (Table 3, Fig. 1). The main features of the cyanobacterial morphotypes isolated are provided in Table 4. Of the six genera identified by direct microscopic inspection (Table 5), only Merismopedia was isolated and the four other genera isolated were not detected by direct microscopic inspection. These results also demonstrate that each method (direct microscope inspection or isolation) underestimated the diversity of the cyanobacterial population.
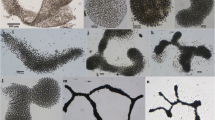
Cyanobacterial morphotypes isolated from the Cajati waste stabilization pond. A Leptolyngbya CENA103, B Leptolyngbya CENA104 (arrows indicate necrid formation and a false branch), C Nostoc CENA105, D Merismopedia CENA106, E Nostoc CENA107, F Synechococcus CENA108, G Limnothrix CENA109 (arrow indicates the gas vacuole), H Limnothrix CENA110 (arrow indicates the gas vacuole), I Limnothrix CENA111 (arrow indicates the gas vacuole), J Leptolyngbya CENA112
Direct counting of preserved cells showed values of 5.1 × 106 and 3.9 × 106 cells ml−1 in summer and autumn, respectively (Table 5). The abundance of the cyanobacterial population (91.7% in summer and 96.4% in autumn) was higher among the total algal population. Direct microscopic inspection of the cyanobacterial population of Cajati WSP showed the presence of six genera: Synechocystis, Aphanocapsa, Merismopedia, Lyngbya, Phormidium, and Pseudanabaena (Table 5). Synechocystis was strongly dominant, representing 82.5% of the summer sample and 92.3% of the autumn sample.
16S rDNA gene analyses and phylogeny
Nearly complete nucleotide sequences (1,409–1,415 bp) of the 16S rDNA spanning base positions 27-1494 (corresponding to E. coli numbering) were established for 10 cyanobacterial strains. The 16S rDNA sequences of the 10 strains isolated from the Cajati WSP showed different degrees of identities when compared to cyanobacterial sequences from public databases, as shown in Table 6.
Phylogenetic relationships of the 16S rDNA sequences from the isolated strains were compared with other cyanobacterial sequences retrieved from GenBank. The trees constructed using NJ and MP methods were largely congruent and thus only the NJ tree is presented, including bootstrap values for both methods (Fig. 2). Heterocystous forms (Nostocales) were grouped together, while unicellular and filamentous non-heterocystous forms (Chroococcales and Oscillatoriales) were dispersed within the tree. The two Chroococcalean isolates (Merismopedia CENA106 and Synechococcus CENA108) were clustered together in a major clade with high bootstrap values (100% NJ and 99% MP) despite morphological differences. However, Merismopedia CENA106 and Synechococcus CENA108 were positioned in distinct internal clades. The phylogenetic analyses showed that filamentous non-heterocystous cyanobacteria (Oscillatoriales) are mixed with unicellular (Chroococcales) strains, which indicates a polyphyletic origin. The sequences of Leptolyngbya sp. CENA103 and Leptolyngbya sp. CENA112 formed a clade with the closest related Antarctic isolate (Leptolyngbya frigida ANT.LH70.1—AY493574). However, the 16S rDNA sequence of Leptolyngbya sp. CENA104 were grouped with two Leptolyngbya sequences from GenBank and other Oscillatoriales members. Interestingly, the filaments of Leptolyngbya CENA104 grown in flasks containing liquid BG-11 medium were attached to the glass, forming a thin biofilm, while filaments from Leptolyngbya CENA103 and CENA112 formed aggregated clusters floating in the medium (Table 4). Nevertheless, both Leptolyngbya clades were clustered together in a major clade with bootstrap values of 62% NJ and 60% MP. Another Oscillatoriales clade, supported by high bootstrap values (100% NJ and 99% MP), was formed by the three Limnothrix strains (CENA109, CENA110, and CENA111). Distance and parsimony methods revealed that all sequences of Limnothrix strains were tightly clustered together with the two sequences of Limnothrix redekei, 165a and 165c, and with Planktothrix sp. FP1. The two Nostoc isolates (CENA105 and CENA107) clustered with Nostocales members, although they were in two distinct internal clades. Both Nostoc strains grouped with their closest relative, e.g., Nostoc CENA105 with Nostoc sp. 8941, and Nostoc CENA107 with Nostoc sp. strain ‘Mollenhauer 1:1-067’. The filamentous heterocystous cyanobacteria formed a monophyletic cluster supported by high bootstrap values (100% NJ and 97% MP).
Phylogenetic relationships between waste stabilization pond cyanobacterial isolates and related cyanobacteria based on 16S rDNA sequences (1,343 bp) with Escherichia coli K12 used as the outgroup. Sequences obtained in the present study are indicated in bold. Numbers near nodes indicate bootstrap values above 50% for NJ and MP analyses
Microcystin analysis
Specific ELISA assays for hepatotoxic microcystins performed on the 10 cyanobacterial isolates gave positive results for the six strains (Synechococcus CENA108, Merismopedia CENA106, Leptolyngbya CENA103, Leptolyngbya CENA112, Limnothrix CENA109, and Limnothrix CENA110) (Table 7). ELISA tests were also positive for wastewater samples collected from raw influent (3.70 μg/l) and treated effluent (3.74 μg/l) during the summer, and no microcystins were detected in the samples from the facultative pond central area.
Discussion
Cyanobacterial strains colonizing a WSP in Brazil have been isolated, cultured in laboratory and genetically characterized for the first time. In this study, two non-heterocystous forms (Limnothrix and Leptolyngbya), two unicellular forms (Synechococcus and Merismopedia), and one filamentous heterocystous-form (Nostoc) were isolated. However, the non-heterocystous forms (Limnothrix sp. CENA109, Limnothrix sp. CENA111, Leptolyngbya sp. CENA103, Leptolyngbya sp. CENA112, and Leptolyngbya sp. CENA104) were more representative than the unicellular and heterocystous forms obtained with our culturing method. In Brazil, the only study available is a floristic investigation of cyanobacterial strains conducted in WSP systems from Minas Gerais State, where Oscillatoria, Phormidium, Microcystis, and Anabaena were found (Von Sperling, 1996). Worldwide, few studies have focused on natural cyanobacteria diversity in WSP systems. Summer blooms containing Spirulina and Oscillatoria have been observed in urban-based sewage stabilization ponds in Delhi, India (Nandini, 1999). In a study conducted in Portugal, Microcystis aeruginosa, Planktothrix mougeotii and Pseudanabaena mucicola were the main species found in two WSPs (Vasconcelos & Pereira, 2001). In a WSP system in Marrakech, Morocco, two Synechocystis sp. strains and Pseudanabaena galeata were isolated (Oufdou et al., 2000; Oudra et al., 2002). Based on these studies, the most common morphotypes in WSPs are filamentous non-heterocyste-forming strains (Pseudanabaena, Planktothrix, Oscillatoria and Phormidium), followed by unicellular forms (Microcystis and Synechocystis) and filamentous heterocyste-forming strains (Anabaena). Therefore, to our knowledge, we provide here the first report of the occurrence of Synechococcus, Merismopedia, Leptolyngbya, Limnothrix, and Nostoc in a WSP system.
Our phylogenetic analysis of 16S rDNA sequences shows that strains belonging to the orders Chroococcales and Oscillatoriales were dispersed within the tree, indicating that they do not form natural clades. This result is consistent with the findings of other studies (Castenholz, 2001; Litvaitis, 2002; Seo & Yokota, 2003; Taton et al., 2006; Willame et al., 2006). The 16S rDNA sequence of Merismopedia CENA106 formed an internal clade with Merismopedia tenuissima 0BB46S01 within a major clade grouping of Synechococcus and Cyanobium strains. Previous studies reported that Merismopedia species are clustered together with Synechocystis (Palinska et al., 1996), and, more recently, with Snowella and Woronichinia. All these genera belong to the family Merismopediaceae (Rajaniemi-Wacklin et al., 2005). Although strain CENA106 forms rectangular colonies typical of Merismopedia (Herdman et al., 2001), it is genetically different from existing sequences and may belong to a different genus. In this study, the Merismopedia group was not phylogenetically coherent as previously reported (Rajaniemi-Wacklin et al., 2005), and requires further revision.
The three Limnothrix sp. 16S rDNA sequences, together with L. redekei 165a, L. redekei 165c, and Planktothrix sp. FP1, formed a separate cluster within the cyanobacterial 16S rDNA gene tree. The FP1 strain originating from a lake in Italy (Pomati et al., 2000) may have been misidentified as Planktothrix. As observed earlier (Gkelis et al., 2005), Planktothrix sp. FP1 trichomes share characteristics in common with typical Limnothrix rather then Planktothrix trichomes.
The coherent phylogenetic lineage of Leptolyngbya strains with members of Oscillatoria and Phormidium suggests that Leptolyngbya is not monophyletic. Previous studies have shown that strains assigned to the genus Leptolyngbya cluster with other members of the Oscillatoriales (Litvaitis, 2002; Marquardt & Palinska, 2006). However, reinvestigation of these members should be conducted since the divergent generic nomenclature employed by different authors can cause confusion and lead to misidentification.
The phylogenetic trees revealed good congruence with morphology-based classifications for the order Nostocales, which is in accordance with the previous findings (Urbach et al., 1992, 1998; Wilmotte, 1994; Nelissen et al., 1996; Garcia-Pichel et al., 1998; Litvaitis, 2002). Both Nostoc strains (CENA105 and CENA107) isolated from the Cajati WSP clustered with symbiotic Nostoc (8941 and ‘Mollenhauer 1:1-067’). Some free-living cyanobacteria may have symbiotic competence, and thus fall into a clade intermixed with symbiotic members (Svenning et al., 2005).
The environmental conditions of the Cajati WSP favor cyanobacterial growth. The high level of nutrients, mainly phosphorus and nitrogen, the alkaline conditions, and the high HRT contributed to cyanobacterial colonization. The abundance of cyanobacteria estimated by the direct microscope counting technique revealed that WSPs support a diverse assemblage of these organisms. Maximum counts were obtained in the summer (5.1 × 106 cells/ml). This estimate was higher than those obtained for a facultative pond from Portugal, where a maximum of 8.8 × 103 cyanobacterial cells/ml was observed (Vasconcelos & Pereira, 2001). It should be pointed out that the analysis of preserved samples collected in the Cajati facultative pond resulted in the identification of six cyanobacterial genera (Synechocystis sp., Aphanocapsa sp., Merismopedia punctata, Merismopedia tenuissima, Lyngbya sp., Phormidium sp. and Pseudanabaena sp.), whereas the isolation technique identified five genera (Synechococcus, Merismopedia, Limnothrix, Leptolyngbya, and Nostoc), with only Merismopedia being detected by both techniques. Therefore, these two techniques were complementary and allowed for a better estimation of the cyanobacterial population. Furthermore, the few representative taxa obtained using the isolation techniques are congruent with literature data, since culture-based methods usually underestimate the cyanobacterial population owing to strain selectivity (Ward et al., 1998). In addition, the low diversity may be dictated by environmental constraints. Despite the limitations of culturing methods, the isolation and characterization of cyanobacterial strains remain extremely important for diversity studies, since the isolates provide a link between genotypic and phenotypic features, allowing for a better understanding of cyanobacterial physiology and autoecology.
The presence of the hepatotoxin microcystin was detected only in the wastewater samples collected in summer. The concentrations of microcystins in the raw influent (3.70 μg/l) and treated effluent (3.74 μg/l) from summer were nearly identical, while no microcystins were found in the central sampling of facultative pond. This result may be partly explained by the tendency of cyanobacterial cells to accumulate along the edges of water bodies due to wind action, and by the existence of microcystin-degrading bacteria. It is well known that several bacteria can degrade microcystins (Jones et al., 1994; Bourne et al., 1996; Park et al., 2001; Tsuji et al., 2005; Amé et al., 2006) and bacterial communities are present in WSP (Daims et al., 2006). Besides the detection of microcystins in the summer wastewater samplings, six microcystin-producing cyanobacterial isolates (Synechococcus CENA108, Merismopedia CENA106, Leptolyngbya CENA103, Leptolyngbya CENA112, Limnothrix CENA109, and Limnothrix CENA110) were identified by ELISA assay. Therefore, despite the negative ELISA assay for microcystins in the WPS samples collected in autumn, the potential for microcystin production still exists, since Leptolyngbya CENA112, Limnothrix CENA109, and Limnothrix CENA110 were isolated from these samples. The presence of microcystins in a Brazilian WSP system has already been observed during a cyanobacterial bloom in a pond at São Lourenço da Serra, São Paulo (Furtado, 2003). Microcystins were detected by ELISA assay at a concentration of 4.49 μg/l and direct microscopic inspection of this sample revealed the presence of two Chroococcaleans, Radiocystis fernandoi and Microcystis panniformis, as the dominant species. An ELISA test specific for microcystins performed in the maturation and outflow ponds of two WSPs in Portugal, where M. aeruginosa was the dominant species, showed microcystin concentrations varying from 2.3 to 56.0 μg/l and 1.7 to 4.6 μg/l, respectively (Vasconcelos & Pereira, 2001). Synechocystis sp. and Pseudanabaena galeata isolated from a WSP system in Marrakech, Morocco, showed microcystin concentrations analyzed by HPLC of 842.5 and 57.0 μg/g, respectively (Oudra et al., 2002).
Most cyanotoxin research has been conducted on planktonic cyanobacteria because of the potential for bloom forming; however, recent studies show that benthic cyanobacteria can also produce toxins (Aboal et al., 2005; Mohamed et al., 2006; Izaguirre et al., 2007; Richardson et al., 2007). Of the five cyanobacterial genera isolated from the Cajati WSP, certain strains of Nostoc (Sivonen et al., 1990), Leptolyngbya (Richardson et al., 2007), and Phormidium (Aboal et al., 2005) have been reported as microcystin producers. Furthermore, two Leptolyngbya strains (NPLJ-34 and NPLJ-35), a Synechococcus elongatus strain (NPLB-1) and a Nostoc muscorum strain (NPBR-3) isolated from Brazilian water bodies, have been reported as hepatotoxic, although the specific toxin was not identified (Azevedo & Magalhães, 2002). The strain CENA106 isolated in this study is the first identified Merismopedia capable of producing microcystin.
This study has provided genetic information on cyanobacteria isolated from a WSP environment for the first time, since only morphological descriptions existed previously. In addition, the novel 16S rDNA sequences established have improved our taxonomic analysis of cyanobacteria by including strains from WSPs. While microcystin-producing cyanobacteria have been identified, treated effluents containing cyanotoxins can be discharged into rivers, irrigation channels, estuaries, or reservoirs representing a human health hazard. This finding emphasizes the need to introduce monitoring activities with regard to toxic cyanobacterial species, including analysis of the cyanobacterial toxins.
References
Aboal, M., M. A. Puig & A. D. Asencio, 2005. Production of microcystins in calcareous Mediterranean streams: the Alharabe River, Segura River basin in south-east Spain. Journal of Applied Phycology 17: 231–243.
Allen, M. M., 1968. Simple conditions for growth of unicellular blue-green algae on plates. Journal of Phycology 4: 1–4.
Altschul, S. F., W. Gish, M. Miller, E. W. Myers & D. J. Lipman, 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410.
Amé, V., R. Echenique, S. Pflugmacher & A. Wunderlin, 2006. Degradation of microcystin-RR by Sphingomonas sp. CBA4 isolated from San Roque reservoir (Córdoba – Argentina). Biodegradation 17: 447–455.
ANZECC & ARMCANZ, 2000. Australian and New Zealand guidelines for fresh and marine water quality; Australian and New Zealand Environmental and Conservation Council (AZECC) and Agriculture and Resource Management Council of Australia and New Zealand (ARMCANZ), Canberra [http://www.rpdc.tas.gov.au/soer/source/718/index.php].
APHA (American Public Health Association). American Water Works Association, and Water Pollution Control Federation, 1998. Standard Methods for the Examination of Water and Wastewater, 20th ed. APHA, Washington, DC.
Azevedo, S. M. F. O. & V. F. Magalhães, 2002. Causes and consequences of the presence of toxic cyanobacteria in Brazilian ecosystems. In Sar, E. A., M. E. Ferrario & B. Reguera (eds), Harmful Algal Blooms in South America. Spanish Institute of Oceanography, Madrid, Spain: 226–228.
Birnboim, H. C. & J. Doly, 1979. Rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research 7: 1513–1523.
Bourne, D. G., G. J. Jones, R. L. Blakeley, A. Jones, A. P. Negri & P. Riddles, 1996. Enzymatic pathway for the bacterial degradation of the cyanobacterial cyclic peptide toxin microcystin LR. Applied and Environmental Microbiology 62: 4086–4094.
Castenholz, R. W., 2001. Phylum BX. Cyanobacteria. Oxygenic photosynthetic bacteria. In Garrity, G., D. R. Boone & R. W. Castenholz (eds), Bergey’s Manual of Systematic Bacteriology. Volume 1: The Archaea and the Deeply Branching and Phototropic Bacteria, 2nd edition. Springer-Verlag, New York: 473–599.
Chernicharo, C. A. L., L. Florencio, R. K. X. Bastos, R. P. Piveli, M. Von Sperling & L. O. Monteggia, 2006. Wastewater treatment and effluent production to adequate different types of water reuse. In Florencio, L., R. K. X. Bastos & M. M. Aisse (eds), PROSAB – Treatment and Utilization of Sanitary Sewage. ABES, Rio de Janeiro: 75–76.
CONAMA (Brazilian National Environmental Council), 2005. Resolution 357, Establishes classes of bodies of water, sets forth guidelines for their classification and imposes conditions and standards for the discharge of effluents, among other measures. Ministry of the Environment.
Daims, H., M. W. Taylor & M. Wagner, 2006. Wastewater treatment: a model system for microbial ecology. Trends in Biotechnology 24: 483–489.
Fiore, M. F., D. H. Moon, S. M. Tsai, H. Lee & J. T. Trevors, 2000. Miniprep DNA isolation from unicellular and filamentous cyanobacteria. Journal of Microbiological Methods 39: 159–169.
Furtado, A. L. F. F., 2003. Assessment by denaturing gradient gel electrophoresis (DGGE) of Microcystis (Cyanobacteria) species in a stabilization ponds system in the city of São Lourenço da Serra (Vale do Ribeira de Iguape)-SP. M.Sc. dissertation, São Carlos Engineering School-Hydraulic and Sanitation Department, University of São Paulo: 108 pp.
Garcia-Pichel, F., U. Nubel & G. Muyzer, 1998. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Archives of Microbiology 169: 469–482.
Gkelis, S., P. Rajaniemi, E. Vardaka, M. Moustaka-Gouni, T. Lanaras & K. Sivonen, 2005. Limnothrix redekei (Van Goor) Meffert (Cyanobacteria) strains from lake Kastoria, Greece form a separated phylogenetic group. Microbial Ecology 49: 176–182.
Herdman, M., R. W. Castenholz, I. Iteman & R. Rippka, 2001. Form-genus XIV. Synechocystis. In Garrity, G., D. R. Boone & R. W. Castenholz (eds), Bergey’s Manual of Systematic Bacteriology. Volume 1: The Archaea and the Deeply Branching and Phototropic Bacteria, 2nd edition. Springer-Verlag, New York: 512–514.
Izaguirre, G., A. D. Jungblut & B. A. Neilan, 2007. Benthic cyanobacteria (Oscillatoriaceae) that produce microcystin-LR, isolated from four reservoirs in southern California. Water Research 41: 492–498.
Jones, G. J., G. Bourne, R. L. Blakeley & H. Doelle, 1994. Degradation of the cyanobacterial hepatotoxin microcystin by aquatic bacteria. Natural Toxins 2: 228–235.
Komárek, J. & K. Anagnostidis, 1989. Modern approach to the classification system of cyanophytes, 4 – Nostocales. Algological Studies 56: 247–345.
Komárek, J. & K. Anagnostidis, 1999. Cyanoprokaryota, Part 1: Chroococcales, Süsswasserflora von Mitteleuropa, Bd 19/1. Gustav Fischer Verlag, Stuttgart.
Komárek, J. & K. Anagnostidis, 2005. Cyanoprokaryota, Part 2: Oscillatoriales, Süsswasserflora von Mitteleuropa, Bd 19/2. Spektrum Akademischer Verlag, Heidelberg.
Kumar, S., K. Tamura & M. Nei, 2004. MEGA3, integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5: 150–163.
Lane, D. J., 1991. 16S/23S rRNA sequencing. In Stackebrandt, E. & M. Goodfellow (eds), Nucleic Acid Techniques in Bacterial Systematics. Wiley, Chichester: 115–175.
Litvaitis, M. K., 2002. A molecular test of cyanobacterial phylogeny: inferences from constraint analyses. Hydrobiologia 468: 135–145.
Marquardt, J. & K. A. Palinska, 2006. Genotypic and phenotypic diversity of cyanobacteria assigned to the genus Phormidium (Oscillatoriales) from different habitats and geographical sites. Archives of Microbiology 187: 397–413.
Mohamed, Z. A., M. H. El-Sharouny & W. S. M. Ali, 2006. Microcystin production in benthic mats of cyanobacteria in the Nile River and irrigation canals, Egypt. Toxicon 47: 584–590.
Nandini, S., 1999. Variations in physical and chemical parameters and plankton community structure in a series of sewage-stabilization ponds. Revista de Biologia Tropical 47: 149–156.
Neilan, B. A., D. Jacobs, T. D. Dot, L. L. Blackall, P. R. Hawkins, P. T. Cox & A. E. Goodman, 1997. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. International Journal of Systematic Bacteriology 47: 693–697.
Nelissen, B., R. De Baere, A. Wilmotte & R. De Wachter, 1996. Phylogenetic relationships of nonaxenic filamentous cyanobacterial strains based on 16S rRNA sequences analysis. Journal of Molecular Evolution 42: 194–200.
Oudra, B., M. Loudiki, V. Vasconcelos, B. Sabour, B. Sbiyyaa, K. H. Oufdou & N. Mezrioui, 2002. Detection and quantification of microcystins from cyanobacteria strains isolated from reservoirs and ponds in Morocco. Environmental Toxicology 17: 32–39.
Oufdou, K., N. Mezrioul, B. Oudra, M. Barakate, M. Loudiki & A. A. Alla, 2000. Relationship between bacteria and cyanobacteria in the Marrakech waste stabilization ponds. Water Science and Technology 42: 171–178.
Palinska, K. A., W. Liesack, E. Rhiel & W. E. Krumbein, 1996. Phenotype variability of identical genotypes: the need for a combined approach in cyanobacterial taxonomy demonstrated on Merismopedia-like isolates. Archives of Microbiology 166: 224–233.
Park, H.-D., Y. Sasaki, T. Maruyama, E. Yanagisawa, A. Hiraishi & K. Kato, 2001. Degradation of the cyanobacterial hepatotoxin microcystin by a new bacterium isolated from a hypertrophic lake. Environmental Toxicology 16: 337–343.
Pomati, F., S. Sacchi, C. Rosseti, S. Giovannardi, H. Onodera, Y. Oshima & B. A. Neilan, 2000. The freshwater cyanobacterium Planktothrix sp. FP1: molecular identification and detection of paralytic shellfish poisoning toxins. Journal of Phycology 36: 553–562.
Rajaniemi-Wacklin, P., A. Rantala, M. A. Mugnai, S. Turicchia, S. Ventura, J. Komárková, L. Lepistö & K. Sivonen, 2005. Correspondence between phylogeny and morphology of Snowella spp. and Woronichinia naegeliana, cyanobacterial commonly occurring in lakes. Journal of Phycology 42: 226–232.
Richardson, L. L., R. Sekar, J. L. Myers, M. Gantar, J. D. Voss, L. Kaczmarsky, E. R. Remily, G. L. Boyer & P. V. Zimba, 2007. The presence of the cyanobacterial toxin microcystin in black band disease of corals. FEMS Microbiology Letters 272: 182–187.
Seo, P.-S. & A. Yokota, 2003. The phylogenetic relationships of cyanobacteria inferred from 16S rRNA, gyrB, rpoC1 and rpoD1 gene sequences. Journal of General and Applied Microbiology 49: 191–203.
Sivonen, K. & G. Jones, 1999. Cyanobacterial toxins. In Chorus, I. & J. Bartram (eds), Toxic Cyanobacteria in Water. A Guide to Their Public Health Consequences Monitoring and Management. E and FN Spon, London: 41–111.
Sivonen, K., W. W. Carmichael, M. Namikoshi, K. L. Rinehart, A. M. Dahlem & S. I. Niemelän, 1990. Isolation and characterization of hepatotoxic microcystin homologues from the filamentous freshwater cyanobacterium Nostoc sp., strain 152. Applied and Environmental Microbiology 56: 2650–2657.
Svenning, M. M., T. Eriksson & U. Rasmussen, 2005. Phylogeny of symbiotic cyanobacteria within the genus Nostoc based on 16S rDNA sequence analysis. Archives of Microbiology 183: 19–26.
Taton, A., S. Grubisic, D. Ertz, D. A. Hodgson, R. Piccardi, N. Biondi, M. R. Tredici, M. Mainini, D. Losi, F. Martinelli & A. Wilmotte, 2006. Polyphasic study of Antarctic cyanobacterial strains. Journal of Phycology 42: 1257–1270.
Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin & D. G. Higgins, 1997. The CLUSTAL_X windows interface, flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Research 25: 4876–4882.
Tsuji, K., M. Asakawa, Y. Anzai, T. Sumino & K. Harada, 2005. Degradation of microcystins using immobilized microorganism isolated in an eutrophic lake. Chemosphere 65: 117–124.
Urbach, E., D. L. Robertson & S. W. Chisholm, 1992. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature 355: 267–270.
Urbach, E., D. J. Scanlan, D. L. Distel, J. B. Waterbury & S. W. Chisholm, 1998. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). Journal of Molecular Evolution 46: 188–201.
Vasconcelos, V. M. & E. Pereira, 2001. Cyanobacteria diversity and toxicity in a wastewater treatment plant (Portugal). Water Research 35: 1354–1357.
Von Sperling, M., 1996. Stabilization Ponds. Principles of Biological Treatment of Wastewater, Volume 3. Universidade Federal de Minas Gerais, Belo Horizonte, Brazil.
Ward, D. M., M. J. Ferris, S. C. Nold & M. M. Bateson, 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiology and Molecular Biology Reviews 62: 1353–1370.
Willame, R., C. Boutte, S. Grubisic, A. Wilmotte, J. Komárek & L. Hoffmann, 2006. Morphological and molecular characterization of planktonic cyanobacteria from Belgium and Luxembourg. Journal of Phycology 42: 1312–1332.
Wilmotte, A., 1994. Molecular evolution and taxonomy of the cyanobacteria. In Briant, D. A. (ed.), The Molecular Biology of Cyanobacteria. Kluwer, Dordrecht: 1–25.
Acknowledgments
We thank the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for financial support (2002/13449-1 to M.C.C. and 2005/56303-5 to M.F.F.). A.L.F.F. Furtado and R.Y. Honda received graduate scholarships from CAPES (Ministry of Education Agency) and A.S. Lorenzi received a graduate scholarship from CNPq (National Council for Scientific and Technological Development—140327/2004-5). D.B. Genuário was supported by FAPESP graduate scholarship (2007/06360-8). M.F. Fiore would also like to thank CNPq for a research fellowship (311094/2006-6). We thank Dr. A.C.P. Miwa for the algae and cyanobacteria counting and Dr. L.C.R. Pessenda (Carbon 14 Laboratory, CENA/USP) for permission to use the Axioskop 40 microscope.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: D. Hamilton
Rights and permissions
About this article
Cite this article
Furtado, A.L.F.F., Calijuri, M.d.C., Lorenzi, A.S. et al. Morphological and molecular characterization of cyanobacteria from a Brazilian facultative wastewater stabilization pond and evaluation of microcystin production. Hydrobiologia 627, 195–209 (2009). https://doi.org/10.1007/s10750-009-9728-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-009-9728-6