Abstract
For wheat improvement, knowledge of diversity patterns and evolutionary relationships among the germplasm is essential for the sustainable conservation of valuable genetic materials. Therefore, this investigation aims to decipher the genetic diversity and population structure of wheat germplasm and identify diverse accessions to expedite the wheat breeding programs. In this study, a diverse set of 81 bread wheat genotypes, including derived doubled haploids, elite exotic lines, indigenous landraces and cultivars were used. Phenotypic and molecular characterization was conducted using 10 morpho-physiological traits and 68 microsatellite (SSR) markers, respectively. Among, the nine morphological descriptors the highest morphological variation (H) was observed for 1000 grain weight (1.06). The genotypes based on phenotypic traits were grouped into three clusters using Euclidian distance. Principal component analysis (PCA), revealed that the first four significant principal components accounted for 78% of the variance. A total of 58 alleles were generated from 27 polymorphic SSR primers with an average of 2.15 alleles per locus. The polymorphic information content (PIC) ranged from 0.26 (Wmc434) to 0.76 (Wmc47), with an average of 0.69. Molecular analyses by NTSYS-PC delineated the genotypes into four clusters, whereas while the cluster tree generated using neighbour-joining analysis identified three major clusters. Structure analysis grouped the genotypes into three sub-populations. When comparing agro-morphological and molecular clusters, seven genotypes (DH 194, DH 207, DH 215, DH 217, CIMMYT line 30 Entry No.10, Chamba landrace 18 and C 306) were identified as common. Notably, these genotypes were identified as distant and belong to different diversity clusters. Therefore, integrating morphological analysis with a polymorphic SSR survey may be effective in selecting diverse parental lines in wheat hybridization programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bread wheat (Triticum aestivum L., 2n = 6x = 42) was one of the first domesticated food crops and is the major cereal crop. It provides the basic staple food for a large portion of the world population, especially in Europe, West Asia and North Africa. Wheat remains the most important source of food and nutrients for humans. It is the second largest cereal crop in terms of global production, following maize, with a worldwide cultivation average of approximately 220.76 million hectares and a production of 770.88 million metric tonnes (FAOSTAT 2021).
Over time monoculture of elite high yield-cultivars has reduced the genetic base of crops, leaving them vulnerable to rapidly evolving disease-pest and changing climate. In contemporary agriculture, rigorous selection for desired traits narrows down the genetic base of cultivars and causes genetic erosion, resulting in a decline in genetic diversity (Tanksley and McCouch 1997). Additionally, climate change intensifies the occurrence of abiotic and biotic stresses, posing a serious threat to agricultural productivity. Modern cultivars are unable to adapt to climate change due to a lack of genetic diversity, while wild relatives of crops may possess novel genes for tolerance to these adverse conditions, making them suitable for use in breeding programs (Maxted et al. 2009). Integrated strategies that could enhance resistance to these stresses include mining a variety of germplasm sources for novel potential alleles and choosing lines with higher adaptability (Karsai et al. 2012; Orabi et al. 2014; Warschefsky et al. 2014). Furthermore, genetic enhancement should be grounded in the exploitation of genetic diversity present in various germplasm resources, thereby making diversity research a crucial component of future breeding programs. Plant breeders utilize the genetic diversity of a crop for its improvement either by subjecting it to selection for developing a variety or by using it as a parent in hybridization programs.
The knowledge of genetic variability and the association of agro-morphological traits with grain yield is a prerequisite for a successful crop breeding program (Sharma et al. 2022). Genetic improvement of wheat relies on the availability of diverse germplasm sources and estimating genetic variability is beneficial for raising productivity in a more efficient way, as well as improving resistance to various stresses (Khodadadi et al. 2011). Diverse germplasm sources, such as landraces, serve as potent reservoirs of diversity. In the case of wheat, the northwestern Himalayas are rich in wheat diversity, as evidenced by the presence of numerous local landraces. Therefore, it is important to sustain local genetic diversity that has evolved over time in response to prevalent biotic and abiotic stresses in genetic amelioration programs. This approach expands the genetic background of the crops, enabling the development of novel and diverse cultivars that can thrive under various stress conditions (Singh et al. 2016; Zampieri et al. 2017).
The genetic diversity among different accessions can be assessed using morpho-physiological or molecular markers. Among these, clustering the genotypes based on agro-morphological descriptors is an easy way to analyze genetic diversity (Khan et al. 2014). However, accurately estimating genetic diversity based solely on agro-morphological traits is challenging because these traits are often influenced by the environment, dependent on the growth stage and show a low degree of variation (Voichita et al. 2011). To overcome these limitations, morpho-physiological markers can be used in conjunction with molecular markers, which provide precise and accurate methods for analyzing genetic diversity (Choudhury et al. 2014; Haun et al. 2011; van de Wouw et al. 2010). If the goal is to assess the genetic diversity among a large number of wheat accessions inexpensively and quickly using microsatellite/ simple sequence repeat (SSR) markers is an excellent strategy. These markers are easily detected through polymerase chain reaction (PCR), have a wide distribution throughout genome, show co-dominant inheritance, have multi-alleles and produce highly reproducible results. They are therefore highly suitable for analyzing diversity, identifying cultivars, determining hybridity and mapping physical structure of the genome (Scott et al. 2000; Roder et al. 2002; Kalia et al. 2011).
The integrated approach of morphological characterization along with their molecular profiling provides information about the genetic diversity in the germplasm more efficiently and precisely. Thus, the current study was conducted to assess the genetic diversity among various north–west Himalayan potential landraces, winter × spring and spring × spring wheat derived doubled haploids, a set of elite exotic collections and popular Indian cultivars. This assessment was done using various morpho-physiological traits and SSR markers.
Materials and methods
The experimental material comprises 81 diverse bread wheat genotypes (Table 1). This includes 11 potential landraces collected from the north-western Himalayas, 23 established doubled haploids by chromosome elimination technique (Chaudhary et al. 2005), 40 elite exotic collections from the International Maize and Wheat Improvement Center (CIMMYT), and seven popular Indian cultivars viz., HPW 360, HPW 368, HPW 373, DH 114, VL 829, VL 892 and C 306.
Phenotyping
The 81 genotypes were assessed for various morpho-physiological traits in lattice design of 9 × 9 with four replications during rabi 2018–19. Two rows of 1 m length for each genotype were grown with a spacing of 20 × 10 cm. A single row of the susceptible check i.e., Sonalika was included after every block (9 genotypes) to ensure the uniform spread of stripe rust and powdery mildew inoculum in the experimental field. The recommended cultural practices were adhered to for cultivating a robust crop. The data was recorded for several morpho-physiological traits namely Plant height (cm), Effective tillers per plant, Spikelets per spike, Grains per spike, 1000-grain weight (g), Biological yield per plant (g), Harvest index (%), Grain yield per plant (g); and for days to 50% flowering and 75% maturity, which were recorded on a plot basis. Reactions to stripe rust and powdery mildew disease were recorded in a scale of 0–100 scale (Peterson et al. 1948) and 0–9 (Bennett and Westcott 1982), respectively.
Genotyping
For molecular analysis, genomic DNA was isolated from 81 bread wheat genotypes using CTAB method (Doyle and Doyle 1987) with certain modifications. A total of 68 SSR primer pairs (Supplementary Table 1) (http://wheat.pw.usda.gov/) were screened for polymorphism and 27 primers showed polymorphism and were scored for further analysis.
PCR amplification and band profiling
PCR (Polymerase chain reaction) reactions were carried out in 0.2 ml PCR tubes with final volume of 12.5 µl. Each tube contained 1.0 µl of template DNA (50 ng/µl), 0.5 µl each of forward and reverse primers (50 ng), 1.25 µl of 10X PCR buffer, 1.0 µl of MgCl2 (25 mM), 1.0 µl of dNTP mix (0.5 mM each of dATP, dGTP, dCTP and dTTP), 7 µl of sterilized distilled water and 0.25 µl of Taq polymerase (5U/µl) for 81 genotypes. The amplifications were conducted using an Eppendorf Thermal-cycler. The PCR conditions for SSRs were optimized with an initial denaturation at 94 °C for 5 min; an annealing temperature of X °C (ranging from 40.3–61.9 °C), extension at 72 °C for 45 s and a final extension at 72 °C for 5 min. The samples were then stored at 4 °C. The PCR products were subjected to electrophoresis in a 4% agarose gel at 100 V for 120–150 min and amplicon size was estimated using a 100 bp DNA ladder and observed under a UV transilluminator using a gel documentation system (UVITEC, Cambridge).
Data analysis
Morphological diversity analysis
For all genotypes nine morphological descriptors (Table 2) were transformed into binary representation based on the presence (‘1’) and absence (‘0’). The binary data of the studied traits was subjected to construct a dendrogram using NTSYS–PC (version 2.02) (Rohlf 2006). PAST (PAleontological STatistics) software (version 4.03) was used to scrutinize the Shannon diversity index (H) for each descriptor (Hammer et al. 2001).
The estimation of genetic diversity was based on 10 morpho-physiological traits which were then subjected to Euclidean Distance cluster analysis. This was followed by grouping of genotypes to construct trees of hierarchical clusters using Ward’s method (1963) with XLSTAT software. Additionally, the clustering pattern of the 81 bread wheat genotypes was determined through principal component analysis (PCA) using the EIGEN procedure.
Molecular analysis
Polymorphic SSR marker profiles were scored and expressed in a binary matrix with ‘1’ indicating the presence of a particular marker allele and 0 for the absence. The data was then used to construct a dendrogram using an unweighted pair group method with arithmetic mean (UPGMA) algorithm in the SAHN program of the NTSYS–PC package (version 2.02) (Rohlf 2006). Neighbor-joining tree were created using statistical software DARWIN version 6 (Perrier and Jacquemoud-Collet 2006) to group the studied germplasm. The summary statistics including Ne (effective number of alleles), Na (number of alleles), E-Ho (observed heterozygosity), E-He (expected heterozygosity), Av-He (average heterozygosity), and I (Shannon’s diversity index) per locus were estimated using POPGEN computer software version 1.32 (Yeh and Boyle 1997). The estimation of polymorphic information contents (PIC) values was performed using Cervus 3.0.3 software (Anderson et al. 1993).
The statistical software GenAlEx version 6.5 (Peakall and Smouse 2012) was used to evaluate the Analysis of molecular variance (AMOVA) and Principal Coordinate Analysis (PCoA). The population structure of the 81 bread wheat genotypes was determined using Bayesian model-based STRUCTURE 2.3.4 program (Pritchard et al. 2000). The examination involved a range of K values from 1 to 10, with the admixture model used for 10,000 replicates with 10,000 burning periods. To determine the optimal K value, the ΔK method, as described by Evano et al. (2005), was used with the online Structure Harvester tool.
Results
Considerable differences were observed among the 81 genotypes in terms of 9 morphological descriptors. The Shannon diversity index (Table 2) ranged from 0.48 (Maturity) to 1.06 (1000 grain weight) with an average value of 0.83 and a range of 0.58, exhibiting a broad range of genetic variation in the studied genotypes. Besides, spike length (0.99) recorded the highest evenness and lowest for maturity (0.54). The cluster analysis conducted using a genetic distance matrix by NTSYS–PC, grouped all the genotypes into 6 clusters (Fig. 1a) at a genetic similarity level of 0.64.
Euclidean distance diversity analysis
The squared Euclidean distances matrix grouped 81 genotypes into three clusters (Fig. 1b, Supplementary Table 2) using Ward’s method (1963). The resulting clusters, labeled I to III, contained 35, 21 and 25 genotypes respectively, indicating a broad range of genetic diversity among genotypes. The average inter-cluster distance ranged from 15.51 to 19.28, with the highest inter-cluster genetic divergence observed between clusters I and II (19.28) and lowest between clusters II and III (15.51) (Table 3).
The cluster means for different traits exhibited significant differences among the clusters for each trait (Supplementary Table 3). Cluster I showed desirable values for days to 50% flowering (133.57), days to 75% maturity (165.60), 1000 grain weight (49.72 g) and biological yield (23.16 g). Cluster II showed desirable values for harvest index (45.43%) and grain yield per plant (8.08 g); cluster III for plant height (91.43 cm), effective tillers per plant (4.38), spikelets per spike (17.69) and grains/spike (62.43). Therefore, clusters I and III are considered the most important with desirable cluster means for most of the traits.
The maximum assistance towards genetic divergence was accounted by days to 50% flowering (22%), followed by 1000 grain weight (20.2%), days to 75% maturity (16.1%), plant height (12.2%), biological yield per plant (10%), grain yield per plant (9.6%), harvest index (3.6%), spikelets per spike (2.9%), grains per spike (2%) and effective tillers per plant (1.5%).
Principal component analysis (PCA)
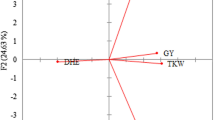
The first four significant principal components (PCs) cumulated to account for 78 percent of the variation. The eigen values range from 2.87 to 0.093 for PC1 to PC8, respectively. The most important component PC1 described 28.7% of overall variance which was mainly attributed to by grains per spike followed by spikelets per spike and days to flowering. However, PC2 contributed 21.6% of the total variation and was primarily influenced by seed yield per plant, biological yield per plant and tillers per plant (Supplementary Table 4). The scatter biplot of PC1 against PC2 showed that the majority of genotypes were distinct, as they were dispersed throughout the diagram (Fig. 2).
The screening for stripe rust (Puccinia striiformis f. sp. tritici) and powdery mildew (Blumeria graminis f. sp. tritici) disease showed that 11 genotypes viz., DH 194, DH 195, DH 202, DH 205, DH 215, DH 219, CIMMYT line 60 Entry no. 37, Chamba landrace 1, Chamba landrace (1)2, Chamba landrace 16, HPW 360 and HPW 368 were found to be moderately resistant to resistant for both the diseases (Supplementary Table 5).
Molecular genetic diversity
A set of 68 SSR primers distributed across the wheat genome, was screened for PCR amplification using 81 bread wheat genotypes (Fig. 3). Subsequently, 27 SSR markers (39.7%) were selected for further characterization due to their ability to produce distinct polymorphism and stable amplification patterns. These 27 SSR markers were then used to assess the gene pool composition of the test genotypes using a suite of genetic diversity analysis software tools.
The parameter values for the number of alleles per locus (Na), effective number of alleles per locus (Ne), Shannon’s diversity index (I), polymorphism information content (PIC), expected heterozygosity (E-Ho), expected heterozygosity (He), and average heterozygosity (Av-He) per locus were used to estimate genetic diversity (Table 4). SSR makers detected a total of 58 alleles, ranging from 2 to 3 with an average of 2.15 alleles per locus. However, the effective number of alleles per locus ranged from 1.41 to 2.60 with a mean of 2.15. The mean value of Shannon’s diversity index (I) was 1.93, with a range of values from 0.48 to 1.01. The polymorphic information content (PIC) varied from 0.26 (Wmc434) to 0.76 (Wmc47), with an average value of 0.69. Expected homozygosity and heterozygosity ranged from 0.38 to 0.70 and from 0.30 to 0.62; with the mean of 0.52and 0.48, respectively.
The coefficient of genetic similarity matrix for all 3,240 pairs of genotypes varied from 0.21 to 0.93 with an average of 0.56. The genetic distance was calculated among 81 wheat genotypes based on SSR primers. The genotypes were grouped into four clusters using UPGMA algorithm A, B, C and D, at 0.56 level of genetic similarity (Fig. 4), comprised of 25, 4, 23 and 29 genotypes, respectively. DARwin software delineated genotypes into 3 clusters A, B and C having 29, 29 and 23 genotypes respectively (Fig. 5).
The genetic variation within and among gene pools was partitioned using analysis of molecular variance (AMOVA). A substantial proportion (96%) of the allelic diversity was credited to genotypes within the gene pool, while only 4% was found among the populations (Table 5, Fig. 6). Principal Coordinate Analysis (PCoA) was used to further confirm the findings of cluster analysis. Wheat genotypes were segregated into four distinct populations (Fig. 7), indicating diverse accessions were based on genetic constitution. The first 3 axes of the PCoA plot collectively accounted for 29.63% of the total variation. The first, second and third PCs explained 13.14%, 9.8% and 6.69% variation respectively (Table 6). A minimal introgression was observed between the gene pools in the axes 1 versus axes 2 plot.
STRUCTURE software confirmed that individuals within a population are genetically related. The Evanno test resulted maximum peak value of ΔK at K = 3 (Fig. 8a). Structure analysis indicated that the population of the 81 bread wheat genotypes under study was a mixed population with three sub-populations, viz., POP 1, POP 2 and POP3 (Fig. 8b). In total, 27 genotypes (33.33%) were assigned to POP 1; POP 2 contained 28 genotypes (34.57%); and POP 3 contained 26 genotypes (32.10%). Additionally, almost all sub-populations have different maturity and grain yield per plant. This depicted the relatedness of genotype within sub-populations and allowed us to determine the introgression in the genotype in the present population of wheat genotypes.
Comparison of genetic diversity between populations
The genetic diversity based on different groups among different populations is presented in Table 7. The molecular diversity based on NTSYS, among the four groups, group ‘D’ showed the highest genetic diversity based on polymorphism information content (PIC = 0.31), the effective number of alleles (Ne = 1.71) and Shannon’s diversity index (I = 0.68). The highest genetic diversity for different maturity populations was exhibited by the medium maturity population based on polymorphism information content (PIC = 0.37), the effective number of alleles (Ne = 1.90) and Shannon’s diversity index (I = 0.68). The genetic diversity among different plant height populations indicated that medium and long plant height groups showed the highest genetic diversity based on polymorphism information content (PIC = 0.36), while medium height group had the highest effective number of alleles (Ne = 1.88) and Shannon’s diversity index (I = 0.67). In case of different yielding populations indicated that medium yielding group had the highest genetic diversity with PIC = 0.37, Ne = 1.92 and I = 0.68, followed by low yielding group with PIC = 0.34, Ne = 1.81 and I = 0.63. However, among different types of germplasm doubled haploids had the highest genetic diversity based on polymorphism information content (PIC = 0.37), the effective number of alleles (Ne = 1.90) and Shannon’s diversity index (I = 0.68); therefore, doubled haploids are the important germplasm sources concerning wheat breeding programs.
Discussion
The comprehensive study of germplasm to understand genetic diversity and population dynamics is crucial for crop improvement. For plant breeders, the wide genetic diversity is vital for selecting germplasm with desired traits and choosing genetically diverse parents (Oliveira et al. 2017). Conventionally, agro-morphological traits have been employed to dissect genetic diversity and to establish associations among genotypes. As morphological traits are vulnerable to environmentally influenced variations, a holistic approach grounded on morphological along with molecular research can be employed to decipher genetic patterns of variations among germplasm (Sharma et al. 2022). This combined approach can be exploited to widen the genetic base by introducing new desired traits in bread wheat. Utilizing molecular markers enables exploration of genetic diversity among populations by identifying individual differences at the nucleotide sequence level, which furthermore is used to unravel the genetic differences of wheat germplasm through genetic mapping research (Gordon et al. 2019).
In the present study, we used morphological and microsatellite SSR markers to analyze the genetic diversity of 81 bread wheat genotypes comprising established doubled haploids, potential landraces from north-western Himalayas, elite genotypes from CIMMYT and improved cultivars of Indian origin.
Morphological markers represent the genotype's expression under natural field conditions following interactions with the environment (Sood et al. 2023). To minimize environmental errors, a simple lattice design was employed for phenotypic evaluation. The study identified significant variation among the 81 genotypes across nine morphological descriptors. The highest Shannon diversity index was observed for 1000 grain weight (1.06) and higher mean values (0.83) indicated that the genotypes possess significant variability for various phenotypic traits under study. Ouaja et al. (2021) obtained greater values of the Shannon index (0.80) indicating a high level of diversity among accessions studied.
Genetic diversity analysis by squared Euclidean distance matrix employing Ward’s method (1963), based on 10 morpho-physiological traits, resulted in the grouping of genotypes into 3 clusters. Different clustering patterns were also observed in bread wheat genotypes by Singh et al. (2018), Santosh et al. (2019), Aashu et al. (2022) and Mohi-Ud-Din et al. (2022). Cluster I possesses the maximum genotypes (35), followed by cluster III (25) and cluster II (21). The greatest inter-cluster distance was found between clusters I and II (19.28). Cluster II showed the highest value for grain yield per plant (8.08 g). Within Cluster II, genotype CIMMYT Entry No.237 demonstrated the highest yield, followed by CIMMYT Entry No. 242 of Cluster III and CIMMYT Entry No. 101 of Cluster II, these genotypes show promising potential as valuable breeding stocks for incorporation into breeding programs. These finding holds promise for future pre-breeding programs, as these genetically diverse clusters could be utilized for enhancing wheat breeding efforts.
Multivariate principal component analysis (PCA) seeks to elucidate the significance of the largest contributor to the total variation at each axis of differentiation. PCA resulted in the first four significant principal components (PCs), accounting for 78% of the variation. Among these significant PCs, spikelets per spike exhibited the highest positive value, followed by grains per spike, harvest index and grain yield per plant. Hence, these traits are likely to be potent contributors towards genetic divergence and effective selection under these characters could lead to improvement in wheat yield. The scattered biplot of all the genotypes throughout the PC1 versus PC2 indicates a diverse germplasm under study. Thus, the categorization of variables into different classes based on uniformity can be obtained through biplot analysis (Ahmed et al. 2022). Zhao et al. (2020) also concluded the importance of PC1 and PC2 in their study.
To overcome the constraints of morphological characterization, SSR markers were used to assess the genetic diversity accurately. Molecular markers have become the method of interest allowing plant breeders to dissect the genetic diversity of a population based on variation among individuals at the genetic level (Gordon et al. 2019).
The 27 polymorphic SSR markers detected 58 alleles among 81 bread wheat genotypes, and the Na-number of alleles per locus (2.15), Ne-effective number of alleles per locus (2.15), I- Shannon’s diversity index (1.93), PIC- polymorphism information content (0.69), E-Ho-expected heterozygosity (0.52), E-He- expected heterozygosity (0.48), and Av-He- average heterozygosity (0.47) were evaluated. The average value of the number of alleles per locus was 2.15 which is comparable with earlier research (Kumar et al. 2022; Meena et al. 2022). The PIC value yields information about the number of alleles and their distribution and assesses the informativeness of studied markers. Indeed, a higher PIC value (> 0.5), indicates a greater level of genetic diversity within the studied genotypes (Nagy et al. 2012). The average PIC value for 27 SSR markers is 0.69. Notably, 3 markers viz., Wmc125, Wmc506 and Wmc47 exhibited higher PIC values (> 0.50) proving that they were highly informative and effective in the study, while the least PIC value (PIC = 0.26) was recorded for Wmc 434. The results of higher PIC value were similar to Abbasov et al. (2018) having a mean PIC of 0.62 in diploid wheat accessions and Mohi-Ud-Din et al. (2022), who got a PIC value of 0.830 in bread-wheat genotypes.
Cluster analysis via molecular markers is a crucial technique to understand the interactions and genetic diversity of parents for effective hybridization programs. Molecular analysis using NTSYS-pc grouped 81 bread wheat genotypes into 4 major groups at a similarity level of 0.56, employing the UPGMA algorithm. In contrast, the cluster tree generated through neighbor-joining analysis identified three major clusters. On comparing both clustering patterns generated by molecular analysis, a high degree of congruency was observed. The differences in clustering patterns observed between two software programs, namely NTSYS-pc v.2.02 and DARwin v.6.0, can be attributed to variations in the clustering methods employed by the two systems. Specifically, NTSYS-pc computes similarity coefficients, while DARwin computes dissimilarity coefficients. These distinctions may be associated with gene mutation, duplication or other genetic aberrations, leading to increased genetic variations. Furthermore, the clustering patterns of genotypes by molecular approaches showed minimal similarities to the clustering based on phenotypic genetic diversity. Only seven genotypes—DH 194, DH 207, DH 215, DH 217, CIMMYT line 30 Entry No.10, Chamba landrace 18, and C 306—were found to be common. This suggests that the divergence may arise from environmental interactions influencing morphological traits. It indicates a greater variation between genotypes, implying that the studied genotypes are more diverse and possess a broad genetic base. Similarly, Haque et al. (2021) grouped 26 genotypes into four major clusters and Farhangian-Kashani et al. (2021) grouped 105 wheat genotypes into six clusters.
The pairwise genetic distance has an inevitable role in accessing the generic diversity of germplasm, lowest pairwise genetic similarity (0.21) between genotypes CIMMYT line 30 Entry no. 30 and DH 215 have high genetic divergence and these pairs could be utilized in further wheat improvement programs. While, the highest genetic similarity distance (0.93) between CIMMYT line 60 Entry no. 24 and CIMMYT Entry no. 240 shows that the genotypes are of similar genetic origin, so selections can’t be made in such pairs. The average genetic distance estimated (0.56) is lower suggesting greater genetic diversity in the germplasm. Consistent results were obtained by Haque et al. (2021) and Phougat et al. (2018) for 44 bread wheat genotypes, while Fu et al. (2016) observed lower average values for pairwise genetic distances in 21 tall fescue cultivars.
The results of AMOVA were useful in the estimation of molecular variation among bread wheat genotypes; the majority (96%) of variation was allocated due to differences within population, while only 4% was due to differences distributed among the population. Mohi-Ud-Din et al. (2022) found that 95% of the variation was within the cluster while only 5% of the total variation was among all clusters. PCoA is a multivariate approach employed to analyze clusters of wheat genotypes, which further reaffirms the genetic relationship among the genotypes. Eighty-one wheat genotypes were classified into four groups. The first 2 PCs explained an ample amount of variance (13.14 and 9.08, respectively). Similar findings were concluded by Sharma et al. (2022).
The analysis of population structure identified three sub-populations among the 81 bread wheat genotypes. This grouping pattern aligns with findings reported by Jabari et al. 2023 for 54 accessions of Triticum and Aegilops. In contrast, Tyagi et al. (2022) reported four main groups within 37 wheat genotypes. The results of structure analysis did not delineate genotypes in a clear classification according to germplasm type, days to maturity and grain yield. This is probably due to the expeditious exchange of germplasm over the globe for various genetic amelioration programs.
An integrative approach, combining both agro-morphological and molecular markers is essential for assessing diversity within germplasm. Upon comparing agro-morphological and molecular clusters, seven genotypes namely, DH 194, DH 207, DH 215, DH 217, CIMMYT line 30 Entry No.10, Chamba landrace 18, and C 306 were identified as common, emphasizing the morphological and microsatellite data are consistent with one another. Plant breeders consider early maturity, high gain yield, biotic and abiotic resistance as traits of greater importance for the pre-breeding programs of bread wheat. All the CIMMYT lines are early to medium maturity with medium to high yielding and showed resistance to stripe rust and powdery mildew. Therefore, these elite exotic lines along with the diverse genotypes from different clusters can be utilized expansively in future wheat breeding programs.
Conclusion
In the current study, a holistic strategy involving agro-morphological and molecular evaluations was followed to assess the genetic diversity of bread wheat genotypes. The findings highlighted noteworthy genetic differences among the accessions under experimentation, attributed to sources of collection. Upon comparing clusters based on phenotypic and molecular data, seven genotypes viz., DH 194, DH 207, DH 215, DH 217, CIMMYT line 30 Entry No.10, Chamba landrace 18, and C 306 were found similar. Also, the majority of genotypes from the elite exotic collection were early to medium maturing with medium to high yielding and were from different diversity clusters and sub-populations. Additionally, these genotypes demonstrated notable resistance to both rust and powdery mildew. Hence, the genetic study involving morpho-physiological traits and molecular SSR markers concluded that there is ample genetic diversity among the studied 81 bread wheat genotypes. This study further facilitated the selection of diverse parental lines to be utilized in the augmentation of wheat germplasm.
References
Aashu SYPS, Divya P, Tanya B (2022) Estimating genetic diversity of bread wheat (Triticum aestivum L.) in different environments. Electron. J. Plant Breed 13(2):399–409. https://doi.org/10.37992/2022.1302.054
Abbasov M, Akparov Z, Gross T, Babayeva S, Izzatullayeva V, Hajiyev E, Rustamov K, Gross P, Tekin M, Akar T, Chao S, Brueggeman R (2018) Genetic relationship of diploid wheat (Triticum spp.) species assessed by SSR markers. Genet Resour Crop Evol 65:1441–1453. https://doi.org/10.1007/s10722-018-0629-2
Ahmed HGM-D, Zeng Y, Shah AN, Yar MM, Ullah A, Ali M (2022) Conferring of drought tolerance in wheat (Triticum aestivum L.) genotypes using seedling indices. Front Plant Sci 13:961049. https://doi.org/10.3389/fpls.2022.961049
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimising parental selection for genetic linkage maps. Genome 36:181–186
Bennett FGA, Westcott B (1982) Field assessment of resistance to powdery mildew in mature wheat plants. Plant Pathol 31:261–268
Chaudhary HK, Sethi GS, Singh S, Pratap A, Sharma S (2005) Efficient haploid induction in wheat by using pollen of Imperata cylindrica. Plant Breed 124:96–98
Choudhury BI, Khan ML, Dayanandan S (2014) Patterns of nucleotide diversity and phenotypes of two domestication related genes (OsC1 and Wx) in indigenous rice varieties in northeast India. BMC Genet 15:71–82
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Evano G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
FAOSTAT (2020) Food and agricultural organization of the United Nations. http://www.FAOstat.fao.org
Farhangian-Kashani S, Azadi A, Khaghani S et al (2021) Association analysis and evaluation of genetic diversity in wheat genotypes using SSR markers. Biologia Futura 72:441–452. https://doi.org/10.1007/s42977-021-00088-y
Fu K, Guo Z, Zhang X, Fan Y, Wu W, Li D, Peng Y, Huang L, Sun M, Bai S, Ma X (2016) Insight into the genetic variability analysis and cultivar identification of tall fescue by using SSR markers. Hereditas 11(153):9. https://doi.org/10.1186/s41065-016-0013-1
Gordon E, Kaviani M, Kagale S, Payne T, Navabi A (2019) Genetic diversity and population structure of synthetic hexaploid-derived wheat (Triticum aestivum L.) accessions. Genet Resour Crop Evol 66:335–348. https://doi.org/10.1007/s10722-018-0711-9(0123456789(),-volV()0123456789().,-volV)
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
Haque MS, Saha NR, Islam MT et al (2021) Screening for drought tolerance in wheat genotypes by morphological and SSR markers. J Crop Sci Biotechnol 24:27–39. https://doi.org/10.1007/s12892-020-00036-7
Haun WJ, Hyten DL, Xu WW, Gerhardt DJ, Albert TJ, Richmond T et al (2011) The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol 155:645–655
Jabari M, Golparvar A, Sorkhilalehloo B, Shams M (2023) Investigation of genetic diversity of Iranian wild relatives of bread wheat using ISSR and SSR markers. J Genet Eng Biotechnol 21:73. https://doi.org/10.1186/s43141-023-00526-5
Kalia RK, Rai MK, Kalia S, Singh RA, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Karsai I, Gy V, Petrovics S, Petcu E, Kobiljski B, Ivanovska S, Bedő Z, Veisz O (2012) Assessment of the spatial genotypic and phenotypic diversity present in the various winter wheat breeding programs in Southeast Europe. Euphytica 186:139–151
Khan SA, Iqbal J, Khurshid H, Saleem N, Rabbani MA, Zia M et al (2014) The extent of intra-specific genetic divergence in Brassica napus L. Population estimated through various agro-morphological traits. Eur Acad Res 2:2255–2275
Khodadadi M, Fotokian MH, Miransari M (2011) Genetic diversity of wheat (Triticum aestivum L.) genotypes based on cluster and principal component analyses for breeding strategies. Aust J Crop Sci 5:17–24
Kumar D, Singh V, Yashveer S, Samita K, Koli GK, Yadav OP (2022) Assessment of genetic diversity among nine bread wheat (Triticum aestivum L.) genotypes using microsatellite markers. J Pharm Innov 11(3):2415–2419
Maxted N, Kell S (2009) CWR in crop improvement: to what extent are they used? CropWild relative. Newsletter 7:7–8
Meena VK, Sharma RK, Chand S, Kumar M, Kumar N, Jain N, Singh A (2022) Elucidating molecular diversity in spring wheat (Triticum aestivum L. em.Thell.) under terminal heat stress environment using morpho-physiological traits and SSR markers. Indian J Genet Plant Breed 82(1):47–55. https://doi.org/10.31742/IJGPB.82.1.7
Mohi-Ud-Din M, Hossain MA, Rohman MM, Uddin MN, Haque MS, Dessoky ES, Alqurashi M, Aloufi S (2022) Assessment of genetic diversity of bread wheat genotypes for drought tolerance using canopy reflectance-based phenotyping and SSR marker-based genotyping. Sustainability 14:9818. https://doi.org/10.3390/su14169818
Nagy S, Poczai P, Cernák I, Gorji AM, Hegedűs G, Taller J (2012) PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem Genet 50:670–672
Oliveira MM, Sousa LB, Reis MC, Silva Junior ES, Cardoso DBO, Hamawaki OT et al (2017) Evaluation of genetic diversity among soybean (Glycine max) genotypes using univariate and multivariate analysis. Genet Mol Res 16:1–10. https://doi.org/10.4238/gmr16029661
Orabi J, Jahoor A, Backes G (2014) Changes in allelic frequency over time in European bread wheat (Triticum aestivum L.) varieties revealed using DArT and SSR markers. Euphytica 197:447–462
Ouaja M, Bahri BA, Aouini L, Ferjaoui S, Medini M, Marcel TC, Hamza S (2021) Morphological characterization and genetic diversity analysis of Tunisian durum wheat (Triticum turgidum var. durum) accessions. BMC Genom Data 22(1):3. https://doi.org/10.1186/s12863-021-00958-3
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin
Peterson RF, Campbell AB, Hannah AE (1948) A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can J Res 26:496–500
Phougat D, Panwar IS, Punia MS, Sethi SK (2018) Microsatellite markers based characterization in advance breeding lines and cultivars of bread wheat. J Environ Biol 39:339–346
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945e959
Roder MS, Wendehake K, Korzun V, Bredemeijer G, Laborie D, Bertrand L, Isaac P, Rendell S, Jackson J, Cooke RJ, Vosmann B, Ganal MW (2002) Construction and analysis of a microsatellite-based database of European wheat cultivars. Theor Appl Genet 106:67–73
Rohlf FJ (2006) NTSYS-pc. numerical taxonomy and multivariate analysis system, Version 2.02. Setauket, New York
Santosh JJP, Singh A, Gahatyari NC (2019) Genetic diversity analysis in bread wheat (Triticum aestivum L.em.Thell.) for yield and physiological traits. Int J Curr Microbiol App Sci 8(02):3059–3068. https://doi.org/10.20546/ijcmas.2019.802.358
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS et al (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Sharma A, Sharma S, Kumar N, Rana RS, Sharma P, Kumar P et al (2022) Morphomolecular genetic diversity and population structure analysis in garden pea (Pisum sativum L.) genotypes using simple sequence repeat markers. PLoS ONE 17(9):e0273499. https://doi.org/10.1371/journal.pone.0273499
Singh RP, Singh PK, Rutkoski J, Hodson DP, He X, Jørgensen LN, Hovmøller MS, Huerta-Espino J (2016) Disease impact on wheat yield potential and prospects of genetic control. Annu Rev Phytopathol 54:303–322
Singh G, Kumar P, Kumar R, Gangwar LK (2018) Genetic diversity analysis for various morphological and quality traits in bread wheat (Triticum aestivum L.). J Appl Nat Sci 10(1):24–29. https://doi.org/10.31018/jans.v10i1.1572
Sood T, Sood S, Sood VK, Badiyal A, Kapoor S, Sood V, Kumar N (2023) Characterisation of bell pepper (Capsicum annuum L. var. grossum Sendt.) accessions for genetic diversity and population structure based on agro-morphological and microsatellite markers. Sci Hortic 321:112308. https://doi.org/10.1016/j.scienta.2023.112308
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Tyagi S, Kumar A, Gautam T, Pandey R, Rustgi S, Mir RR (2021) Development and use of miRNA-derived SSR markers for the study of genetic diversity, population structure, and characterization of genotypes for breeding heat tolerant wheat varieties. PLoS ONE 16(2):e0231063. https://doi.org/10.1371/journal.pone.0231063
van de Wouw M, van Hintum T, Kik C, van Treuren R, Visser B (2010) Genetic diversity trends in twentieth-century crop cultivars: a meta-analysis. Theor Appl Genet 120:1241–1252
Voichita H, Rodica P, Ioan H, Ana C (2011) Genetic variability in a set of early maize inbred lines. Bull Univ Agric Sci Veter Med CLUJ-NAPOC Agric 68(1):155–164
Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244
Warschefsky E, Penmetsa RV, Cook DR, vonWettberg E (2014) Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am J Bot 101:1791–1800
Yeh FC, Boyle TJB (1997) Population genetic analysis of codominant and dominant markers and quantitative traits. Belg J Bot 129:157
Zampieri M, Ceglar A, Toreti DF, A (2017) Wheat yield loss attributable to heat waves, drought and water excess at the global, national and subnational scales. Environ Res Lett 12(6):064008
Zhao D, Li X, Zhao L, Li L, Zhang Y, Zhang Z, Liu L, Xu H, Zhao W, Wu T, Siddique KH (2020) Comparison of zinc and iron uptake among diverse wheat germplasm at two phosphorus levels. Cereal Res Commun. https://doi.org/10.1007/s42976-020-00081-6
Acknowledgements
The authors are grateful to the Department of Genetics and Plant Breeding, CSK HPKV, Palampur for providing facilities and financial support during the research.
Author information
Authors and Affiliations
Contributions
HKC conceptualized and planned the experiments; SV executed the experiments and drafted the manuscript; SV and NK analyzed the data & HKC, SV, NK, KS, MS and VKS provided suggestions and polished the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Verma, S., Chaudhary, H.K., Singh, K. et al. Genetic diversity dissection and population structure analysis for augmentation of bread wheat (Triticum aestivum L.) germplasm using morpho-molecular markers. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-023-01851-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-023-01851-x