Abstract
The genetic diversity present in the breeding programs of southeast Europe was assessed in a set of 114 winter wheat (Triticum aestivum L.) cultivars using AFLP and SSR markers. The average genetic diversity characterised with the Jaccard’s distance coefficient was 0.605 with an interval of 0.053 and 0.889. The wheat cultivars originating from the four countries differed from each other in their clustering patterns, including the numbers of clusters and the most prevalent cluster, which was breeding program-specific. Hungarian and Romanian cultivars showed closer relationships, and Serbian and Macedonian cultivars grouped together more frequently. The phenotypic variability of the same cultivars was assessed under diverse ecological conditions of the four growing sites, measuring the disease resistance against two foliar diseases, and several agronomic traits. Of the phenotypic traits, powdery mildew and leaf rust responses showed significant associations with genetic diversity, whereas heading date, plant height and yield components did not. Through parallel assessment of genotypic and phenotypic diversity it was possible to separate winter wheat cultivars with similar genotype but diverse phenotype from those with similar phenotype but diverse genotype. This information will allow breeders to make informed decisions in selecting parents for new crosses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In breeding for enhanced adaptation, adequate genetic diversity is a prerequisite for any crop improvement program. Genetic progress through selection is directly related to the variability present in the gene pool, and the quality of the genes contributed by the parents. There is concern that erosion of genetic variability might result in reduced ability of crops to respond to changes in climate, pathogen and pest populations, agricultural practices, or quality requirements (Smale 1997; Tester and Langridge 2010; van de Wouw et al. 2010). This aspect is receiving more attention due to the predicted consequences of global climate change, which may make it necessary to rapidly develop new genotypes with the ability to cope better with altered environmental conditions. Of the agronomically important traits, those connected with better adaptation are of the highest priority, such as resistance to various abiotic and biotic stresses, water use efficiency, nitrogen use efficiency and yield stability (Tester and Langridge 2010). If these traits are to be improved, not only must their genetic bases be revealed, but the genetic diversity present in different breeding materials must also be determined. This information will be very useful in assessing the need to intensify the exchange programs between breeding teams, to apply the modern tools of biotechnology and plant genomics and to incorporate exotic germplasm into the respective breeding pools.
Genetic diversity can be quantified indirectly by estimating genetic distance using pedigree information to calculate the coefficient of parentage (COP) or by determining morphological and phenotypic traits including those of agronomic importance, or directly, using molecular markers to compare variation in DNA sequences between genotypes (Almanza-Pinzón et al. 2003; Dreisigacker et al. 2004; Fufa et al. 2005). Indirect measurements have several drawbacks. The COP method requires complete and correct pedigree information, which is not always available. In addition, it is based on several simplifying assumptions, ignoring natural or artificial selection and the effects of mutation and genetic drift (Parker et al. 2002; Soleimani et al. 2002; Almanza-Pinzón et al. 2003; Reif et al. 2005). Morphological traits are limited in number and, like various phenotypic traits, they can be significantly modified by the environment and may be controlled by epistatic and pleiotropic gene effects (Maric et al. 2004; Roy et al. 2004; Fufa et al. 2005). Molecular marker systems, on the other hand, overcome these shortcomings, thus representing the best way of assessing genetic diversity.
With the development of robust, reliable marker systems such as AFLP, SSR and DArT markers, the number of genetic diversity studies has increased to a great extent in various plant species over the last decade (Röder et al. 2002; Landjeva et al. 2006; Chao et al. 2007; van de Wouw et al. 2010). In the case of wheat, detailed experiments have covered various aspects of spatial and temporal genetic diversity, evaluating the changes in different areas of the world over the last century (Donini et al. 2000; Manifesto et al. 2001; Christiansen et al. 2002; Roussel et al. 2004, 2005; Reif et al. 2005; Hao et al. 2006; Balfourier et al. 2007; White et al. 2008; Fu and Somers 2009; van de Wouw et al. 2010). Assessments on the genetic diversity within European winter wheat germplasm pools showed that differences between European accessions are a function of their geographical origin rather than their date of registration (Huang et al. 2002; Roussel et al. 2005; Balfourier et al. 2007). There was a clear separation between materials originating from geographical areas located north or south of the arc formed by the Alps and the Carpathian mountains. In addition, genetic diversity between the southeast European wheat genotypes proved to be larger than between the north European wheat accessions, underlining the fact that the Carpathian Basin represents a special agro- and socio-ecological area with highly diversified, unique climate conditions (e.g. temperature, rainfall) and soil characteristics (Stachel et al. 2000; Huang et al. 2002; Roussel et al. 2005). However, these studies included only a limited number of southeast European varieties covering a wide time period in the last century.
Taking into account the special agro-ecological characteristics of the Carpathian Basin and the need to prepare for predicted climate changes, a southeast European winter wheat collection consisting of 114 genotypes was set up and model experiments were initiated by breeding groups from four southeast European countries, viz. Hungary, Serbia, Romania and Macedonia. The majority of accessions involved in the experiment were bred, registered and/or used as crossing parents in the four countries in the 1990s, thus representing a contemporary set of winter wheat germplasm. The long-term aim of the joint research activities was to exploit genetic diversity in current winter wheat breeding programs of the southeast European region with the purpose of improving abiotic and biotic stress tolerance and developing new breeding materials with better adaptability to changing environments. This paper presents the first results of a one-year experiment to assess (1) the phenotypic diversity of the collection, covering several agronomic traits and biotic stress tolerance under diverse ecological conditions, and (2) the genetic diversity of the collection using AFLP and SSR markers. Based on these results, suggestions are made as to how this information could be used for breeding purposes.
Materials and methods
In order to assess the level of genetic and phenotypic diversity in the breeding programs in central and southeastern Europe a collection of 114 winter wheat cultivars was set up. These genotypes were bred, registered and/or used as crossing parents in the four countries. They were contributed by the following institutions: 31 wheat cultivars were contributed by the Agricultural Research Institute of the Hungarian Academy of Sciences, Martonvásár, Hungary (MV), 32 cultivars by the National Agricultural Research and Development Institute, Fundulea, Romania (FU), 35 cultivars by the Institute of Field and Vegetable Crops, Novi Sad, Serbia (NS), and 12 cultivars by the Institute of Agriculture, Skopje, Macedonia (MC) (Supplementary Table 1). In addition, four other genotypes were included in the experiment: two cultivars from Germany (Tommi and Ellvis), one from Italy (Ravenna) and Plainsman V, a drought tolerant U.S.A. cultivar. This wheat collection was then used for a series of genotypic and phenotypic analyses.
Assessment of genotypic diversity in the winter wheat collection
Two marker technologies were applied: AFLP, which identifies a higher number of marker loci randomly distributed over the whole genome, and SSR, which identifies a higher number of alleles within marker loci with known chromosomal locations. For AFLP analysis, the restriction enzymes Sse8371 and MseI were used, and after ligating the adapters a pre-selective PCR was conducted using primers containing no selective nucleotides. Fragments multiplied in this way were further amplified with primer pairs labelled with IRDye, each containing two selective nucleotides (Buerstmayr et al. 2002), after which the products were separated on 6% acrylamide gels using a LI-COR4300 DNA analyser. The three AFLP reactions performed on the wheat collection were designated by the abbreviations of the four selective nucleotides (TCAT, TCGA, TCTC). For SSR analysis, four primer pairs were tested on the whole germplasm collection: gwm46, gwm95, gwm261 and gwm680, each identifying a single marker locus on chromosomes 7B, 2A, 2D and 6B, respectively, but known to distinguish a large number of alleles at those loci (Huang et al. 2002).
For the SSR method each fragment generated by a primer pair was considered to be an allele of the same marker locus, whereas for AFLP polymorphic fragments were considered to represent distinct marker loci randomly distributed over the whole genome. Thus the total number of alleles (SSR) and loci (AFLP), together with the number of rare and single alleles (SSR) and loci (AFLP) were obtained. An allele (SSR) or marker locus (AFLP) was considered to be rare if its frequency in the wheat genotypes was lower than 0.05. The polymorphism information content (PIC) value was calculated for each locus (SSR) and for each selective primer pair (AFLP) (Anderson et al. 1993) as \( PIC = 1 - \mathop \sum \nolimits_{i = 1}^{n} \mathop p\nolimits_{i}^{2} \) where p i is the frequency of the ith allele or locus.
The dissimilarity matrix was constructed from the binary data using Jaccard’s distance coefficient (JDC = 1−a/(n−d), where a = the number of marker fragments two individuals have in common, n = total number of marker fragments and d = number of marker fragments both individuals lack). The scale for each pairwise comparison ranges from 0 to 1, where 0 represents complete similarity and 1 complete difference. The hierarchical clustering protocol was carried out using the un-weighted pair-group method using the arithmetic mean (UPGMA) grouping of the SPSS 16.0 software package on the matrix of JDC values. The groupings thus achieved were verified using the discriminant function of the SPSS 16.0 software package.
Phenotypic assessment of genetic diversity
To assess the phenotypic variability of the varieties under diverse ecological conditions, field experiments were carried out in the multiple environments of the four participating research teams, applying the same experimental design. The experimental fields of the four breeding groups are located in and around the Carpathian Basin (Table 1). The genotypes were sown in head rows with two replications; the sowing dates at the various locations were between October 10 (FU) and October 18 (NS) 2007. Heading date (HD) as days from sowing, plant height (PH), the average seed number/spike (SEED), thousand kernel weight (TKW) and leaf diseases were scored by the teams as listed in Table 3. The leaf diseases (powdery mildew and leaf rust) were scored when the symptoms were at their highest levels, with an evaluation of severity (ratio of the diseased leaf area to the whole plant area as a %; Stubbs et al. 1986). In the case of powdery mildew, symptoms resulting from natural infection were recorded at each location. In the case of leaf rust, plots were artificially inoculated at Martonvásár with a mixture of pathogen races, whereas the other three teams evaluated naturally occurring disease symptoms. The following categories were used: 0–5% resistant, 5–20% moderately resistant, 20–40% moderately susceptible, and 40–80% susceptible. The data were processed using the Windows Excel and Statistica 6 for Windows programs. Cluster analysis was carried out by applying the UPGMA method on the Euclidean distance matrix of the phenotypic data set, excluding the disease measurements. The groupings thus achieved were verified and further analysed using the discriminant function and the principal component analysis of the Statistica 6 software package.
Results
Genotypic diversity in the winter wheat collection
One of the major aims of the research was to determine the level of genetic diversity of wheat cultivars bred by the collaborating teams and/or grown in the central and southeast European region. In addition to overall genetic diversity, the diversity levels among cultivars contributed by each group were also examined and compared.
The three AFLP analyses resulted in 90 polymorphic fragments (markers) in the overall group of 114 wheat genotypes (Supplementary Table 2). The PIC value from each analysis was high, leading to an average value of 0.784. The average r value between individual marker pairs was 0.004 with a range of −0.876 to 0.899. As 97.6% of all possible 4,095 pairwise r values were located between −0.4 and 0.4, the 90 polymorphic fragments were considered to represent 90 independent marker loci. When the separate cultivar groups were examined, slight decreases in the levels of polymorphism were apparent in FU and MC cultivars. AFLP analysis did not identify any marker loci characteristic of a single cultivar group, but there were significant differences between groups in the ratio of complete absence or presence of marker loci. These values were the highest in MC cultivars, followed by FU cultivars.
The four SSR primers made it possible to examine the level of polymorphism at four loci. On average, 10.25 alleles/marker locus were detected in the 114 wheat genotypes; 53.7% of these alleles were rare. The average PIC value of the SSR was 0.50, with a larger variation between individual loci. There was also considerable variation in the PIC of the SSR primers within individual cultivar groups. For three of the four SSR primers the most frequent allele was the same in all four wheat groups. Thirteen of the 22 rare alleles were characteristic of only one cultivar group; the occurrence of rare alleles was the highest in the NS group (9), followed by MV (2), FU (1) and MC (1).
The data matrix of the AFLP and SSR markers was used to establish the genetic diversity present in the 114 wheat cultivars (Fig. 1) and in the various breeding programs (Table 2). The average JDC value was 0.605 with an interval of 0.053–0.889. The cultivars were placed in 8 clusters (Supplementary Table 1). Based on the results of AMOVA the between-group difference was highly significant, but it explained only a small portion of the total variance (7.2%), thus underlining the large variance within each cluster. Of the 8 clusters, two contained only one genotype; Plainsman V was the only member of Geno_clu1, and Martonvásár 17 was the only entry in Geno_clu5. All other clusters contained between 4 and 41 cultivars. The two largest clusters were Geno_clu2 and Geno_clu3, which contained 27% and 36% of the wheat cultivars, respectively, but even within these clusters the variance was relatively high. For Geno_clu3 the average JDC value was 0.453 with a range of 0.053–0.769, whereas for Geno_clu2 it was 0.528 with a range of 0.095–0.821.
The wheat cultivar groups originating from the four countries differed from each other in distribution patterns among the 8 clusters, including both the number of clusters and the most prevalent cluster (Table 2). The majority of cultivars in the Hungarian breeding program grouped in Geno_clu2, the majority of Romanian cultivars was in Geno_clu3, the majority of Serbian cultivars was in Geno_clu6, and the majority of MC cultivars was in Geno_clu8. Based on the clustering patterns, MV and FU cultivars exhibited closer relationships, whereas NS and MC cultivars tended to group together.
Location effects
Weather conditions in the experimental period varied in the four locations (Table 1). The winter was the coldest in FU, followed by MV, whereas the spring and early summer period was the warmest in MC, followed by NS. The average monthly-minimum temperature was the lowest in January with a value of −5.3°C in FU, but was not enough to differentiate the winter hardiness of the wheat genotypes. The amount of rainfall during the growing season was the highest in NS and the lowest in MV, but the distribution showed similar tendencies at all four locations; January and February were the driest, and the highest amount of precipitation was in October and November. Of the four locations, spring was the driest and warmest in MC, but even at this location, there was 84 mm rain in April and May, preventing evaluation of drought tolerance under natural field conditions.
The location and genotype main effects were highly significant in at least two locations for each trait measured (Table 3). Variances in HD and PH were mostly derived from location and genotype main effects, which together explained 98.3% of the total variance for HD, and 88.4% of the variance in PH. For both traits, the location effect contributed the largest portion of variance, showing that environmental differences similarly influenced the wheat genotypes. The pair wise correlation coefficients between the individual locations were in the range of 0.57–0.87 for HD, and 0.52–0.74 for PH. Correlation was the strongest between MV and FU for both traits. Location × genotype interaction had a more significant role in determining leaf rust and powdery mildew responses. Accordingly, correlations between locations were lower, in the range of 0.41 (NS-FU)–0.53 (MV-NS) for powdery mildew, and 0.01 (FU-MC)–0.41 (NS-FU) for leaf rust response.
Phenotypic diversity assessment in the wheat collection
Based on the results of the one-season field test there were no significant differences in the average values of agronomic traits (HD, PH and yield components) between the cultivars of the four breeding programs (Supplementary Table 3). Medium early cultivars were the most frequent in all the breeding programs. The widest maturity ranges were characteristic of MV and FU cultivars, which included both the earliest and the latest heading genotypes. MV and NS cultivars showed considerable variations in PH, average number of kernels/spike and TKW.
With respect to resistance to leaf diseases, there was substantial variation between cultivar groups that was apparent not only in the group means, but also in the distribution patterns. For powdery mildew, more than half of the 114 wheat cultivars (55.2%) were moderately susceptible, averaged over the four locations. The average levels of resistance in MV and FU cultivars were higher, and the 10 resistant cultivars originated from these breeding programs (5 from each). Not only were there no resistant cultivars in the NS and MC groups, but the proportions of moderately susceptible cultivars were also highest for these two groups (68.6 and 66.7%, respectively). In the case of leaf rust, there was a more even distribution of cultivars across the response categories; 21.9% of the 114 cultivars were resistant, 30.7% moderately resistant, 30.7% moderately susceptible and 16.7% susceptible. The average level of resistance was highest in NS cultivars followed by MV and FU cultivars. Of the 25 resistant cultivars 56.0% were of NS origin, 28.0% of MV and 16.0% of FU origin. The disease resistance levels of the MC cultivars were the lowest for both diseases.
These phenotypic observations were also well reflected in the genetic diversity level. When the average phenotypic values of the 8 Geno_clu groups were compared, there were no significant differences between groups in the average values of HD, PH, SEED and TKW at the various locations. On the other hand, the most pronounced differences between the Geno_clu groups were recorded for powdery mildew response measured at three locations (all being significant at the P = 0.001). Geno_clu3 was the most resistant (the group value averaged over the locations was 16.9), with 11 cultivars having a value lower than 10.0. The best resistance sources in this group were Mv Regiment (MV), Liman (FU) and Crina (FU). The powdery mildew resistance of Geno_clu2 was second (with an average value of 24.0), with three cultivars, Mv Béres, Mv Hombár, and Mv Táltos, having values of less than 10.0. These two genotypic groups contained the majority of Hungarian and Romanian cultivars. There were no resistant cultivars in the other Geno_clu groups. The most susceptible groups were Geno_clu7 (30.3), Geno_clu6 (31.2) and Geno_clu4 (33.0), containing mostly NS and MC cultivars. In the case of leaf rust resistance, only values measured at the MV location in artificial inoculation tests showed significant associations with genotype groupings. Geno_clu8 was the most resistant group (average value 12.0) with 50% of its genotypes having scores of less than 10.0. These resistant cultivars all originated from the NS breeding programme, the best being Etida. The most susceptible group was Geno_clu2, with an average value of 38.0. Each Geno_clu group however, contained a few leaf rust-resistant genotypes, unlike to the situation with powdery mildew. The most resistant cultivars in each Geno_clu group listed in decreasing order of the average group resistance level were the following: NS3-3836 (NS) from Geno_clu6, NS119/05 (NS) from Geno_clu4, Gruia (FU) from Geno_clu3, and Mv Toborzó (MV) from Geno_clu2.
In addition to examining the variance in response to the two diseases, multivariate analysis was also carried out on the data matrices of HD, PH, SEED and TKW, taking into account all local measurements. The aim of this analysis was to further characterize the phenotypic diversity in the wheat collection and to establish specific phenotypic categories within this set of genotypes. In the course of cluster analysis, the 114 wheat cultivars were placed into 8 major groups, and the positions of 97.4% of the cultivars were verified by discriminant analysis. The phenotypic groups contained between 7 and 28 cultivars, the smallest being Pheno_clu1 and the largest Pheno_clu2 (Table 4, Supplementary Table 1).
As expected, phenotypic groupings were mostly independent of the genotypic groupings of the cultivars (Table 4); the Spearman rank order correlation was 0.002 (non significant). The only exception was Geno_clu7, containing 4 NS cultivars, which were also similar in phenotypic characteristics. The other Geno_clusters were quite diverse, with the two largest groups (Geno_clu2 and Geno_clu3) covering the whole range of phenotypic diversity.
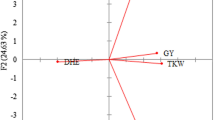
Based on the PCA the first four factors having Eigen values higher than 1 cumulatively explained 78.8% of the total variance. The first factor (with a 32.6% individual value) indicated the importance of heading date measurements at the four locations in discriminating the cultivars, the second factor (22.7%) was correlated with PH measured at three locations, while in the case of the third factor (13.2%) SEED was the main determinant. TKW made only small contributions to the first and second factors, but was a major contributor to the fourth factor (10.2%). Five phenotypic groups were distinctly separated from each other on the plane of the 1st and 2nd factors (Fig. 2a); these were Pheno_clu1, Pheno_clu2, Pheno_clu6, Pheno_clu7 and Pheno_clu8. In the case of the remaining three groups, each showed overlaps with one other group; Pheno_clu3 overlapped with Pheno_clu8, Pheno_clu4 overlapped with Pheno_clu2, and Pheno_clu5 overlapped with Pheno_clu7. However, these three overlapping pairs were distinctly separated from each other based on the plane of the 1st and 3rd factors (Fig. 2b).
The phenotypic characteristics of each Pheno_clu listed in Table 5 corresponded well with the results of PCA analysis. Pheno_clu1 was quite distinct from all the other groups because it contained late heading cultivars with the tallest plant stature. The other fairly separate group was Pheno_clu6, to which the latest heading cultivars belonged. Their TKW were the smallest, and they showed the strongest sensitivity to environment in regard to SEED. Of the three overlapping group pairs, Pheno_clu2 and Pheno_clu4 contained the earliest heading cultivars, which also showed the largest variation in heading date across the four environments. They were also similar in PH and TKW, with the only difference between them being that the SEED was significantly higher for the Pheno_clu4 group irrespective of environment. The group pair Pheno_clu3 and Pheno_clu8 contained mid-late to late flowering cultivars with similar TKW. The cultivars in Pheno_clu3 were somewhat shorter and had significantly lower SEED than those in Pheno_clu8. The SEED of the Pheno_clu8 group was not only the highest, but also showed the strongest stability over environments. Early HD and similar TKW were characteristic of the third pair of Pheno_clu5 and Pheno_clu7. Of the two groups, the cultivars in Pheno_clu5 were significantly taller and had significantly larger SEED. These characteristics of Pheno_clu5 proved to be relatively independent of environment compared to all other groups. Cultivars in Pheno_clu7 were the shortest and belonged to groups with the smallest SEED. In addition, their plant heights and average seed numbers showed larger variation depending strongly on environment.
Discussion
The major aim of this model experiment was to assess the genetic and phenotypic diversity in wheat breeding programs in southeast Europe, with direct reference to breeding practice. This involved a search for marker systems which could be carried out with the facilities available to the breeding groups, and which would make genetic diversity studies effective and fast, whereas having low cost and labour demands. The choice thus fell on a combination of AFLP, which provides high polymorphic locus information content/reaction but unknown chromosomal location, and SSR, which has high polymorphic allele information content/locus and known chromosomal location. These two systems are often used for diversity studies (e.g. Donini et al. 2000; Manifesto et al. 2001; Altintas et al. 2008). In some experiments several hundreds of markers were applied reaching levels of marker coverage that facilitated genome-wide linkage disequilibrium mapping and association analysis (Chao et al. 2007; White et al. 2008; Fu and Somers 2009). However, a much lower number of markers is sufficient for measuring genetic diversity in a germplasm collection. Similar tendencies in the genetic diversity of European wheat accessions were shown by Röder et al. (2002), who used 19 SSR markers and by Balfourier et al. (2007), who used 38 SSR markers. In the case of AFLP, a relatively high level of consistency can be achieved even with a few primer pairs, if these produce an adequate number of markers over the whole genome, although the nature and genomic locations or distribution are unknown (Soleimani et al. 2002). Hazen et al. (2002) used AFLP markers for diversity studies, with parallel investigation on a bi-parental mapping population to determine chromosomal location. They found that the AFLP loci covered the genome fairly evenly, and that clustering only occurred at very low frequencies. As more than 97% of AFLP loci showed no or only weak correlations with each other in the present study, it was evident that they fairly well assessed the genetic diversity in this southeast European winter wheat collection.
A vast amount of information has been published on the temporal and spatial genetic diversity existing in both European and worldwide wheat germplasm collections (Huang et al. 2002; Röder et al. 2002; Roussel et al. 2005; Balfourier et al. 2007), but most of these collections contained landraces, and old or new cultivars covering most of the last century. In addition, as the main aim was to establish major temporal and spatial trends in genetic diversity, they contained only limited numbers of samples from individual geographic regions. In some cases, the purpose was to set up a core collection covering most of the genetic diversity present in gene banks (Balfourier et al. 2007). Even if such research includes hundreds of genotypes, they still represent a relatively small-sample size, in which materials specifically adapted to local constraints and uses will not be represented, thus requiring renewed sampling outside the existing collection (Glaszmann et al. 2010). The present work is the first attempt to set up and evaluate an active winter wheat collection for the southeast European region. Winter wheat production is very important in this geographical region, and vulnerability to global climate change makes it imperative for breeders to prepare in good time. For this reason the cultivar collection consists mainly of wheat cultivars bred, grown and/or used as breeding parents in the four countries during the last two decades. Of the 114 cultivars, very few genotypes overlapped with those used in earlier studies, so most of the data is new to the wheat breeding community, while also strengthening overall tendencies.
Based on molecular marker studies, there is significant genetic diversity between the breeding materials of the four regions, despite the fact that they are located in a macro agro-ecological area (south or southeast of the Carpathian Mountains) identified as a single unit in the course of previous diversity studies (Kim and Ward 2000; Roussel et al. 2005; Balfourier et al. 2007). Nevertheless diversity between the genotype groups was highly significant, while in all cases the most prevalent genotypic cluster was breeding program specific. The macro area of southeastern Europe is characterised by diversified and unique climatic conditions evidenced by data from the four breeding locations even in a single growing season. In addition, the weather is very variable from season to season, due to the random interactions of three climatic zones: Atlantic, Continental and Mediterranean. These factors lead to specific regional agro-ecological conditions, and the need to breed wheat cultivars adapted to the unique combinations of local conditions. The varying breeding priorities, strategies and methods used in the four programs have also significantly influenced the diversity of the cultivars developed by the different breeding teams.
Small though varying ratios of genotypes from each breeding program clustered with materials from other breeding programs. Based on these clustering patterns Hungarian cultivars had greater genetic similarity to the Romanian group, whereas Serbian cultivars grouped more frequently with those from Macedonia. Similar results were achieved by Roussel et al. (2005) and, to a certain extent, by Balfourier et al. (2007) for a small number of genotypes originating from these countries and released during the last century. As the majority of the wheat accessions studied in the present work was bred in the 1990s, this extends the validity of the findings of Roussel et al. (2005) to modern southeast European cultivars. The clustering tendencies could be due to the fact that the agro-ecological conditions around Fundulea (Romania) and Martonvásár (Hungary) are more similar to each other than those prevalent around Novi Sad (Serbia) and Skopje (Macedonia), leading to the breeding of similar genotypes adapted to the specific environmental conditions. The similarity in the meteorological data, the greater prevalence of the two foliar diseases, especially powdery mildew, and the stronger correlations between plant developmental patterns (HD, PH) in Martonvásár and Fundulea all support this assumption.
There has been concern about decreasing genetic diversity caused by modern plant breeding and agricultural practices (Smale 1997; Hoisington et al. 1999; Roussel et al. 2004; Tester and Langridge 2010). Temporal genetic diversity studies carried out at country level or on larger geographical units have resulted in controversial findings (Manifesto et al. 2001; Parker et al. 2002; Roussel et al. 2004, 2005). The general tendency, however, can be summarised as follows: there was a marked decrease in genetic diversity in the 1970s, but since then the level has increased due to breeder awareness of its importance (Smale 1997; Reif et al. 2005; Tester and Langridge 2010; van de Wouw et al. 2010). In addition, an increase in genetic diversity was caused by a significant change in input technologies. This fact, together with climate change, necessitated significant changes in breeding programs as well. Instead of breeding high yielding cultivars, the breeding of cultivars with better adaptability and specific nutritional quality became a major priority, further increasing genetic diversity.
In the present study the within-group portion explained more than 90% of the total variance in the molecular analysis of variance, showing that there is a high level of genetic diversity within each genotypic cluster. Both the average level and range of genetic diversity within and between genotypic groups of the southeast European breeding material are high when compared to other results (Roussel et al. 2005; Balfourier et al. 2007). The four breeding groups have clearly been able to maintain germplasm diversity within their nurseries and collections, different parts of which can be used in breeding programs, thus allowing different sub-sets of the overall diversity to be released in the form of commercial varieties. This diversity will permit the breeding of new varieties better adapted to a changing environment.
Varying success has been achieved when using morphological and phenotypic traits to measure genetic diversity. In general, there was little or no correlation between genetic diversity based on phenotypic traits and that based on molecular markers (Maric et al. 2004; Roy et al. 2004; Fufa et al. 2005; Krystkowiak et al. 2009). Several explanations have been given for these findings: (1) the genome coverage represented by phenotypic traits is likely to be poor; (2) it may be burdened with error resulting from the dependence of trait expression on environmental conditions; (3) alleles of different origin may have the same phenotype; and (4) the number and choice of morphological traits and sample size may also affect the correlation. When selecting phenotypic traits for the examination of genetic diversity in a set of genotypes, it is important to be aware of how the type and direction of natural selection is affected by the agro-ecological areas, and how the influence of breeding priorities combines with purposeful selection in forming distinguishable gene pools. The effects of these factors are not always known (Stachel et al. 2000). Surprisingly in the present work, one significant phenotypic factor proved to be resistance to two foliar diseases, particularly to powdery mildew, which was closely correlated with genetic diversity. The high natural levels of powdery mildew infection occurring in the Martonvásár and Fundulea regions compared to those observed in Novi Sad and Skopje are probably due to the similarities in the prevailing meteorological conditions during spring, and to the similarity in plant developmental processes. This phenomenon, however, necessitates the continual introduction of novel disease resistance genes/alleles/QTLs, which then have a significant impact on genetic diversity. When, on the other hand, HD, PH and some yield components were analysed in the same wheat collection, no association was found between the genetic and phenotypic diversity, indicating the similarity in breeding practice for these traits, irrespective of the agro-ecological area.
The major aim in assessing the phenotypic diversity in the active southeast European wheat collection in this study was not to use the information as an indirect measurement of genetic diversity, but rather to demonstrate how it could be used directly for breeding purposes. The parallel assessment of genotypic and phenotypic diversity made it possible to distinguish cultivars with similar genotype but diverse phenotype from those with similar phenotype but diverse genotype. This knowledge can be directly utilised by breeders to make informed decisions when selecting parents for new crosses. When the aim is to improve a phenotypic trait, crossing partners can be selected from those with similar genotype but different phenotype. When the aim is to increase genetic diversity, parents can be selected from those with diverse phenotype and diverse genotype or from those with similar phenotype but diverse genotype. In the latter case, there is higher probability that the genotypes will carry dispersed sets of alleles which, although manifested as phenotypic similarity in the parents, lead to transgression in the progeny (Krystkowiak et al. 2009). In the present work, these categories were established for HD, PH and yield components, but the method can be applied for any agronomic trait, including end use quality.
References
Almanza-Pinzón MI, Khairallah M, Fox PN, Warburton ML (2003) Comparison of molecular markers and coefficients of parentage for the analysis of genetic diversity among spring bread wheat accessions. Euphytica 130:77–86
Altıntas S, Toklu F, Kafkas S, Kilian B, Brandolini A, Özkan H (2008) Estimating genetic diversity in durum and bread wheat cultivars from Turkey using AFLP and SAMPL markers. Plant Breed 127:9–14
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimising parental selection for genetic linkage maps. Genome 36:181–186
Balfourier F, Roussel V, Strelchenko P, Exbrayat-Vinson F, Sourdille P, Boutet G, Koenig J, Ravel C, Mitrofanova O, Beckert M, Charmet G (2007) A worldwide bread wheat core collection arrayed in a 384-well plate. Theor Appl Genet 114:1265–1275
Buerstmayr H, Lemmens M, Hartl L, Doldi L, Steiner B, Stierscheider M, Ruckenbauer P (2002) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. resistance to fungal spread (Type II resistance). Theor Appl Genet 104:84–91
Chao S, Zhang W, Dubcovsky J, Sorrells M (2007) Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci 47:1018–1030
Christiansen MJ, Andersen SB, Ortiz R (2002) Diversity changes in an intensively bred wheat germplasm during the 20th century. Mol Breed 9:1–11
Donini P, Law JR, Koebner RMD, Reeves JC, Cooke RJ (2000) Temporal trends in the diversity of UK wheat. Theor Appl Genet 100:912–917
Dreisigacker S, Zhang P, Warburton ML, van Ginkel M, Hoisington D, Bohn M, Melchinger AE (2004) SSR and pedigree analyses of genetic diversity among CIMMYT wheat lines targeted to different megaenvironments. Crop Sci 44:381–388
Fu YB, Somers DJ (2009) Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci 49:161–168
Fufa H, Baenziger PS, Beecher BS, Dweikat I, Graybosch RA, Eskridge KM (2005) Comparison of phenotypic and molecular marker-based classifications of hard red winter wheat cultivars. Euphytica 145:133–146
Glaszmann JC, Kilian B, Upadhyaya HD, Varshney RK (2010) Assessing genetic diversity for crop improvement. Curr Opin Plant Biol 13:167–173
Hao CY, Zhang XY, Wang LF, Dong YS, Shang XW, Jia JZ (2006) Genetic diversity and core collection evaluations in common wheat germplasm from the northwestern spring wheat region in China. Mol Breed 17:69–77
Hazen SP, Leroy P, Ward RW (2002) AFLP in Triticum aestivum L.: patterns of genetic diversity and genome distribution. Euphytica 125:89–102
Hoisington D, Khairallah M, Reeves T, Ribaut JM, Skovmand B, Taba S, Warburton M (1999) Plant genetic resources: what can they contribute toward increased crop productivity? Proc Nati Acad Sci USA 96:5937–5943
Huang XQ, Börner A, Röder MS, Ganal MW (2002) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor Appl Genet 105:699–707
Kim HS, Ward RW (2000) Patterns of RFLP-based genetic diversity in germplasm pools of common wheat with different geographical or breeding program origins. Euphytica 115:197–208
Krystkowiak K, Adamski T, Surma M, Kaczmarek Z (2009) Relationship between phenotypic and genetic diversity of parental genotypes and the specific combining ability and heterosis effects in wheat (Triticum aestivum L.). Euphytica 165:419–434
Landjeva S, Korzun V, Ganeva G (2006) Evaluation of genetic diversity among Bulgarian winter wheat (Triticum aestivum L.) varieties during the period 1925–2003 using microsatellites. Genet Res Crop Evol 53:1605–1614
Manifesto MM, Schlatter AR, Hopp HE, Suarez EY, Dubcovsky J (2001) Quantitative evaluation of genetic diversity in wheat germplasm using molecular markers. Crop Sci 41:682–690
Maric S, Bolaric S, Martincic J, Pejic I, Kozumplik V (2004) Genetic diversity of hexaploid wheat cultivars estimated by RAPD markers, morphological traits and coefficients of parentage. Plant Breed 123:366–369
Parker GD, Fox PN, Langridge P, Chalmers K, Whan B, Ganter PF (2002) Genetic diversity within Australian wheat breeding programs based on molecular and pedigree data. Euphytica 124:293–306
Reif JC, Zhang P, Dreisigacker S, Warburton ML, van Ginkel M, Hoisington D, Bohn M, Melchinger AE (2005) Wheat genetic diversity trends during domestication and breeding. Theor Appl Genet 110:859–864
Röder MS, Wendehake K, Korzun V, Bredemeijer G, Laborie D, Bertrand L, Isaac P, Rendell S, Jackson J, Cooke RJ, Vosman B, Ganal MW (2002) Construction and analysis of a microsatellite-based database of European wheat varieties. Theor Appl Genet 106:67–73
Roussel V, Koenig J, Beckert M, Balfourier F (2004) Molecular diversity in French bread wheat accessions related to temporal trends and breeding programmes. Theor Appl Genet 108:920–930
Roussel V, Leisova L, Exbrayat F, Stehno Z, Balfourier F (2005) SSR allelic diversity changes in 480 European bread wheat varieties released from 1840 to 2000. Theor Appl Genet 111:162–170
Roy JK, Lakshmikumaran MS, Balyan HS, Gupta PK (2004) AFLP-based genetic diversity and its comparison with diversity based on SSR, SAMPL, and phenotypic traits in bread wheat. Biochem Genet 42:43–59
Smale M (1997) The green revolution and wheat genetic diversity: some unfounded assumptions. World Dev 25:1257–1269
Soleimani VD, Baum BR, Johnson DA (2002) AFLP and pedigree-based genetic diversity estimates in modern cultivars of durum wheat [Triticum turgidum L. subsp. durum (Desf.) Husn.]. Theor Appl Genet 104:350–357
Stachel M, Lelley T, Grausgruber H, Vollmann J (2000) Application of microsatellites in wheat (Triticum aestivum L.) for studying genetic differentiation caused by selection for adaptation and use. Theor Appl Genet 100:242–248
Stubbs RW, Prescott JM, Saari EE, Dubin HJ (1986) Cereal disease methodology manual. Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT), Mexico
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327:818–822
van de Wouw M, van Hintum T, Kik C, van Treuren R, Visser B (2010) Genetic diversity trends in twentieth century crop cultivars: a meta analysis. Theor Appl Genet 120:1241–1252
White J, Law JR, MacKay I, Chalmers KJ, Smith JSC, Kilian A, Powell W (2008) The genetic diversity of UK, US and Australian cultivars of Triticum aestivum measured by DArT markers and considered by genome. Theor Appl Genet 116:439–453
Acknowledgment
This research was financed by a SEE-ERAnet project (No: 10237/2007-2008).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karsai, I., Vida, G., Petrovics, S. et al. Assessment of the spatial genotypic and phenotypic diversity present in the various winter wheat breeding programs in Southeast Europe. Euphytica 186, 139–151 (2012). https://doi.org/10.1007/s10681-011-0510-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-011-0510-2