Abstract
Satellite cells, muscle-specific stem cells, are anatomically identified as the mononuclear cells residing external to the myofiber plasma membrane and beneath the basal lamina. Skeletal muscle has great regenerative potential, and the regeneration process depends absolutely on satellite cells. In uninjured muscle, satellite cells are maintained in a quiescent state, and some genes are expressed in a quiescent-specific manner. Here we show that Odz4/Ten-m4, a mouse homolog of the Drosophila pair-rule gene odd Oz (odz or Ten-m), is expressed in quiescent satellite cells on the protein level, but not in activated/proliferating myoblasts. Intriguingly, the timing of the reappearance of Odz4 and calcitonin receptor (another quiescence molecule) on Pax7-positive cells was different during the regeneration process. In addition, almost all neonatal satellite cells express Odz4, but only some of them express calcitonin receptor. These results indicate that Odz4 may be useful as a new marker of satellite cells and that quiescence molecules are differently expressed in regenerating and neonatal muscle.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Satellite cells are located beneath the basal lamina and maintained in an undifferentiated quiescent state in adult skeletal muscle (Mauro 1961; Schultz et al. 1978). When a muscle is damaged, satellite cells exit from quiescence and start to proliferate (Charge and Rudnicki 2004). Proliferating satellite cells, called myoblasts, then differentiate, fuse with each other or with injured myofibers, and eventually regenerate mature myofibers. During this process, satellite cells self-renew in anticipation of future damage.
Satellite cells were first identified by their specific location in frog skeletal muscle by electronic microscopy (Mauro 1961). Now we easily detect satellite cells using specific antibodies and microscopy. Among the antibodies, Pax7 is the most accepted marker for satellite cells (Seale et al. 2000). Satellite cells specifically express Pax7 in skeletal muscle, and they require it during postnatal development but not in adulthood (Lepper et al. 2009). In addition to Pax7, many other molecules (including CD34, CXCR4, c-met, m-cadherin, NCAM, integrin-β1, integrin-α7, SM/C-2.6, Syndecan3, and 4) have been used to identify satellite cells by microscopy or flow cytometry (Beauchamp et al. 2000; Cornelison et al. 2001; Fukada et al. 2004; Irintchev et al. 1994; Sherwood et al. 2004; Tatsumi et al. 1998). However, none of these markers is specific to quiescent satellite cells, and myoblasts also express these markers (Kuang and Rudnicki 2008). The identification of a quiescent satellite cell-specific molecule will allow us to elucidate the cellular and molecular mechanisms of satellite cell self-renewal and maintenance.
Previously we compared quiescent satellite cells with activated/proliferating satellite cells (myoblasts) and non-myogenic cells in skeletal muscle, and identified quiescent satellite cell-specific genes including calcitonin receptor and Odz4 (Fukada et al. 2007). Odz is the vertebrate homologue of the Drosophila odd Oz, which is known as a pair-rule gene (Levine et al. 1994). Odz family proteins belong to the type II transmembrane protein family (Oohashi et al. 1999). The functions of the Odz family are largely unknown, but Bagutti et al. indicated that the intracellular domain of teneurin2 (an ortholog of Odz2 in birds) localizes in the nucleus and functions as a transcription repressor like Notch (Bagutti et al. 2003). Here, we show that Odz4 is expressed in quiescent satellite cells but not in activated/proliferating myoblasts on a protein level. Intriguingly, in the middle stage of regeneration, Pax7-positive cells express Odz4, but not calcitonin receptor. In addition, almost all neonatal satellite cells express Odz4, but only some of them express calcitonin receptor. These results indicate that Odz4 is a marker of satellite cells, and that quiescence genes are differently expressed during the regeneration process and in neonatal muscle.
Materials and methods
Mice
Eight- to 12-week-old C57BL/6 mice were purchased from Charles River, Japan. All procedures for experimental animals were approved by the Experimental Animal Care and Use Committee at Osaka University.
Muscle injury
Muscle was injured by injecting 50, 150, and 100 μl cardiotoxin (10 μM in saline, Wako Pure Chemical Industries, Tokyo, Japan) into the tibialis anterior (TA), gastrocnemius (GC), and quadriceps (Qu) muscle, respectively.
Antibodies
Rabbit anti-calcitonin receptor polyclonal antibody was purchased from AbD Serotec Ltd. (Oxford, UK). Rabbit anti-Odz4 polyclonal antibody was kindly provided by Dr. Reinhard Faessler (Zhou et al. 2003). Rat anti-laminin α2 monoclonal antibody (clone 4H8-2) was purchased from Alexis Biochemical (Lausen, Switzerland). Hybridoma of mouse anti-Pax7 antibody was purchased from the Developmental Studies Hybridoma Bank (Iowa City, IA), and the cell supernatant was prepared for immunostaining in our laboratory. FITC-conjugated anti-mouse CD31, FITC conjugated anti-mouse CD45, and PE (phycoerythrin)-conjugated anti-mouse Sca-1 antibodies were purchased from BD PharMingen (San Diego, CA, USA). Biotinylated SM/C-2.6 antibody was produced in our laboratory (Fukada et al. 2004).
Preparation and FACS analysis of skeletal muscle-derived mononuclear cells
Mononuclear cells from uninjured limb muscles were prepared using 0.2 % collagenase type II (Worthington Biochemical Corp., Lakewood, NJ, USA) as previously described (Uezumi et al. 2006).
Mononuclear cells derived from skeletal muscle were stained with FITC-conjugated anti-CD31, CD45, PE-conjugated anti-Sca-1, and biotinylated-SM/C-2.6 antibodies. Cells were then incubated with streptavidin-labeled allophycocyanin (BD Biosciences, San Diego, CA, USA) on ice for 30 min, and resuspended in PBS containing 2 % FCS and 2 μg/ml propidium iodide (PI). Cell sorting was performed using a FACS Aria II™ flow cytometer (BD Immunocytometry Systems, Mountain View, CA, USA). Debris and dead cells were excluded by forward scatter, side scatter, and PI gating. Data were collected using FACSDiva™ software (BD Biosciences). Myogenic cells from regenerating muscle are also highly enriched in the SM/C-2.6(+)CD31(−)CD45(−)Sca-1(−) cell fraction (Segawa et al. 2008).
RT-PCR
Total RNA was extracted from sorted cells using a Qiagen RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany) and then reverse-transcribed into cDNA by using TaqMan Reverse Transcription Reagents (Roche Diagnostics, Mannheim, Germany). The polymerase chain reaction (PCR) was performed with cDNA and specific primers. The PCR primers were calcitonin receptor forward, 5′-GAC CCT CAA AAT CTC CT CTC AC; calcitonin receptor reverse, 5′-ACA TCC TTC CTT CAA TCC GTC C; Odz4 foward, 5′-GGA CAA CAG TGG ATG AGG AAA AGG; Odz4 reverse, 5′-TCA CAA AGA AGC CGT CGT AGC C; Pax7 forward, 5′-GAA AGC CAA ACA CAG CAT CGA; Pax7 reverse, 5′-ACC CTG ATG CAT GGT TGA TGG; Hprt forward, 5′-CTT TGC TGA CCT GCT GGA TTA CAT; and Hprt reverse, 5′-GTC AAG GGC ATA TCC AAC AAC AAA.
Immunohistochemistry
For immunohistochemical examinations, transverse cryosections (6 μm) were fixed in 4 % paraformaldehyde for 10 min. After blocking with 5 % skim milk, sections were then stained with primary antibodies. For Pax7 staining, an M.O.M. kit (Vector Laboratories Inc., Burlingame, CA, USA) was used to block endogenous mouse IgG. After the first staining at 4 °C overnight, sections were reacted with secondary antibodies conjugated with Alexa 488, Alexa 568, or Alexa 647 (Molecular Probes, Eugene, OR, USA). Coverslips were mounted using Vectashield (Vector Laboratories Inc).
Results
Odz4 is expressed on quiescent satellite cells
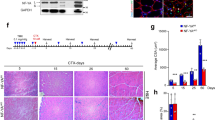
Our previous study demonstrated the mRNA expression of Odz4 in quiescent satellite cells (Fukada et al. 2007). To examine the mRNA expression of Odz4 during skeletal muscle regeneration, myogenic cells were isolated by flow cytometry and RT-PCR was performed. As shown in Fig. 1, the Odz4 transcript was detected in quiescent satellite cells, but not in myogenic cells derived from regenerating muscle 2–3 days after cardiotoxin (CTX) injection. Odz4 mRNA was again detected at 5 days after injury. Consistent with our previous results, the reappearance of calcitonin receptor mRNA occurred in the later stage of muscle regeneration. Although both calcitonin receptor and Odz4 were identified as ‘quiescence genes’, the timing of the reappearance of the two genes was completely different during the regeneration process.
mRNA expression of Odz4 and calcitonin receptor in myogenic cells during skeletal muscle regeneration. RT-PCR was performed to examine the expression levels of Odz4 and calcitonin receptor (CTR) in quiescent satellite cells (QSC), myogenic cells derived from injured muscle 2–14 days after cardiotoxin injection (CTX). H2O was used as a negative control
To reveal the expression of Odz4 on a protein level, uninjured muscle sections were stained with anti-Odz4, laminin α2, and TOTO3. As shown in Fig. 2a, Odz4 protein was detected at the surface of mononuclear cells located beneath the basal lamina. Combined immunostaining of Odz4 and Pax7, a satellite cell marker, also showed that satellite cells expressed Odz4 on a protein level (Fig. 2b).
Odz4 protein expressed on quiescent satellite cells. Uninjured TA muscles were dissected, and cross sections were stained with an antibody to Odz4 (green). A section was co-stained with laminin α2 (LNα2; red) (a) or anti-Pax7 (red) (b) antibodies. Nuclei were stained with TOTO3 (blue). Scale bar 20 μm. (Color figure online)
We next stained a cross section of injured muscle with anti-Odz4 antibody. Three day after cardiotoxin injection, many activated satellite cells were stained with anti-M-cadherin antibody (Fig. 3a), but Odz4 was not detected in activated satellite cells on the serial sections (Fig. 3b). These results indicate that Odz4 is down-regulated after satellite cell activation/proliferation like calcitonin receptor (Fukada et al. 2007).
Odz4 protein not expressed on activated/proliferating myoblasts. a 3 days after cardiotoxin (CTX) injection, muscles were dissected, and cross sections were stained with antibodies to M-cadherin (green) and LNα2 (red). b Serial sections were stained with anti-Odz4 (green) and LNα2 (red) antibodies. Nuclei were stained with TOTO3 (blue). Scale bar 40 μm. (Color figure online)
Odz4 and calcitonin receptor expression in middle stage of muscle regeneration
To examine the reappearance of Odz4 during skeletal muscle regeneration, cross sections of the muscle 7 days after cardiotoxin injection were stained with anti-Odz4 antibody. As shown in Fig. 4, Pax7+ Odz4+ cells were found in the middle stage of muscle regeneration. In contrast, calcitonin receptor was not observed on Pax7+ cells located at small regenerating myofibers, as described previously (Fukada et al. 2007). These results indicated that both Odz4 and calcitonin receptor are expressed in quiescent satellite cells, but the regulation and timing of their reappearance are different.
Reappearance of Odz4+, but not CTR+ satellite cells at 7 days after cardiotoxin injection. TA muscles were sampled at 7 days after cardiotoxin injection. Sections were co-immunostained with anti-CTR (green) or Odz4 (green), Pax7 (red), and LNa2 (white) antibodies. Nuclei were stained with DAPI. Arrowheads indicate Pax7+ Odz4+ cells. Scale bar 50 μm. (Color figure online)
Expression of Odz4 and calcitonin receptor in neonatal stage
During postnatal development, satellite cells actively proliferate and contribute to myofiber growth. Pallafacchina et al. reported that the gene expression profiles of neonatal satellite cells and activated proliferating myoblasts are different (Pallafacchina et al. 2010). To examine the expression of Odz4 and calcitonin receptor in the neonatal period, hind limb skeletal muscles of mice 7 days after birth were fixed and stained with anti-Odz4 or calcitonin receptor antibodies. As shown in Fig. 5, more than 95 % of Pax7+ cells expressed Odz4. On the other hand, approximately 30 % of Pax7+ cells expressed calcitonin receptor in this stage. These results also indicate that Odz4 and calcitonin receptor are differently expressed in satellite cells, and that neonatal satellite cells are different from early proliferating myoblasts during skeletal muscle regeneration.
Expression of CTR and Odz4 in neonatal skeletal muscle. Sections of neonatal muscles were co-immunostained with antibodies to CTR (green), Odz4 (green), Pax7 (red), and LNα2 (white). Nuclei were stained with DAPI. Arrowheads indicate Pax7+ Odz4+ or Pax7+ CTR+ cells. Arrows indicate Pax7+ CTR−/low cells. Scale bar 30 μm. (Color figure online)
Discussion
Here, we showed that Odz4 is expressed in quiescent satellite cells, like calcitonin receptor, but the timing of the reappearance of these genes is different during regeneration. Odz4 is expressed in the middle stage of muscle regeneration, but calcitonin receptor is not. On the other hand, neither Odz4 or calcitonin receptor is expressed in activated/proliferating satellite cells. These results suggest that the characteristics of myogenic cells in the middle stage are different from quiescent and activated/proliferating satellite cells. The characterization of myogenic cells in the middle stage will afford a better understanding of skeletal muscle regeneration.
The Odz family belongs to a type II transmembrane protein family that possesses a C-terminal extracellular domain, transmembrane region, and intracellular domain. Three postulated protease cleavage sites exist in the extra- and transmembrane domains of Odzs (Tucker and Chiquet-Ehrismann 2006). Functional studies of Drosophila indicated that Odz functions as a pair-rule gene (Levine et al. 1994). This is a somewhat surprising because Odz is a cell-surface protein, whereas all other pair-rule genes are transcriptional factors. Intriguingly, the intracellular domain of Odz2 was detected in a human cell line, HT1080 cells (Bagutti et al. 2003). In C. elegans, localization of the intracellular domain of Ten-1 (the C. elegans orthlog of odz) was also demonstrated by using a specific antibody (Drabikowski et al. 2005). Furthermore, it is reported that Odz2 is cleaved at the intracellular domain and works as transcriptional regulator of target genes (Bagutti et al. 2003). Such a mechanism is well known for other signaling molecules, the most prominent example of which is Notch. When Notch is activated, the intracellular domain of Notch is cleaved by γ-secretase and translocates to the nucleus where it activates transcription of target genes through interaction with RBP-J (recombination signal binding protein for immunoglobulin kappa J region, also known as CBF1). Odzs are prominently found in the nervous system in adult mice (Zhou et al. 2003). Although the roles of Odzs are not fully understood, overexpression studies have indicated that Odz3 can promote neurite outgrowth and cell adhesion in vivo (Leamey et al. 2008). In this study, we used an antibody that recognized the extracellular domain of Odz4. Therefore, it is unclear whether Odz4 acts as a transcriptional regulator in satellite cells. The functional analysis of Odz4 might reveal the importance of Odz4 as a transcriptional regulator like Notch signaling in satellite cells.
Recently, it has been reported that microRNA-489, resides in intron 4 of the gene encoding calcitonin receptor, maintains the quiescent state of satellite cells (Cheung et al. 2012). As well as calcitonin receptor, the expression of microRNA-489 is dramatically decreased after satellite cell activation. Intriguingly, in satellite cells, the expression of microRNA-708 is also downregulated after activation, and it is encoded in the intron 1 of Odz4. Therefore, Odz4 and microRNA-708 might play some roles in satellite cells as well as calcitonin receptor and microRNA-489.
In conclusion, we demonstrated the expression of Odz4 in satellite cells in addition to calcitonin receptor. However, the staining patterns of both neonatal and regenerating muscle were different. These finding suggest that the Odz4+ CTR-Pax7+ cell population might have a specific function in the skeletal muscle regeneration process, and further investigation will increase the depth of understating of satellite cell biology.
References
Bagutti C, Forro G, Ferralli J, Rubin B, Chiquet-Ehrismann R (2003) The intracellular domain of teneurin-2 has a nuclear function and represses zic-1-mediated transcription. J Cell Sci 116:2957–2966
Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS (2000) Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol 151:1221–1234
Charge SB, Rudnicki MA (2004) Cellular and molecular regulation of muscle regeneration. Physiol Rev 84:209–238
Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA (2012) Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482:524–528
Cornelison DD, Filla MS, Stanley HM, Rapraeger AC, Olwin BB (2001) Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev Biol 239:79–94
Drabikowski K, Trzebiatowska A, Chiquet-Ehrismann R (2005) ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev Biol 282:27–38
Fukada S, Higuchi S, Segawa M, Koda K, Yamamoto Y, Tsujikawa K, Kohama Y, Uezumi A, Imamura M, Miyagoe-Suzuki Y, Takeda S, Yamamoto H (2004) Purification and cell-surface marker characterization of quiescent satellite cells from murine skeletal muscle by a novel monoclonal antibody. Exp Cell Res 296:245–255
Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S (2007) Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25:2448–2459
Irintchev A, Zeschnigk M, Starzinski-Powitz A, Wernig A (1994) Expression pattern of M-cadherin in normal, denervated, and regenerating mouse muscles. Dev Dyn 199:326–337
Kuang S, Rudnicki MA (2008) The emerging biology of satellite cells and their therapeutic potential. Trends Mol Med 14:82–91
Leamey CA, Glendining KA, Kreiman G, Kang ND, Wang KH, Fassler R, Sawatari A, Tonegawa S, Sur M (2008) Differential gene expression between sensory neocortical areas: potential roles for Ten_m3 and Bcl6 in patterning visual and somatosensory pathways. Cereb Cortex 18:53–66
Lepper C, Conway SJ, Fan CM (2009) Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460:627–631
Levine A, Bashan-Ahrend A, Budai-Hadrian O, Gartenberg D, Menasherow S, Wides R (1994) Odd Oz: a novel Drosophila pair rule gene. Cell 77:587–598
Mauro A (1961) Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol 9:493–495
Oohashi T, Zhou XH, Feng K, Richter B, Morgelin M, Perez MT, Su WD, Chiquet-Ehrismann R, Rauch U, Fassler R (1999) Mouse ten-m/Odz is a new family of dimeric type II transmembrane proteins expressed in many tissues. J Cell Biol 145:563–577
Pallafacchina G, Francois S, Regnault B, Czarny B, Dive V, Cumano A, Montarras D, Buckingham M (2010) An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res 4:77–91
Schultz E, Gibson MC, Champion T (1978) Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J Exp Zool 206:451–456
Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA (2000) Pax7 is required for the specification of myogenic satellite cells. Cell 102:777–786
Segawa M, Fukada S, Yamamoto Y, Yahagi H, Kanematsu M, Sato M, Ito T, Uezumi A, Hayashi S, Miyagoe-Suzuki Y, Takeda S, Tsujikawa K, Yamamoto H (2008) Suppression of macrophage functions impairs skeletal muscle regeneration with severe fibrosis. Exp Cell Res 314:3232–3244
Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ (2004) Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell 119:543–554
Tatsumi R, Anderson JE, Nevoret CJ, Halevy O, Allen RE (1998) HGF/SF is present in normal adult skeletal muscle and is capable of activating satellite cells. Dev Biol 194:114–128
Tucker RP, Chiquet-Ehrismann R (2006) Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development. Dev Biol 290:237–245
Uezumi A, Ojima K, Fukada S, Ikemoto M, Masuda S, Miyagoe-Suzuki Y, Takeda S (2006) Functional heterogeneity of side population cells in skeletal muscle. Biochem Biophys Res Commun 341:864–873
Zhou XH, Brandau O, Feng K, Oohashi T, Ninomiya Y, Rauch U, Fassler R (2003) The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr Patterns 3:397–405
Acknowledgments
We thank Dr. Reinhard Faessler for anti-Odz4 antibody. We thank Katherine Ono for reading this manuscript. This work was supported by a JSPS KAKENHI grant (18800023 to S.F.), MEXT KAKENHI grants (20700358 to S.F), Intramural Research Grant (22-1 to S.F.) for Neurological and Psychiatric Disorders of NCNP, and the Nakatomi Foundation (to S.F.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yamaguchi, M., Ogawa, R., Watanabe, Y. et al. Calcitonin receptor and Odz4 are differently expressed in Pax7-positive cells during skeletal muscle regeneration. J Mol Hist 43, 581–587 (2012). https://doi.org/10.1007/s10735-012-9421-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-012-9421-3