Abstract
The family of basic leucine zipper (bZIP) transcription factors plays diverse crucial roles in numerous biological processes. Despite the identification of bZIP genes in several plants, to our knowledge, bZIP members in watermelon and melon are yet to be comprehensively investigated. The genomes of watermelon and melon encode 59 ClabZIP and 75 CmbZIP putative genes, respectively. Both bZIP protein family members were phylogenetically grouped into seven subfamilies. The majority of bZIP genes in the same subfamily shared similar gene structures and conserved motifs. Chromosome distribution and genetic analysis revealed that 21 duplication events between ClabZIP genes and 106 duplication events between CmbZIP genes have occurred. Further, the three-dimensional structure and functional annotation of bZIP proteins was predicted. For evaluating the expression patterns of ClabZIP and CmbZIP genes, RNA-seq data available in public databases were analyzed. The expression profiles of selected ClabZIP and CmbZIP genes in root and leaf tissues of drought-stressed watermelon and melon were also examined using qRT-PCR. ClabZIP-57, CmbZIP-52, and CmbZIP-31 genes exhibited the highest expression levels after stress exposure in leaf and root tissues. Gene identification studies like the present study offer new perspectives in the analysis of bZIP protein family members and their functions in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Melon (Cucumis melo) and watermelon (Citrullus lanatus) belong to Cucurbits or the Cucurbitaceae family. They are economically highly valuable as fresh produce worldwide; approximately 111 and 29 million tons of watermelons and melons have been produced, respectively, and their cultivated area is measured at a total of approximately 4.7 million hectares (ha) throughout the world (Food and Agriculture Organization 2014). According to FAO reports, Turkey is the second-largest producer of watermelons (3.9 million tons) and melons (1.7 million tons) after China.
Transcription factors (TFs) play regulatory roles for their downstream target genes, which they may induce or repress. They comprise of two structures, namely sequence-specific DNA-binding and activation domains. Based on their 3D structures and DNA-binding sequence properties, TFs can be classified into 40–60 families in plants (Wingender et al. 2001; Yilmaz et al. 2009). The basic leucine zipper (bZIP) TF family is one of the most diverse TFs families in plants (Wang et al. 2015). They are described as containing a highly conserved bZIP domain, which is 60–80 amino acids in length, and which comprises a basic region and a leucine zipper dimerization motif (Wang et al. 2011). The basic region is responsible for nuclear localization and DNA binding, whereas the leucine zipper controls the homo- and/or hetero-dimerization of bZIP proteins (Jakoby et al. 2002; Nijhawan et al. 2008).
Like other TFs, bZIP TFs play significant roles in developmental and differentiation processes in plants. The roles of the family members encompass organ and tissue differentiation (Shen et al. 2007; Silveira et al. 2007), cell elongation (Fukazawa et al. 2000), nitrogen/carbon and energy metabolism (Baena-Gonzalez et al. 2007; Weltmeier et al. 2006), unfolded protein response (Iwata and Koizumi 2005; Liu et al. 2007), seed storage protein gene regulation (Lara et al. 2003) and somatic embryogenesis (Guan et al. 2009). In addition, bZIP TFs regulate signaling processes and play a role in the response to biotic/abiotic stresses. Reports have indicated that expression analyses of bZIP TFs have been examined in various plant species under different stress conditions, such as abscisic acid (ABA) signaling, hypoxia, drought, high salinity, cold stress, hormone and sugar signaling, light responses, osmotic stresses, and pathogen defense (Hsieh et al. 2010; Kobayashi et al. 2008; Lee et al. 2006; Lu et al. 2009; Rodriguez-Uribe and O’Connell 2006; Wang et al. 2015; Yang et al. 2009; Yoshida et al. 2010; Yun et al. 2010). The importance of bZIPs in biological processes and the regulation of stress mechanism in plants have led many researchers to focus on the bZIP gene family. Thus, the presence of many members of the bZIP TF family has been predicted in different plant species. bZIPs have been reportedly identified in Arabidopsis (Jakoby et al. 2002), rice (Nijhawan et al. 2008), soybean (Liao et al. 2008), carrot (Guan et al. 2009), sorghum (Wang et al. 2011), maize (Wei et al. 2012), cucumber (Baloglu et al. 2014), castor bean (Jin et al. 2014b), grapevine (Liu et al. 2014), Brachypodium (Liu and Chu 2015), tomato (Li et al. 2015a), barley (Pourabed et al. 2015), six legumes, (Glycine max, Medicago truncatula, Phaseolus vulgaris, Cicer arietinum, Cajanus cajan, and Lotus japonicus) (Wang et al. 2015), apple (Li et al. 2016), Chinese cabbage (Bai et al. 2016), cassava (Hu et al. 2016).
Recently, genome sequencing projects for members of the Cucurbitaceae family have been completed for cucumber (Huang et al. 2009), melon (Garcia-Mas et al. 2012) and watermelon (Guo et al. 2013). In addition to genome sequences, this family is an important model for sex determination and plant vascular biology studies (Lucas 2006). Although the complete genome sequences of these members elevate our understanding of important gene families, there is still not enough information about the identification and characterization of some TF families in Cucurbits. To our knowledge, our previous study was the first to perform genome-wide identification and expression analysis of bZIPs in cucumber (Baloglu et al. 2014). Due to the current unavailability of the descriptions of bZIPs in watermelon and melon, the present study aimed to examine all bZIP gene family members in all known genomes of Cucurbitaceae family members. With the availability of genome sequences of watermelon and melon, bZIP TF family genes were systematically investigated, compared, and analyzed in this study. Herein, all members of bZIP genes were determined in watermelon and melon genomes, and these genes were then analyzed for motif identification, phylogenetic classification, duplication, and determination of orthologs in different plant species. Furthermore, their expression analysis in different tissues and conditions were examined using both RNA-seq data available in public databases and qRT-PCR.
Materials and methods
Identification of bZIP genes in melon and watermelon genomes
Based on previous reports by the author (Baloglu 2014; Baloglu et al. 2014; Celik Altunoglu et al. 2017; Celik Altunoglu et al. 2016; Kavas et al. 2015, 2016; Yer et al. 2015), a variety of approaches were applied to elucidate bZIP genes from genomes of melon and watermelon. First, bZIP amino acid sequences from several organisms (Arabidopsis thaliana, Cucumis sativus, Gossypium hirsitum, Oryza sativa, G. max, Sorghum bicolor, Triticum aestivum, Triticum durum, Pisum sativum, Zea mays, Hordeum vulgare, Brassica napus, M. truncatula, Nicotiana tabacum, Vitis vinifera) were retrieved from TF database 3.0 (http://plntfdb.bio.uni-potsdam.de/v3.0/) (Jin et al. 2014a). Utilizing these sequences, a BLASTP search at Melonomics database (https://melonomics.net/tools/blast/run/) and Cucurbit Genomics database (http://cucurbitgenomics.org/blast) were performed against melon and watermelon genomes with an e-value cut-off of e−50, respectively. Second, the keyword “bZIP” was also sought in the databases. In addition, HMM profiles of bZIP proteins in the Pfam database (http://pfam.sanger.ac.uk/) were checked. Finally, the expressed sequence tag sequences of bZIP genes of cucumber, melon, and watermelon were used for TBLASTN search in the NCBI database to ensure the inclusion of all possible bZIP proteins.
Chromosomal location, gene structure and distribution of bZIP genes in genomes
The specific gene positions of bZIP genes on watermelon chromosomes were determined using the Cucurbit Genomics database (http://cucurbitgenomics.org/blast). Genes were individually mapped onto 11 watermelon chromosomes on the basis of their ascending order of physical position (bp) and presented using MapChart (Voorrips 2002). Gene structure analysis was performed using Gene Structure Display Server v2.0 (http://gsds.cbi.pku.edu.cn/) (Hu et al. 2015). Intra- and crossgenome relationships of bZIP genes were discovered using Plant Genome Duplication Database (Tang et al. 2008).
Identification of the conserved motifs and construction of phylogenetic tree
The specific motifs found in bZIP protein sequences were determined using MEME (http://meme.nbcr.net/meme3/meme.html) (Bailey et al. 2009). MEME motifs were also checked in the InterPro database with InterProScan, which provides the functional analysis of proteins by predicting their domains and important sites. The amino acid sequences were transferred to MEGA7, and multiple sequence alignments were conducted with CLUSTALW with gap open and gap extension penalties of 10 and 0.1, respectively (Kumar et al. 2016). The alignment file was then utilized for constructing the phylogenetic tree in accordance with the ML method, with bootstrap analysis for 1000 iterations. Finally, the tree was displayed using Interactive tree of life (iTOL; http://itol.embl.de/index.shtml) (Letunic and Bork 2011).
Gene ontology (GO) annotation
For functional annotation identification of bZIP proteins, bZIP protein sequences were imported into the Blast2GO server (http://www.blast2go.com) (Conesa and Götz 2008). First, BLASTP analysis and then mapping of GO terms on the basis of BLAST results were performed. Finally, the annotation of GO terms of each query were determined. The program classifies results into three categories: biological processes, cellular components, and molecular functions.
Comparison of bZIPs between melon–watermelon and other species
For understanding the orthologous relationships between bZIP genes among melon–watermelon and other species, amino acid sequences of melon–watermelon bZIPs were blasted against peptide sequences of Arabidopsis, cucumber, maize, rice, grape, and poplar in Phytozome with an e-value cutoff of e−50 and identity/positive cut-off of 60%.
Calculation of synonymous and non-synonymous substitution rates
The amino acid sequences of duplicated bZIP proteins and orthologous pairs between melon–watermelon and poplar, rice, Arabidopsis, and maize were arranged with CLUSTALW using multiple sequence alignment tool. The CODEML program (http://www.bork.embl.de/pal2nal/) was utilized for calculating synonymous (Ks) and non-synonymous (Ka) change ratios through the alignment of the amino acid sequences and their respective original cDNA sequences of bZIP genes (Suyama et al. 2006). Time (MYA) of the duplication and divergence of each bZIP gene was calculated using the formula T = Ks/2λ (λ = 6.5 × 10−9), and using mutation ratios corresponding with every synonymous area and every year (Lynch and Conery 2000; Yang et al. 2008).
3D protein homology modeling of bZIPs
For the identification of the best template match between sequence and known 3D structure, all bZIP proteins were blasted in PDB using default parameters (Berman et al. 2000). For predicting the 3D structure of bZIP proteins, the obtained data was fed into Phyre2 database (Protein Homology/AnalogY Recognition Engine; http://www.sbg.bio.ic.ac.uk/phyre2) (Kelley and Sternberg 2009); homology modeling analysis was conducted in “intensive” mode in Phyre2. Samples with more than 70% of amino acid residues modeled were considered significant with and confidence cut-off of 90%.
Expression analysis of bZIPs using transcriptome data
For evaluating bZIP gene expression profiles in different tissues and conditions for melon and watermelon, SRA (https://www.ncbi.nlm.nih.gov/sra), an open, public database, was utilized for the retrieval of Illimuna HiSeq reads for RNA-seq analysis. The accession numbers of raw sequencing data for watermelon are listed as follows: SRR1724899, SRR1724900, SRR1724901, SRR1724902, SRR1724903, SRR1724943, WM-UR-1/SRR1001435, WM-UR-2/SRR1001436, WM-IM-1/SRR1001437, WM-IM-2/SRR1001438, WM-PM-1/SRR1001439, WM-PM-2/SRR1001440, WM-MA-1/SRR1001441, WM-MA-2/SRR1001442, SRR494474, SRR518988, SRR518988, SRR494479, SRR518992, SRR518993; and for melon as follows: SRR411102, SRR411100, SRR411106, SRR411104, SRR1033647, SRR1033646, SRR2082958, SRR2082965, SRR2082865, SRR2082935, SRR2082943, SRR2082953, SRR2082831, SRR2082832, SRR2082790, SRR2082791, SRR2082796, SRR2082813.
All reads were downloaded in “.sra” format (raw data), and were converted to “fastq” format using the NCBI SRA Toolkit fastq-dump command. After the removal of low-quality reads (Q score < 20) and trimming adapters using CLC Genomics Workbench 7.5, the quality of the fastq files was checked using FastQC analysis in terms of per-base sequence qualities, per-sequence quality scores, per-base nucleotide content, and sequence duplication levels (Celik Altunoglu et al. 2017). A pipeline for RNA-seq analysis was constructed using CLC Genomics Workbench 7.5. Briefly, unique mapped reads were normalized, standardized, and the exon model reads per kilobase per million mapped reads (RPKM) were calculated to obtain gene expression values. For identifying differentially expressed bZIP genes, FDR value ≤ 0.001, fold change (RPKM-tr/RPKM-cont) ≥ 2, and the absolute ratio of log2 (RPKM-tr/RPKM-cont) ≥ 1 were used as threshold values (Kavas et al. 2016). Finally, the heat maps of hierarchical clustering were visualized using PermutMatrix (Caraux and Pinloche 2005).
Plant growth conditions and drought stress treatment
Melon and watermelon seeds were obtained from Monsanto Gıda ve Tarım Tic. Ltd. Şti (Antalya). After removing the shells and washing the seeds in distilled water, the seeds were transferred into plastic pots for growth. They were irrigated using Hoagland solution for 14 days at 400 µmol m−2 s−1 light intensity at 24 °C ± 2 °C and 16 h light/8 h dark photoperiod. For mimicking drought stress conditions, 10% polyethylene glycol 6000 (PEG-6000) was prepared and added to the Hoagland solution. Similar to other drought stress-related studies conducted by the authors (Baloglu et al. 2014; Celik Altunoglu et al. 2017; Celik Altunoglu et al. 2016), samples were collected from stressed plants and control plants at 0, 3rd, 12th, and 24th hours following the application of the stress condition. For measuring tissue-specific bZIP gene expression analysis, root and leaf samples of the grown plants were used and sampled three times for replication.
RNA isolation and quantitative real-time PCR analysis
Total RNA was extracted using TRIzol reagent (Life Technologies Corporation, Grand Island, NY, USA). All RNA samples were treated with DNase I (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA) for the removal of DNA contaminants. The quality and integrity of the isolated RNA was checked using agarose gel electrophoresis and the Multi Scan Go device (Thermo Fisher Scientific, USA). For gene expression analysis, bZIP gene-specific primers were designed and checked using the NCBI Primer BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primers used in this study are listed in Supplementary Table S6. The bZIP genes were selected based on their expression levels obtained from other reports. Therefore, highly expressed and differently expressed bZIP genes under drought stress conditions were used for qRT-PCR. A cucumber 18S rRNA gene (GenBank ID: X51542.1), amplified with primers 5′-GTGACGGGTGACGGAGAATT-3′ and 5′-GACACTAATGCGCCCGGTAT-3′ was utilized as a control (Baloglu et al. 2014). Three biological replicates and three technical replicates were taken from each sample. SYBR Green master mix and a Light Cycler 480 Real-Time PCR System (Roche Applied Science, Germany) were used for specific gene expression analysis. Using the 18S rRNA gene as an internal control, relative gene expression analysis was performed. The ΔCT and ΔΔCT were calculated as indicated in the authors’ previous studies (Baloglu et al. 2014; Celik Altunoglu et al. 2016, 2017; Kavas et al. 2015; Yer et al. 2015). In addition, the standard errors of mean among replicates were calculated. Analysis of Variance was used for obtaining the statistical significance of the difference between stress-treated samples and control samples. If the P-value was < 0.01, we considered the bZIP genes as differentially expressed genes.
Results and discussion
Identification and phylogenetic classification of the bZIPs in the watermelon and melon genomes
Various strategies such as BLAST, PFAM domains, hidden Markov model (HMM) and key word searches were used for the extensive identification of bZIP genes in target genomes. In addition, last version of CLC Genomics Workbench v11 was used for crosscheck for putative bZIP genes. So, all known bioinformatics techniques were applied to the determination of bZIP genes. As a result of these comprehensive analysis, a total of 59 and 75 bZIP proteins were characterized from watermelon and melon, respectively. Confirmation of the conserved bZIP domain was performed among all identified bZIPs. Watermelon and melon bZIP genes were abbreviated as CmbZIP and ClabZIP, respectively, based on their Latin names. The full length of ClabZIP proteins varied from 85 (ClabZIP-44) to 767 (ClabZIP-49) amino acid residues, with molecular masses ranging from 10 to 82.8 kDa. The full length of CmbZIP proteins varied from 57 (CmbZIP-68) to 721 (CmbZIP-26) amino acid residues, with molecular masses ranging from 6.5 to 78.6 kDa. Physical position, isoelectric points, instability indexes, and phylogeny groups of ClabZIP and CmbZIP protein sequences are shown in Supplementary Table S1. Several reports have shown that bZIP gene numbers are different among plant species based on their genome size and chromosome numbers. The predicted bZIP gene numbers for watermelon and melon were consistent with that in previous research on cucumbers conducted by the authors, in which 64 CsbZIP genes were identified (Baloglu et al. 2014). Consultation with other genome-wide studies of bZIPs revealed that bZIP genes were found in higher numbers in monocots when compared with dicots. For example, 170 in maize (Wei et al. 2012), 141 in barley (Pourabed et al. 2015), 131 in soybean (Liao et al. 2008), 96 in Brachypodium (Liu and Chu 2015), 92 in sorghum (Wang et al. 2011) and 89 in rice (Nijhawan et al. 2008). However, the numbers were ranged between 55 and 75 bZIP genes in dicots (Jakoby et al. 2002; Liu et al. 2014). It can thus be suggested that large genome size, high chromosome number, duplication events, and polyploidy in monocots may cause these elevated numbers.
For investigating the genomic distribution of the ClabZIP genes, they were first arranged on the 11 chromosomes of watermelon and then named from ClabZIP-01 to ClabZIP-59 according to their order of appearance on the chromosomes (Fig. 1). The ClabZIP gene density on the chromosomes was also examined. Chromosome 8 contained the highest number of ClabZIP genes (19%), while the lowest number of genes was found on chromosome 3 (5%) (Supplementary Fig. S1). Some similarities between the characteristic gene distributions of ClabZIP genes and CsbZIP genes in cucumbers were observed. For example, ClabZIP genes were primarily distributed on chromosomes 2 as well as on chromosomes 8, 10, and 11, where they appear to congregate at the upper and lower ends of the arms, respectively (Fig. 1). In the cucumber genome, CsbZIP genes were located on different chromosomes (chromosomes 2, 3 and 7), but the same gene arrangement patterns on chromosome arms were observed (Baloglu et al. 2014). In a watermelon genome study, chromosome-to-chromosome relationships within the Cucurbitaceae family members, including watermelon, cucumber, and melon were examined. A total of 3543 orthologous relationships were noted, which cover approximately 60% of the watermelon genome. A complicated syntenic pattern on these chromosomes was suggested. As a result, chromosomal evolution and rearrangement were examined among these three members of the Cucurbitaceae family (Guo et al. 2013). Because of high orthologous ratios between watermelon, melon, and cucumber, bZIP gene positions showed a similar pattern on each chromosome. In contrast to ClabZIP genes, the physical position of CmbZIPs could not be detected on melon chromosomes because the locations of these genes were still in the scaffolding level.
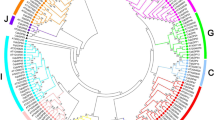
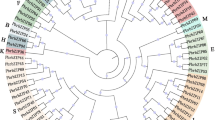
Phylogenic analysis was carried out to understand the evolutionary relationships of bZIP genes among these three important Cucurbit members. Unrooted Maximum Likelihood (ML) trees were separately created using 59 ClabZIP and 75 CmbZIP proteins, together with 64 CsbZIP proteins. The findings of the phylogenetic analysis of ClabZIP proteins indicated that seven discrete groups (Clusters I–VII) were obtained (Fig. 2a). The largest group, Cluster VII, and the second-largest group, Cluster IV, contain 23 and 15 ClabZIP proteins, respectively. CmbZIP proteins were phylogenetically divided into seven clusters (Clusters I–VII), similar to that in watermelon. Cluster VII was divided into four sub-clusters, comprising 49 CmbZIP proteins (Fig. 2b). Lastly, a combined phylogenetic tree was constructed using protein sequences of ClabZIPs from watermelon, CmbZIPs from melon, and CsbZIPs from cucumber for investigating the evolutionary relationships within the bZIP genes in the Cucurbitaceae family (Supplementary Fig. S2). The results revealed that a total of 198 Cucurbit bZIPs were assigned to ten clusters in the phylogenetic tree (Clusters I–X). No distinct discrimination among bZIPs of Cucurbitaceae family members was observed in these clusters—ClabZIPs, CmbZIPs, and CsbZIPs were distributed in all ten clusters. A total of 6, 7, and 10 groups of bZIP TFs have now been classified in cucumber (Baloglu et al. 2014), watermelon/melon (in this study), sorghum (Wang et al. 2011) and Arabidopsis (Jakoby et al. 2002), respectively. Furthermore, ten different clades of bZIP proteins were also observed in different plant species including rice (Nijhawan et al. 2008), six legumes (G. max, M. truncatula, P. vulgaris, C. arietinum, C. cajan, and L. japonicus) (Wang et al. 2015), apple (Li et al. 2016) and cassava (Hu et al. 2016). These results are consistent with data obtained in this study, in which all bZIP proteins from members of the Cucurbitaceae family were divided into the same ten groups, as indicated in these studies. Overall, it seems that plant bZIP proteins may be phylogenetically classified into ten groups based on their gene structure and motif compositions. It can, therefore, be suggested that the interspecies clustering shows a parallel evolution of bZIP TFs among different plant species.
In this study, gene structure profiles and motifs of ClabZIP genes were also examined for verifying the reliability of the phylogeny analysis. The exon–intron profiles of watermelon bZIP genes were determined for the comparison of their gene structures. A total of 17 ClabZIP genes with no intron were detected, accounting for 28.80% of all ClabZIP genes. Most were found in Clusters IV and V. The highest number of introns was observed in Clusters I and VIIa, and their coding regions (cds) were noted to vary from 1 to 12 (Fig. 3). In addition, a correlation was discovered between phylogenetic tree and the exon–intron organization of ClabZIP genes. All ClabZIP proteins in same clusters were classified into same exon–intron groups. In Fig. 3, the phylogenetic tree of ClabZIP proteins was redrawn to indicate this relationship. Similar studies indicated that different numbers of introns were identified in castor bean (11 introns) (Jin et al. 2014b), Arabidopsis (Jakoby et al. 2002) and rice (Nijhawan et al. 2008) (12 introns each), sorghum (14 introns) (Wang et al. 2011) and cucumber (12 introns) (Baloglu et al. 2014). The gene structures of all 585 bZIPs were also examined in six legumes, and similar structural patterns were observed among them (Wang et al. 2015). These results together with those of the present study further support the notion that the overall pattern of exon/intron profiles could be considered as an index for group classification and phylogenetic relationships in plant bZIP gene families.
Additional structural features, such as motifs and the bZIP domains of ClabZIP and CmbZIP proteins were also identified in the present study. Multiple EM for motif elicitation (MEME) analysis determined 20 motifs; all ClabZIP and CmbZIP proteins contain bZIP domains (Pfam number: PF00170), which are Motif 1–5 for watermelon bZIP proteins (Table 1A), and Motif 3-4-5-7 for melon bZIP proteins (Table 1B). In addition to bZIP domains, different domains, such as seed dormancy control (Pfam number: PF14144, DOG1) and CAMP-response element binding domains (Motif 2 for melon and Motif 6 for watermelon) were detected in ClabZIP and CmbZIP protein sequences. Some uncharacterized conserved motifs were also found (Supplementary Fig. S3). Watermelon and melon bZIP proteins within the same phylogenetic cluster had similar motif compositions suggesting that, like gene structure profiles, motif compositions also supported group classification and resulted in conserved evolution. These results match those observed in earlier studies. In one such study, a total of 50 conserved motifs and domains were identified in bZIP proteins from six legume species (Wang et al. 2015). A total of 50 conserved motifs and domains were identified in six legumes of bZIP proteins in that study. It was also proposed that most of the motifs appeared specific to each group for each legume species. They also suggested that the group-specific motifs are also useful for determination of specific functions and groups of bZIP members.
Duplication and evolutionary analysis of the bZIPs
Tandem or segmental gene duplication events lead to the formation of numerous copies of genes, which may result in the evolution of gene families. Divergence and duplication events for ClabZIP and CmbZIP genes were analyzed. Non-synonymous (Ka) versus synonymous (Ks) substitution ratios (Ka/Ks) were first evaluated for duplicated ClabZIP and CmbZIP genes to search for the association of Positive Darwinian Selection. A total of 18 pairs of segmentally and three pairs of tandemly duplicated genes were identified among ClabZIP genes (Fig. 4a). Average Ka/Ks ratios were 0.12 for segmentally duplicated and 0.16 for tandemly duplicated genes. Predicted divergence times of segmentally and tandemly duplicated ClabZIP genes ranged between 7 and 413 million years ago (MYA), with an average of 85 MYA; and between 3 and 12 MYA, with an average of 6 MYA, respectively (Supplementary Table S2). In addition, results indicated the presence of orthologous bZIP gene pairs between watermelon and monocot plants; maize (114 pairs of genes), rice (86 pairs of genes). Also orthologous bZIP gene pairs between watermelon and eudicot plants; arabidopsis (120 pairs of genes), poplar (81 pairs of genes), and grape (38 pairs of genes). 55 pairs of orthologous bZIP gene pairs were determined with cucumber which is also a member of cucurbiticae family. The earliest divergence time was noted between watermelon and maize bZIP genes, with an average of 150–170 MYA, which was followed by rice (155–165 MYA), Arabidopsis (130–150 MYA), grape (35–40 MYA), poplar (25–30 MYA), and cucumber (1–3 MYA) (Fig. 4b) (Supplementary Table S3).
a Estimation of duplication and divergence times of ClabZIP and CmbZIP genes. b Estimation of divergence times of ClabZIP genes with orthologous bZIP gene pairs between melon and rice, Arabidopsis, maize, poplar, cucumber, and grape. c Estimation of divergence times of CmbZIP genes with orthologous bZIP gene pairs between watermelon and rice, Arabidopsis, maize, poplar, cucumber, and grape
According to the duplication analysis of CmbZIP genes, 106 pairs of bZIP genes displayed duplication events, with an average divergence time of 120–130 MYA (Fig. 4a) (Supplementary Table S4). In addition, a total of 107, 106, 105, 63, 41, 27 and 385 pairs of orthologous bZIP genes were identified between melon and cucumber, watermelon (cucurbiticae members); poplar, grape, Arabidopsis (eudicot plants); maize and rice (monocot plants), respectively. The latest divergence time was observed between melon and Arabidopsis or maize with an average of 1–3 MYA. Divergence times between melon and watermelon, cucumber, poplar, grape, rice were 5–10 MYA, 5–15 MYA, 25–35 MYA, 45–55 MYA, and 30–35 MYA respectively (Fig. 4c) (Supplementary Table S5). According to the divergence times and gene pairs, especially the divergence time between C. sativus and Z. mays, it looks like ClabZIP and CmbZIP genes are conserved between monocots and dicots. bZIP family appeared before the divergence of monocots and dicots (Nijhawan et al. 2008). This data shows that the structure and function of most bZIP genes remained conserved during angiosperm evolution (Li et al. 2015b). In the light of this information, our findings are consistent with our knowledge of the evolution of bZIP genes so far and will act as a valuable information in the understanding of monocot/eudicot evolution of bZIP genes.
In previous studies for the genome-wide determination of bZIP genes in other plants, four pairs of tandemly and 12 pairs of segmentally duplicated genes were observed in cucumber bZIP genes (Baloglu et al. 2014). Moreover, in Arabidopsis, sorghum, and rice, an average of three pairs of tandemly duplicated genes were monitored, which was consistent with results for watermelon obtained in this study. Segmental duplication of bZIP genes in Arabidopsis, sorghum, and rice were 39, 49, and 52 pairs of genes, respectively (Jakoby et al. 2002; Nijhawan et al. 2008; Wang et al. 2011). These results indicate that segmental duplication events may dominantly lead to that gene family expansion of bZIP genes. Based on divergence time analysis of bZIP genes, cucumber and poplar bZIP genes were the latest diverged genes, with a predicted time of 10–15 MYA (Baloglu et al. 2014). Similar results were obtained in the present study, which indicate that watermelon bZIP genes diverged last with poplar bZIP genes, after cucumber bZIP genes. Moreover, bZIP genes in the cucumber genome were separated from rice bZIP genes, with a predicted time of 26–38 MYA (Baloglu et al. 2014), whereas bZIP genes in the watermelon genome showed divergence from maize and rice, with a predicted time of 150–170 MYA and 155–165 MYA, respectively. However, the latest divergence time rate was calculated between bZIP genes from melon and those from Arabidopsis and maize. These findings indicate that that cucumber and watermelon bZIP genes are phylogenetically closer to poplar bZIP genes than to melon bZIP genes. Moreover, shared orthologous gene numbers were similar between melon–cucumber (107 pairs of genes) and melon–watermelon (106 pairs of genes), and these numbers were higher than that of orthologous genes from other plants. This can be explained by the membership of these plants in the same family.
Functional annotation and homology modeling of bZIPs proteins
The gene ontology (GO) annotation of bZIP proteins was performed using Blast2GO analysis for predicting their functional properties. Both ClabZIP and CmbZIP proteins were shown to play roles in developmental, metabolic, and cellular processes. In addition, the regulation of biological process and responses to stimuli were also observed roles of bZIP proteins. These findings are consistent with the known regulatory roles of bZIP proteins in various biological processes, including nitrogen/carbon and energy metabolism, pathogen defense, organ and tissue differentiation, flower growth, and seed maturation as well as light and stresssignaling (Ciceri et al. 1999; Jakoby et al. 2002; Silveira et al. 2007; Walsh et al. 1998). Also, these results can be verified with these studies which indicated the involvement of bZIP proteins in normal and stressed growth conditions. Based on molecular function prediction, ClabZIP and CmbZIP proteins had primarily binding or TF activities, which reflects their regulatory role. Lastly, both bZIP proteins generally existed in cell parts, organelles, and membranes in melon and watermelon (Fig. 5). Cucumber bZIP proteins were also found in cell parts and organelles, which were similar locations to melon or watermelon bZIP proteins (Baloglu et al. 2014). Moreover, according to the subcellular localization of some bZIP proteins in tomato, five proteins were nuclear-localized transcriptional activators (Li et al. 2015a). Detailed location analysis of melon or watermelon bZIP proteins indicated that these cell parts mainly comprised the membrane or nucleus, which is concordant with their roles as TFs. Results of these previous studies were also consistent with the findings of this study (Huang et al. 2010; Takahashi et al. 2012; Yang et al. 2013).
BLASTP search was applied for estimating the predicted three-dimensional (3D) structure of bZIP proteins from melon and watermelon in Protein Data Bank (PDB), a repository of information regarding the 3D structures of large biological molecules, including proteins and nucleic acids. Protein Homology/AnalogY Recognition Engine (Phyre2) database was utilized for homology modeling via HMM–HMM search using the detection rates of proteins (Söding 2005). According to these estimations, ClabZIP-04-05-08-25-29-30-40-41-44-47-54-59 and CmbZIP-02-06-11-16-18-19-31-38-43-49-50-51-55-63-67-68 were selected for homology ratio evaluation (PDB ID numbers for each bZIP proteins were indicated in parenthesis). Modeled residues ranged between 17 and 44% for watermelon and between 27 and 42% for melon bZIP proteins, with a confidence interval of > 95% (Fig. 6). Selected proteins for homology modeling from melon and watermelon displayed only an alpha helical structure, which was consistent with the structures of cucumber bZIP proteins (Baloglu et al. 2014). The bZIP domain is located on a contiguous alpha helical structure that includes a basic region of 16 amino acid residues that are responsible for interaction with DNA, and a heptad repeat of leucine residues or bulky hydrophobic amino acids. These two subunits adhere via interactions between the hydrophobic sides of their helices to bind DNA. These interactions lead to the construction of the coil–coil structure, which gives this group of proteins the “zipper” name (Jakoby et al. 2002). The determination of reliable 3D structures of melon and watermelon bZIP proteins may be valuable for clarification of their mode of action.
Genome-wide expression analysis and drought stress responses of melon and watermelon bZIP genes
To explore the expression profiles of bZIP genes in watermelon and melon in different tissues, an RNA-seq approach was utilized with Sequence Read Archive (SRA) data sets. Heat maps were constructed to indicate expression patterns of bZIP genes in phloem and vascular tissues as well as the various days of fruit development after pollination in watermelon (Supplementary Fig. S4). According to the heat map, ClabZIP-11 was expressed in vascular tissues, whereas ClabZIP-15 and ClabZIP-57 were only expressed in phloem tissues. However, ClabZIP-50 and ClabZIP-55 expression was present in both the vascular and phloem tissues. Expression patterns were similar to ClabZIP-11 and ClabZIP-57 genes, which were upregulated after the 18th day following pollination. In addition, ClabZIP-11 and ClabZIP-57 genes exhibited a similar upregulated high expression pattern on the 34th day of fruit development. Review of the data sets, including fruit development stages and after pollination, revealed that expression levels of ClabZIP-11 and ClabZIP-57 genes were upregulated from 18th to 50th day. Therefore, for validating the expression patterns of watermelon bZIP genes under drought stress conditions, ClabZIP-11, ClabZIP-15, ClabZIP-50, ClabZIP-55 and ClabZIP-57 genes were selected on the basis of their upregulated expression levels in the heat map. The quantitative real time PCR (qRT-PCR) data from stressed leaf and roots of watermelon indicated that all genes except ClabZIP-55 displayed an upregulated expression pattern in the 1st hour of stress exposure in leaf and root tissues (Fig. 7). However, the expression levels of these genes (except ClabZIP-11 and ClabZIP-55) were downregulated in the 12th hour of stress exposure in comparison with the control. ClabZIP-57 gene exhibited the highest expression level in the 6th hour of stress exposure compared with other hours in leaf and root tissues. In addition, the highest expression level was observed for ClabZIP-57 gene in the 6th hour of stress exposure among other studied watermelon bZIP genes. Moreover, all genes except ClabZIP-50 demonstrated maximum expression levels in the 6th hour in leaf tissues. Generally, a trend of increasing expression levels was observed in the 1st, 3rd, and 6th hours of stress exposure in all studied genes in leaf tissues. Except for ClabZIP-15 and ClabZIP-50, other bZIP genes again displayed an increased expression pattern in the 1st, 3rd, and 6th hours of stress exposure in root tissues. ClabZIP-15 and ClabZIP-50 genes were upregulated in the 1st hour of stress exposure.
In addition, using the SRA database, a heat map was constructed in which expression profiles of bZIP genes in different varieties of melon, fruit stages, and salt stress conditions were also evaluated (Supplementary Fig. S5). According to the transcriptome profile of melon bZIP gene expression under salt stress, all genes studied using qRT-PCR (except for CmbZIP-31) showed an increased expression profile under salt stress conditions. Increased response of CmbZIP-52 and CmbZIP-63 genes after the 1st hour of drought stress measured using qRT-PCR analysis was consistent with the transcriptome profile of these genes under salt stress conditions. These genes may be considered a class of abiotic stress responsive genes. According to the gene expression profile during different fruit development stages, CmbZIP-31 expression was augmented in some white fruit stage samples (20 days after anthesis) and yellow fruit stage samples (10 and 20 days after anthesis). The expression of CmbZIP-52, CmbZIP-53, and CmbZIP-72 were upregulated only in yellow fruit 20 days after anthesis, while expression levels of CmbZIP-63 were elevated only in white fruit 20 days after anthesis. These results indicate that that CmbZIP-31, CmbZIP-52, CmbZIP-53, CmbZIP-63, and CmbZIP-72 genes may also play a role in fruit development. In addition, CmbZIP-31-52-53-72 genes presented different expression patterns in Piel de sapo pinonet, Piel de sapo-t111, and Cantalupo vedrantais varieties of melon. CmbZIP-63 was only expressed in Piel de sapo-t111 and Conomon SC varieties of melon. These differences in gene expression may be explained by differences between varieties of melon.
According to the qRT-PCR analysis of CmbZIP-31, CmbZIP-52, CmbZIP-53, CmbZIP-63, and CmbZIP-72 genes in melon, a statistically significant upregulation was observed in the 1st hour of drought stress treatment in root tissues for all genes (Fig. 7). In addition, all genes except CmbZIP-53 displayed a statistically sign3ificant upregulated pattern in the 1st hour of stress exposure. This rapid increase can be attributable to the mode of response of TFs. A good example of this event was the increased expression pattern of CmbZIP-52 and CmbZIP-63 genes in the 1st hour of stress exposure. CmbZIP-53 gene expression also increased in the 1st, 3rd, and 6th hours both in leaf and root tissues. The expression profiles of CmbZIP-52 and CmbZIP-63 genes were similar and displayed an increased expression profile in the 1st, 3rd, and 6th hours in root and leaf tissues compared with the control. The leaf response with highest expression was observed in the 1st hour of stress exposure by CmbZIP-52 and CmbZIP-63 genes. In addition, CmbZIP-31 and CmbZIP-72 genes reflected the best root response in the 3rd hour of stress exposure. The most active expression periods of determined genes in qRT-PCR analysis in melon were the 1st, 3rd, and 6th hours after stress treatment. This may demonstrate the necessity of rapid TF response. All studied melon bZIP gene expressions decreased after the 12th hour of stress exposure in both leaf and root tissues compared with the control, and this decreasing trend was also observed in the studied watermelon bZIP genes.
Regarding orthologous genes between melon or watermelon bZIP genes and those of other plants, one of the bZIP genes from maize (bZIP-62, GRMZM2G00017), which is orthologous to ClabZIP-11, showed a significantly decreased expression profile during an infection of Colletotrichum graminicola in maize. On the contrary, maize bZIP-62 was upregulated in a drought-resistant line of maize (Bt-1) under Fusarium moniliforme infection (Wei et al. 2012). In this study, the ClabZIP-11 gene was upregulated in different vascular tissues under drought stress conditions, as well as during fruit development. These results suggest a significant role of these genes during biotic and abiotic stress conditions. A ClabZIP-50 ortholog in grape—VvbZIP-25 (GSVIVT01033531001)—plays a role in seed development, and is upregulated under heat stress exposure (45 °C) based on microarray and real-time data. It may be inferred that ClabZIP-50 plays role in normal tissue development and responses to drought and heat stress conditions. Moreover, grape bZIP genes, such as VvbZIP-23 and VvbZIP-42 (which are orthologous to the CmbZIP-53 gene) were involved in leaf-stem development and berry maturity, respectively (Liu et al. 2014). These grape genes were also upregulated under drought stress conditions. These results were consistent with the findings of this study, in which upregulation of the CmbZIP-53 gene was observed under drought or salt stress conditions and in different fruit development stages. Cucumber bZIP transcripts including CsbZIP-06, CsbZIP-08, CsbZIP-12, CsbZIP-15, CsbZIP-29, CsbZIP-30, CsbZIP-44, CsbZIP-53, CsbZIP-55, and CsbZIP-59 accumulated in root tissues, whereas their suppression was detected using qRT-PCR under drought stress conditions in cucumber leaf tissues for all studied bZIP genes (Baloglu et al. 2014). However, the ClabZIP-11 gene (which is orthologous to cucumber CsbZIP59) was upregulated in both leaf and root tissues in the 1st, 3rds and 6th hours of stress application in watermelon. Moreover, melon CmbZIP-63 (orthologous to cucumber CsbZIP06) and CmbZIP-72 (orthologous to cucumber CsbZIP-59) gene transcripts accumulated in the 1st hour of water deficiency in both root and leaf tissues of melon. The accumulation of these gene transcripts in root tissues under drought stress conditions were consistent with orthologous cucumber bZIP genes. Cucumber, melon, and watermelon are all members of the Cucurbitaceae family, which may explain these results. However, the lack of expression data in the 1st hour for cucumber bZIP genes rendered it impossible to compare the expression patterns between bZIP genes in watermelon and melon. In this study, the response of ClabZIP and CmbZIP genes under drought stress conditions were especially observed in the 1st hour of stress.
Conclusion
The bZIP TF family plays various important roles in plant developmental and physiological processes in and biotic/abiotic stress responses. We identified 59 ClabZIP and 75 CmbZIP TF-encoding genes in watermelon and melon genomes, respectively. The combined phylogenetic tree was constructed using bZIP protein sequences from watermelon, melon, and cucumber for investigating the evolutionary relationships within the bZIP genes in the Cucurbitaceae family. A total of 198 Cucurbit bZIP proteins were assigned to 10 clusters in the phylogenetic tree. The highest shared orthologous bZIP gene numbers were obtained between melon–cucumber (107 pairs of genes) and watermelon–Arabidopsis (120 pairs of genes). Divergence time calculations indicate that cucumber and watermelon bZIP genes were closely related to poplar bZIP genes. We also examined the expression patterns of ClabZIP and CmbZIP genes in root and leaf tissues of watermelon and melon under drought stress conditions using qRT-PCR and RNA-seq data available in public databases. We found some ClabZIP and CmbZIP genes which may be considered early response genes for drought conditions in watermelon and melon. Gene identification studies like the present study open new perspectives in the analysis of bZIP protein family members and their functions in plants.
References
Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448:938–942
Bai Y et al (2016) Genome-wide analysis of the bZIP gene family identifies two ABI5-like bZIP transcription factors, BrABI5a and BrABI5b, as positive modulators of ABA signalling in Chinese cabbage. PLoS ONE. https://doi.org/10.1371/journal.pone.0158966
Bailey TL et al (2009) MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Baloglu MC (2014) Genome-wide in silico identification and comparison of Growth Regulating Factor (GRF) genes in Cucurbitaceae family. Plant Omics 7:260–270
Baloglu MC, Eldem V, Hajyzadeh M, Unver T (2014) Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE. https://doi.org/10.1371/journal.pone.0096014
Berman HM et al (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Caraux G, Pinloche S (2005) PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 21:1280–1281. https://doi.org/10.1093/bioinformatics/bti141
Celik Altunoglu Y, Baloglu P, Yer EN, Pekol S, Baloglu MC (2016) Identification and expression analysis of LEA gene family members in cucumber genome. Plant Growth Regul 80:225–241. https://doi.org/10.1007/s10725-016-0160-4
Celik Altunoglu Y, Baloglu MC, Baloglu P, Yer EN, Kara S (2017) Genome-wide identification and comparative expression analysis of LEA genes in watermelon and melon genomes. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-016-0405-8
Ciceri P, Locatelli F, Genga A, Viotti A, Schmidt RJ (1999) The activity of the maize Opaque2 transcriptional activator is regulated diurnally. Plant Physiol 121:1321–1327
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. https://doi.org/10.1155/2008/619832
Food and Agriculture Organization of the United Nations (FAO) (2014) FAOstat, statistical databases. Last updated 15 Aug 2014. http://www.fao.org
Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12:901–915. https://doi.org/10.2307/3871218
Garcia-Mas J et al (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci 109:11872–11877. https://doi.org/10.1073/pnas.1205415109
Guan Y, Ren H, Xie H, Ma Z, Chen F (2009) Identification and characterization of bZIP-type transcription factors involved in carrot (Daucus carota L.) somatic embryogenesis. Plant J 60:207–217. https://doi.org/10.1111/j.1365-313X.2009.03948.x
Guo S et al (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45:51–58
Hsieh T-H, Li C-W, Su R-C, Cheng C-P, Tsai Y-C, Chan M-T (2010) A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231:1459–1473. https://doi.org/10.1007/s00425-010-1147-4
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hu W et al (2016) Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci Rep 6:22783. https://doi.org/10.1038/srep22783
Huang S et al (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281
Huang X-S, Liu J-H, Chen X-J (2010) Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol 10:230–230. https://doi.org/10.1186/1471-2229-10-230
Iwata Y, Koizumi N (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102:5280–5285. https://doi.org/10.1073/pnas.0408941102
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111. https://doi.org/10.1016/S1360-1385(01)02223-3
Jin J, Zhang H, Kong L, Gao G, Luo J (2014a) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:D1182–D1187. https://doi.org/10.1093/nar/gkt1016
Jin Z, Xu W, Liu A (2014b) Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 239:299–312. https://doi.org/10.1007/s00425-013-1979-9
Kavas M, Kizildogan A, Gokdemir G, Baloglu MC (2015) Genome-wide investigation and expression analysis of AP2-ERF gene family in salt tolerant common bean. EXCLI J 14:1187–1206. https://doi.org/10.17179/excli2015-600
Kavas M, Baloglu MC, Atabay ES, Ziplar UT, Dasgan HY, Unver T (2016) Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Genet Genomics 291:129–143. https://doi.org/10.1007/s00438-015-1095-6
Kelley LA, Sternberg MJE (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371
Kobayashi F, Maeta E, Terashima A, Takumi S (2008) Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol Plant 134:74–86. https://doi.org/10.1111/j.1399-3054.2008.01107.x
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278:21003–21011. https://doi.org/10.1074/jbc.M210538200
Lee SC, Choi HW, Hwang IS, Choi DS, Hwang BK (2006) Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 224:1209–1225. https://doi.org/10.1007/s00425-006-0302-4
Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. https://doi.org/10.1093/nar/gkr201
Li D, Fu F, Zhang H, Song F (2015a) Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genomics 16:771. https://doi.org/10.1186/s12864-015-1990-6
Li X et al (2015b) Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genomics 16:976. https://doi.org/10.1186/s12864-015-2196-7
Li Y-Y, Meng D, Li M, Cheng L (2016) Genome-wide identification and expression analysis of the bZIP gene family in apple (Malus domestica) Tree Genet Genomes. https://doi.org/10.1007/s11295-016-1043-6
Liao Y et al (2008) Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228:225–240. https://doi.org/10.1007/s00425-008-0731-3
Liu X, Chu Z (2015) Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics 16:227. https://doi.org/10.1186/s12864-015-1457-9
Liu J-X, Srivastava R, Che P, Howell SH (2007) Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 51:897–909. https://doi.org/10.1111/j.1365-313X.2007.03195.x
Liu J et al (2014) Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15:281. https://doi.org/10.1186/1471-2164-15-281
Lu G, Gao C, Zheng X, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229:605–615. https://doi.org/10.1007/s00425-008-0857-3
Lucas TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking annual. Rev Plant Biol 57:203–232. https://doi.org/10.1146/annurev.arplant.56.032604.144145
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. https://doi.org/10.1126/science.290.5494.1151
Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146:333–350. https://doi.org/10.1104/pp.107.112821
Pourabed E, Ghane Golmohamadi F, Soleymani Monfared P, Razavi SM, Shobbar Z-S (2015) Basic leucine zipper family in barley: genome-wide characterization of members expression analysis. Mol Biotechnol 57:12–26. https://doi.org/10.1007/s12033-014-9797-2
Rodriguez-Uribe L, O’Connell MA (2006) A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris). J Exp Bot 57:1391–1398. https://doi.org/10.1093/jxb/erj118
Shen H, Cao K, Wang X (2007) A conserved proline residue in the leucine zipper region of AtbZIP34 and AtbZIP61 in Arabidopsis thaliana interferes with the formation of homodimer. Biochem Biophys Res Commun 362:425–430. https://doi.org/10.1016/j.bbrc.2007.08.026
Silveira AB, Gauer L, Tomaz JP, Cardoso PR, Carmello-Guerreiro S, Vincentz M (2007) The Arabidopsis AtbZIP9 protein fused to the VP16 transcriptional activation domain alters leaf and vascular development. Plant Sci 172:1148–1156. https://doi.org/10.1016/j.plantsci.2007.03.003
Söding J (2005) Protein homology detection by HMM–HMM comparison. Bioinformatics 21:951–960. https://doi.org/10.1093/bioinformatics/bti125
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612. https://doi.org/10.1093/nar/gkl315
Takahashi H, Kawakatsu T, Wakasa Y, Hayashi S, Takaiwa F (2012) A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol 53:144–153. https://doi.org/10.1093/pcp/pcr157
Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science 320:486–488. https://doi.org/10.1126/science.1153917
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Walsh J, Waters CA, Freeling M (1998) The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade–sheath boundary. Genes Dev 12:208–218
Wang J, Zhou J, Zhang B, Vanitha J, Ramachandran S, Jiang S-Y (2011) Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on SorghumF. J Integr Plant Biol 53:212–231. https://doi.org/10.1111/j.1744-7909.2010.01017.x
Wang Z et al (2015) Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics 16:1053. https://doi.org/10.1186/s12864-015-2258-x
Wei K et al (2012) Genome-wide analysis of bZIP-encoding genes in maize. DNA Res 19:463–476. https://doi.org/10.1093/dnares/dss026
Weltmeier F et al (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J 25:3133–3143. https://doi.org/10.1038/sj.emboj.7601206
Wingender E et al (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res 29:281–283. https://doi.org/10.1093/nar/29.1.281
Yang Z, Gu S, Wang X, Li W, Tang Z, Xu C (2008) Molecular evolution of the CPP-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol 67:266–277. https://doi.org/10.1007/s00239-008-9143-z
Yang O, Popova OV, Süthoff U, Lüking I, Dietz K-J, Golldack D (2009) The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 436:45–55. https://doi.org/10.1016/j.gene.2009.02.010
Yang Y-G, Lv W-T, Li M-J, Wang B, Sun D-M, Deng X (2013) Maize membrane-bound transcription factor Zmbzip17 is a key regulator in the cross-talk of ER quality control and ABA signaling. Plant Cell Physiol 54:2020–2033. https://doi.org/10.1093/pcp/pct142
Yer EN, Baloglu MC, Ziplar UT, Ayan S, Unver T (2015) Drought-responsive Hsp70 gene analysis in Populus at genome-wide level plant. Mol Biol Rep 34:483–500. https://doi.org/10.1007/s11105-015-0933-3
Yilmaz A, Nishiyama MY, Fuentes BG, Souza GM, Janies D, Gray J, Grotewold E (2009) GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol 149:171–180. https://doi.org/10.1104/pp.108.128579
Yoshida T et al (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685. https://doi.org/10.1111/j.1365-313X.2009.04092.x
Yun K-Y et al (2010) Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. https://doi.org/10.1186/1471-2229-10-16
Author information
Authors and Affiliations
Contributions
YCA and MCB conceived the study. FC, NMU and YK performed the experiments and carried out the analysis. YCA and MCB wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Unel, N.M., Cetin, F., Karaca, Y. et al. Comparative identification, characterization, and expression analysis of bZIP gene family members in watermelon and melon genomes. Plant Growth Regul 87, 227–243 (2019). https://doi.org/10.1007/s10725-018-0465-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-0465-6