Abstract
Potentilla alba L. is a valuable medicinal plant widely used in folk and traditional medicine and particularly promising in complex treatment of thyroid pathology. Natural resources of this species are insufficient due to ever-growing use in contemporary medicine. Comprehensive investigations of different P. alba populations are essential for the successful extension of P. alba plantings. Aiming for a better understanding of karyotype structure, chromosome behaviour in meiosis and developing new diagnostic characters, we performed molecular cytogenetic characterization and leaf structure and ultrastructure analyses of two introduced P. alba samples originating from different habitats. Based on chromosome morphology, distribution of 45S/5S rDNA and DAPI-banding patterns, all chromosomes in the karyotypes were identified and the P. alba chromosomal idiogram was constructed. Our findings confirmed P. alba karyotype stability and also revealed several diagnostic characters of this species: the features of cells of upper and lower leaf epidermis, the presence of calcium oxalate druses and three types of leaf indumentum, essential for evaluation of genetic diversity in different populations, validation of raw materials and further selection progress. The meiotic abnormalities were detected probably related to low pollen activity and indicated the advantages of vegetative propagation in the development of a P. alba plantation system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The large and polymorphic genus Potentilla L. (Rosaceae) consists of more than 500 species including valuable medicinal plants (Heywood 2007). They are annual and perennial herbs or shrubs distributed in arctic and temperate regions of the northern hemisphere (Wolf 1908; Yuzepchuk 1941; Heywood 2007). The phylogenetic relationships within the genus Potentilla are rather intricate as its speciation is a continuous process. Inter- and intraspecific hybridization events as well as crossings with various species from other genera of the family Rosaceae (e.g., Fragaria, Sibbaldia) result in the appearance of polyploids and/or new morphobiotypes with chromosome number polymorphism (Stebbins 1950; Asker and Frost 1970; Asker 1971; Grant 1981; Masterson 1994; Eriksson et al. 2003; Soltis and Soltis 2009). Besides, the capability of Potentilla species for different types of propagation (seed and/or vegetative) also complicates the taxonomy of the genus (Dickinson et al. 2007; Dobeš et al. 2015). Currently, for clarification of phylogenetic relationships within the genus and a better understanding of plant adaptation mechanisms to different environmental conditions (Shimono et al. 2009; Ma et al. 2015), the genus Potentilla is being intensively investigated at the morphological, biochemical and molecular levels (Eriksson et al. 1998; Kolodziejek and Gabara 2007; Dobes and Paule 2010; Paule et al. 2011; Rani et al. 2012; Faghir et al. 2014; Bogacheva et al. 2016).
Potentilla alba L. is one of the most valuable species of the genus, widely used in folk and traditional medicine. It contains broad range of biologically active compounds (iridoids, saponins, phenolcarbonic acids, quercetin, gallotannin, phytosterols) providing the vast variety of its pharmacologic properties (Smyk and Krivenko 1975; Kovalenko et al. 2004; Matkowski et al. 2006; Ossipov et al. 2017). In pharmaceutical industry, raw material of this medicinal plant is included as a component in various remedies used in the complex treatment of thyroid pathology, hepatotherapy, coronary heart diseases and gastrointestinal tract problems (Kovalenko et al. 2004; Dorman et al. 2011; Kvacheniuk and Kvacheniuk 2012; Kaminskiĭ et al. 2013; Turchaninova 2014).
Nowadays, despite the rather wide distribution area of P. alba (Yuzepchuk 1941), it is considered to be a depleted and threatened species, and its poor natural resources are inadequate for the increasing demand for various biopharmaceuticals (Smyk and Krivenko 1975). This requires extensive use of the raw material obtained from introduced P. alba populations. Comprehensive morphological and genomic investigations of different P. alba populations are essential for successful extension of P. alba plantings and selecting new promising varieties with desired characteristics adapted to environments. Morphological and anatomical studies of P. alba allowed the establishment of some specific characters (Yuzepchuk 1941; Bogacheva et al. 2016). The genomic peculiarities of P. alba are still poorly investigated as only chromosome number and sizes (1–2 µm) have been determined for this species (and for most species of the genus Potentilla as well) (Asker 1985a; Iwatsubo and Naruhashi 1991; Delgado et al. 2000; Tomasz and Kołodziejek 2008).
In the present study, aiming for a better understanding of karyotype structure, chromosome behaviour in meiosis and developing new diagnostic characters essential for evaluation of genetic diversity in populations of P. alba, we performed its molecular cytogenetic characterization and studied the leaf structure and ultrastructure of two introduced samples originating from different habitats.
Materials and methods
Plant material
In the present study, we investigated two introduced P. alba samples (К 127-02) and (К 328-02) which were derived from promising wild morphotypes in Ivanovo (56°59′48″N, 40°58′55″E) and Penza (53°12′00″N, 45°00′00″E) regions (Russian Federation), respectively. Since 2016, these samples have been cultivated in the Botanic Gardens of the All-Russian Institute of Medicinal and Aromatic Plants (VILAR), Moscow region, Russian Federation with a view to developing a new cultivar.
Chromosome spread preparation
In FISH assays, mitotic chromosome spreads were prepared from plant root meristem according to the technique developed previously for plant species with small chromosomes (Muravenko et al. 2009; Amosova et al. 2014).
For meiotic chromosome preparation, young floral buds were fixed in Carnoy’s solution for 30 min at 4 °C and then chromosome spreads were prepared as previously described (Samatadze et al. 2014). The slides were stored in 96% ethanol at − 20 °C until used.
FISH procedure
Following probes were used for FISH:
-
pTa71 containing a 9 kb long repeated DNA sequence of common wheat encoding 18S, 5.8S and 26S rRNA genes including spacers (Gerlach and Bedbrook 1979);
-
pTa794 containing a 420 bp long repeated DNA sequence of wheat containing the 5S rRNA gene and the intergenic spacer (Gerlach and Dyer 1980);
-
The rDNA probes were labelled directly with SpectrumAqua (45S rDNA) and SpectrumRed (5S rDNA) fluorochromes (Abbott Molecular, Wiesbaden, Germany) according to the manufacturer’s protocol. FISH procedure was carried out according to Muravenko et al. (2009). After overnight hybridization, the slides were washed as described previously (Amosova et al. 2017).
DAPI-banding
After FISH procedure chromosome slides were stained with 0.1 μg/mL DAPI (4′,6-diamidino-2-phenylindole) (Serva, Heidelberg, Germany) dissolved in Vectashield medium (Vector laboratories, Peterborough, UK).
Chromosome analysis
Chromosome slides were examined using an Olympus BX61 epifluorescence microscope (Olympus, Tokyo, Japan) combined with a monochrome CCD camera (Cool Snap, Roper Scientific Inc., Tucson, USA). The images were captured in grayscale channels. Then they were pseudocoloured and processed with Adobe Photoshop 10.0 (Adobe Systems Inc., Birmingham, USA) and VideoTest-Kario 1.5 (Ista-VideoTest, St Petersburg, Russia) software. Based on the results of measurements of 15 metaphase plates from 10 individual plants of each sample, total chromosome length, length of the short arm and the centromeric index (100 × length of short arm/total chromosome length) were calculated. Identification of P. alba mitotic chromosomes was based on their morphology and distribution of chromosome markers. The cytological numerical designation of P. alba chromosomes was by the decreasing order of size rather than by centromeric index (Levan et al. 1964). For meiosis analysis, at least 100 cells (10 plants) of each sample were analyzed.
Phytotomy and ultrastructure of leaves
Sample preparation for light and scanning electron microscopy (SEM) was performed according to the standard techniques described earlier (Dolgova and Ladygina 2003).
The study of the ultrastructure of P. alba leaf blades was conducted with the use of JEOL JSM – 6490LV scanning electron microscope (3.0 nm at 30 kV high vacuum mode) (Jeol, Tokyo, Japan). For this, the samples were coated with 20 nm (40 s at 40 mA) platinum film in the JEOL auto fine coater JFC – 160 (Jeol, Tokyo, Japan).
Results
DAPI-banding
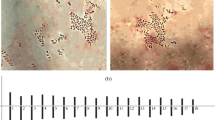
Karyotypes of both P. alba samples had 14 pairs of rather small (1.5–3 μm) chromosomes. Karyotype formula was К = 2n = 28 = 2(7 m + 5sm + 2st). The analysis of DAPI-banding patterns showed similar distribution of DAPI-bands in chromosomes of the P. alba specimens: large DAPI-bands were mostly located in the pericentromeric regions of chromosomes while polymorphic small and middle-sized bands were detected in the subtelomeric and intercalary regions (Fig. 1).
FISH-based localization of 45S (green) and 5S (red) rDNA and DAPI-banding patterns on P. alba chromosomes. Metaphase spreads of the samples from the Penza (A, B) and Ivanovo (C, D) regions after FISH and DAPI-banding (inverted images); (E, F)—Karyograms (the same metaphase plates as in A–D) of the specimens from the Penza and Ivanovo regions after DAPI-banding (inverted image) and FISH (only chromosomes with the hybridizations sites are presented). (Color figure online)
Chromosomal localization of 45S and 5S rDNA
In karyotypes of both P. alba samples, 45S rDNA sites were localized in the distal region of the short arms of two satellite (SAT) chromosome pairs. Double 5S rDNA sites were detected in the short arm (proximal and telomeric position) of one metacentric chromosome pair (Fig. 1). Based on chromosome morphology, DAPI-banding patterns and distribution of 45S and 5S rDNA sites, all chromosome pairs in the karyotypes were identified (Fig. 1) and chromosome idiograms of P. alba were constructed (Fig. 2). In the P. alba karyotypes, chromosomal abnormalities were not revealed.
Analysis of meiosis
In both P. alba samples, analysis of meiosis indicated fourteen rod bivalents. Besides, at metaphase I (M-I), chromosome associations (trivalents and quadrivalents) were found (Fig. 3A). At anaphase I (A-I) and anaphase II (A-II), most cells had normal chromosome disjunction (14:14) (Fig. 3B) but in 2.07–3.12% of the studied cells, various abnormalities (chromosome lagging, fragments, etc.) were also observed (detailed in Fig. 3C–G). At A-I and A-II, the most frequent aberration was lagging of several chromosomes behind the others resulting in their non-uniform distribution within a cell (Fig. 3C–G) as most lagging chromosomes could not reach the cellular poles and remained in the cell cytoplasm. In both studied P. alba samples, at the tetrad stage of meiosis II, normal tetrads (Fig. 3H) as well as few polyads (Fig. 3I) were observed.
Meiosis in maternal pollen cells of P. alba. A M-I: (n = 10II + 1IV); B A-I: normal distribution of chromosomes in the cell (14:14); C, D A-I: chromosome lagging; E A-I: non-uniform chromosome distribution within the cell; F A-II: non-uniform chromosome distribution within a cell; G A-II: chromosome lagging; H tetrad; I hexad
Phytotomy and ultrastructure of leaves
In most P. alba plants, palmately compound leaf blades with five leaflets were observed though polymorphism in number of leaflets (from 4 to 6) was also detected (Fig. 4).
In both P. alba samples, the upper leaf epidermis was represented by cells with straight polygonal or slightly flexuous walls (Fig. 5A). In the mesoderm, calcium oxalate druses located along the leaf veins were detected (Fig. 5A). The cells of the lower leaf epidermis had flexuous walls (Fig. 5B). The glandular hairs were pigmented, and inside the head as well as in the stalk cells, brown secreted material was visible (Fig. 5B).
Optical sections of leaf blades in P. alba. A Upper leaf epidermis cells with straight polygonal or slightly flexuous walls. Long arrows indicate calcium oxalate druses. B Lower leaf epidermis cells with flexuous walls. Long arrows indicate glandular hairs with brown secreted content. (Color figure online)
SEM analysis revealed numerous stomata on the lower leaf epidermis (Fig. 6). The stomata were anomocytic, and they were not detected on the upper epidermis of the leaves. Along the leaf edges, numerous simple trichomes were observed. The lower leaf epidermis was covered with simple unicellular hairs as well as glandular hairs consisting of a one- or two-celled round head continuous with a two- or three-celled stalk, and one basal cell. The simple unicellular hairs observed on the lower leaf epidermis were longer and thinner than the simple hairs on the leaf edges. It was found that the lower leaf epidermis of P. alba from the Penza region was more densely pubescent with simple unicellular trichomes than in P. alba plants from the Ivanovo region (Fig. 6).
Leaf ultrastructure in P. alba. SEM images of leaf blades of the specimens from the Ivanovo region (A, C, E) and Penza region (B, D, F). Long white arrows indicate simple trichomes; short white arrows indicate simple unicellular hairs; arrow heads indicate stomata; blue arrows indicate glandular hairs. (Color figure online)
Discussion
Karyotype analysis
For most species of the genus Potentilla, chromosome numbers and sizes (1–2 µm) were revealed (Müntzinc 1958; Asker 1985a; Iwatsubo and Naruhashi 1991; Delgado et al. 2000; Tomasz and Kolodziejek 2008). Within the genus, a wide variation in chromosome number ranging from 2n = 14 to 2n = 112 was found; most common is 2n = 28 with the basic (monoploid) chromosome number x = 7 supporting a paleopolyploid origin (Delgado et al. 2000; Tomasz and Kolodziejek 2008; Jeelani et al. 2012). The species within the genus Potentilla are usually subdivided into two groups: the species having constant chromosome number and the ones with variable chromosomal numbers which can vary even within the same species (Stebbins 1950; Mesicek and Sojak 1993).
Similar to most Potentilla species, P. alba has 2n = 28 (Delgado et al. 2000; Rani et al. 2012). In the present study, the karyotype analysis of two introduced P. alba populations confirmed chromosome number stability as well as a possible tetraploid origin of the genome of this species. It is known that the detailed investigation of karyotypes of the species with small chromosomes needs special approaches (Muravenko and Zelenin 2009). The application of DNA intercalator 9-AMA, which slowed down the process of chromosome condensation, allowed us to accumulate prometaphase chromosomes and obtain longer chromosomes in the spreads (1.5–3 μm) resulting in rather informative chromosome DAPI-banding patterns. In P. alba karyotypes, we observed large pericentromeric and also small polymorphic telomere and intercalary DAPI bands, and such patterns are typical for plant species having small chromosomes (Guerra 2000; Pinto-Maglio 2006; Muravenko et al. 2009; Samatadze et al. 2012; Yurkevich et al. 2013; Amosova et al. 2014).
It is common knowledge that eukaryotic ribosomal DNA is highly conserved and consists of tandem repeat units with thousands of copies which are clustered in one or several chromosome pairs (Pedersen and Linde Laursen 1994). For this reason, sites of rDNA are easily mapped on chromosomes by FISH and used as chromosomal markers essential for genomic investigations as well as clarification of phylogenetic relationships among species (Liu et al. 2006; Moraes et al. 2007; Las Peñas et al. 2008; Amosova et al. 2015; Bolsheva et al. 2016). In the present study, for the first time, we performed localization of 45S and 5S rDNA loci in chromosomes of P. alba and found 45S rDNA hybridization signals in two chromosome pairs whereas double 5S rDNA sites were detected in only one chromosome pair. These findings agreed with the hypothesis on a possible tetraploid origin of the P. alba genome, and localization of two 5S rDNA loci in one chromosome pair could indicate chromosome reorganization occurred during speciation.
Analysis of meiosis
Meiotic abnormalities (cytomixis, chromatin stickiness, the presence of unoriented bivalents, chromatin bridges, etc.) were early revealed in some Potentilla species (Rani et al. 2012). These abnormalities were described in other plant species and can be related to the intraspecific genetic variability (Baptista-Giacomelli et al. 2000; Sheidai et al. 2003). They are considered to be the result of some genetic factors (Bellucci et al. 2003; Ghaffari 2006; Fadaei et al. 2010), environmental factors (Nirmala and Rao 1996) as well as genomic responses to environmental variation (Baptista-Giacomelli et al. 2000). It is known that in tetraploid, allo- and autopolyploids, meiotic abnormalities can lead to pollen sterility and decreasing seed production (Stebbins 1950).
Serious problems during the introduction of different P. alba populations are the lack of planting material and also lower seed production which slow efforts on plantation system development. P. alba seeds are characterized by a low germination ability and a long period of seedling emergence. Besides, the seedlings grow very slowly. Consequently, vegetative propagation is considered to be more effective (compared to seed propagation) for increasing plantation areas of this valuable medicinal plant (Smyk and Krivenko 1975; Kosman et al. 2013). The analysis of meiosis is a rather effective tool for clarification of the cause of low seed production as it enables to reveal abnormalities in early ontogenesis, which can lead to pollen sterility and, thus, decreasing seed production.
In meiosis of the P. alba samples, normal chromosome disjunction (14:14) typical for both diploids and paleopolyploids was observed. Besides, small number cells with various abnormalities were also detected. Chromosomal lagging represented considerable proportion among the revealed aberrations, and this is illustrative of the spindle assembly defects. As the result of such chromosomal aberrations, gametes with unbalanced chromosome number were formed (Sybenga 1996). Misorient tetrads and polyads observed at the tetrad stage of meiosis II were probably caused by defects in achromatic spindle orientation.
Phytotomy and ultrastructural characterization of leaves
The identification and quality of the raw material of P. alba are especially important for its medicinal use. Morphological analysis is one of the most universal and available approach. For specification within the genus Potentilla, the features of the leaf blades are important diagnostic characters (Faghir et al. 2010). The type of leaf is usually characteristic of a species (monomorphic), although some species produce more than one type of leaf (dimorphic or polymorphic). Polymorphic leaves were early observed in several Potentilla species (Müntzinc 1958; Chitwood and Sinha 2016). In P. alba, palmately compound leaves with five leaflets are considered to be a characteristic feature (Yuzepchuk 1941). In the present study, P. alba plants with polymorphic leaf blades were found in both studied samples. As leaf shape polymorphism can be related to ontogenetic factors and/or environmental plasticity (Chitwood and Sinha 2016), it cannot be considered as a constant diagnostic character for the species, and accordingly, other diagnostic features (epidermis structure, stomata types, trichome characters, the presence and forms of crystal inclusions, etc.) are needed for species characterization (Faghir et al. 2010).
Our study of the leaf structure and ultrastructure of the P. alba samples revealed several common characters in the P. alba samples: the structure of cells of upper and lower leaf epidermis, the presence of calcium oxalate druses as well as three types of leaf indumentum. Similar features were earlier detected in the cells of P. alba rhizomes and roots (Bogacheva et al. 2016). Moreover, the distinctive differences in leaf indumentum ultrastructure between the P. alba samples were also observed. This finding could be related to the species environmental plasticity as ontogenetic polyvariety, which is considered to be a species adaptation mechanism, was early detected within representatives of the genus Potentilla (Müntzinc 1958; Asker 1985b).
Conclusion
In this study, for the first time, we performed molecular cytogenetic characterization and analyzed leaf structure and ultrastructure of two introduced P. alba samples from different habitats and distant regions. Based on chromosome morphology and distribution of chromosomal markers, all chromosomes in karyotypes were identified and the chromosomal idiogram of P. alba was constructed. Our findings confirm P. alba karyotype stability and also reveal several diagnostic characters of this species essential for evaluation of genetic diversity in different populations, validation of raw materials as well as further progress in selection. The various meiotic abnormalities together with the low seed production of most P. alba populations, hindering efforts on development of a plantation system, suggest the advantages of vegetative propagation for the successful extension of plantings for this valuable medicinal plant.
References
Amosova AV, Zemtsova LV, Grushetskaya ZE, Samatadze TE, Mozgova GV, Pilyuk YE, Volovik VT, Melnikova NV, Zelenin AV, Lemesh VA, Muravenko OV (2014) Intraspecific chromosomal and genetic polymorphism in Brassica napus L. detected by cytogenetic and molecular markers. J Genet 93:123–143
Amosova AV, Bolsheva NL, Samatadze TE, Twardovska MO, Zoshchuk SA, Andreev IO, Badaeva ED, Kunakh VA, Muravenko OV (2015) Molecular cytogenetic analysis of Deschampsia antarctica Desv. (Poaceae), Maritime Antarctic. PLoS ONE 10(9):e0138878. https://doi.org/10.1371/journal.pone.0138878
Amosova AV, Zemtsova LV, Yurkevich OY, Zhidkova EN, Książczyk T, Shostak NG, Muravlev AA, Artemyeva AM, Samatadze TE, Zoshchuk SA, Muravenko OV (2017) Genomic changes in generations of synthetic rapeseed-like allopolyploid grown under selection. Euphytica 9:213–217
Asker S (1971) Some viewpoints on Fragaria × Potentilla intergeneric hybridization. Hereditas 67:181–190
Asker S (1985a) Chromosome studies in Potentilla. Hereditas 102:289–292
Asker S (1985b) Polymorphism of Potentilla tabentaemontani and related taxa on Gotland. Hereditas 102:39–45
Asker S, Frost S (1970) The “Potentilla collina problem”—a chemotaxonomic approach. Hereditas 66:49–70
Baptista-Giacomelli FR, Pagliarini MS, Almeida JL (2000) Meiotic behavior in several Brazilian oat cultivars (Avena sativa L.). Cytologia 65:371–378
Bellucci M, Roscini C, Mariani A (2003) Cytomixis in pollen mother cells of Medicago sativa L. J Hered 94:512–516
Bogacheva NG, Meshkov AI, Konyaeva EA, Alent’eva OG (2016) Pharmocognostic study of the rhizomes and the roots Potentilla alba L. Prob Biol Med Pharm Chem 1:28–32
Bolsheva NL, Dyachenko OV, Samatadze TE, Rachinskaya OA, Zakharchenko TV, Shevchuk NS, Amosova AV, Muravenko OV, Zelenin AV (2016) A karyotype of Mesembryanthemum crystallinum (Aizoaceae) studied by chromosome banding, FISH with rDNA probes and immunofluorescence detection of DNA methylation. Plant Biosyst 160:916–922
Chitwood DH, Sinha NR (2016) Evolutionary and environmental forces sculpting leaf development. Curr Biol 26(7):297–306
Delgado L, Gallego F, Rico E (2000) Karyosystematic study of Potentilla L. subgen. Potentilla (Rosaceae) in the Iberian Peninsula. Bot J Linn Soc 132:263–280
Dickinson TA, Lo E, Talent N (2007) Ploidy, reproductive biology, and Rosaceae: understanding evolution and making classifications. Plant Syst Evol 266:59–78
Dobes C, Paule J (2010) A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): implications for its geographic origin, phylogeography and generic circumscription. Mol Phylogenet Evol 56:156–175
Dobes C, Luckl A, Kausche L, Scheffknecht S, Prohaska D, Sykora C, Paule J (2015) Parallel origins of apomixis in two diverged evolutionary lineages in tribe Potentilleae (Rosaceae). Bot J Linn Soc 177:214–229
Dolgova AA, Ladygina EY (2003) Manual of practical pharmacognosy. Meditsina, Moscow
Dorman DHJ, Shikov AN, Pozharitskaya ON, Hiltunen R (2011) Antioxidant and pro-oxidant evaluation of Potentilla alba L. rhizome extract. Chem Biodivers 8:1344–1356
Eriksson T, Donoghue MJ, Hibbs MS (1998) Phylogenetic analysis of Potentilla using DNA sequences of nuclear ribosomal internal transcribed spacers (ITS), and implications for the classification of Rosoideae (Rosaceae). Plant Syst Evol 211:155–179
Eriksson T, Hibbs MS, Yoder AD, Delwiche CF, Donoghue MJ (2003) The phylogeny of Rosoideae (Rosaceae) based on sequences of the internal transcribed spacers (ITS) of nuclear ribosomal DNA and the TRNL/F region of chloroplast DNA. Int J Plant Sci 164:197–211
Fadaei F, Sheidai M, Asadi M (2010) Cytological study of the genus Arenaria L. (Caryophyllaceae). Caryologia 63:149–156
Faghir MB, Attar F, Farazmand A, Ertter D, Eriksen B (2010) Leaf indumentum types in Potentilla (Rosaceae) and related genera in Iran. Acta Soc Bot Pol 79:139–145
Faghir MB, Attar F, Farazmand A, Kazempour Osaloo S (2014) Phylogeny of the genus Potentilla (Rosaceae) in Iran based on nrDNA ITS and cpDNA trnL-F sequences with a focus on leaf and style characters’ evolution. Turk J Bot 38:417–429
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res 8:4851–4865
Ghaffari SM (2006) Occurrence of diploid and polyploid microspores in Sorghum bicolor (Poaceae) is the result of cytomixis. Afr J Biotechnol 5:1450–1453
Grant V (1981) Plant speciation. Columbia University Press, New York
Guerra M (2000) Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol 23(4):1029–1041
Heywood VH (2007) Flowering plants of the world. Mayflower Books. Elsevier, New York
Iwatsubo Y, Naruhashi N (1991) Karyomorphological and cytogenetical studies in Potentilla (Rosaceae) I. Karyotypes of nine Japanese species. Cytologia 56:1–10
Jeelani SM, Kumari S, Gupta RC (2012) Meiotic studies in some selected angiosperms from the Kashmir Himalayas. J Syst Evol 50:244–257
Kaminskiĭ AV, Kiseleva IA, Teplaia EV (2013) Clinical application of Potentilla alba for prevention and treatment of thyroid gland pathologies. Likars’ka Sprava 8:99–108
Kołodziejek J, Gabara B (2007) Characteristics of achenes in Potentilla collina group (Rosaceae). Acta Soc Bot Pol 76(1):35–42
Kosman VM, Faustova NM, Pozharitskaya ON, Makarov VG (2013) Accumulation of biologically active compounds in underground parts of composition of Potentilla alba L. after various cultivation terms. Russ J Bioorg Chem 2:139–146
Kovalenko PG, Antonjuk VP, Maliuta SS (2004) Secondary metabolites production from transformed cells of Glycyrrhiza glabra and Potentilla alba as producers of radio-protective compounds. Ukr Bioorg Acta 1–2:13–22
Kvacheniuk AN, Kvacheniuk EL (2012) The use of phytotherapy for treatment of thyroid diseases. Likars’ka Sprava 3:99–104
Las Peñas ML, Bernardello G, Kiesling R (2008) Karyotypes and fluorescent chromosome banding in Pyrrhocactus (Cactaceae). Plant Syst Evol 272:211–222
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220
Liu B, Chen C, Li X, Qi L, Han S (2006) Karyotype analysis and physical mapping of 45S rDNA in eight species of Sophora, Robinia, and Amorpha. Front Biol China 3:290–294
Ma L, Sun X, Kong X, Galvan JV, Li X, Yang S, Yang Y, Yang Y, Hu X (2015) Physiological, biochemical and proteomics analysis reveals the adaptation strategies of the alpine plant Potentilla saundersiana at altitude gradient of the Northwestern Tibetan Plateau. J Proteom 112:63–82
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264:421–423
Matkowski A, Świąder K, Ślusarczyk S, Jezierska-Domaradzka A, Oszmiański J (2006) Free radical scavenging activity of extracts obtained from cultivated plants of Potentilla alba L. and Waldsteinia geoides L. Herva Pol 52(4):91–97
Mesicek J, Sojak J (1993) Annotated chromosome numbers of selected Asiatic Potentilla species. Folia Geobot Phytotax 28(4):437–446
Moraes AP, Filho WS, Guerra M (2007) Karyotype diversity and the origin of grapefruit. Chromosome Res 15:115–121
Müntzinc A (1958) Heteroploidy and polymorphism in some apomictic species of Potentilla. Hereditas 44:280–329
Muravenko OV, Zelenin AV (2009) Chromosomal organization of the genomes of small chromosome plants. Russ J Genet 45:1338–1350
Muravenko OV, Yurkevich OYu, Bolsheva NL, Samatadze TE, Nosova IV, Zelenina DA, Volkov AA, Popov KV, Zelenin AV (2009) Comparison of genomes of eight species of sections Linum and Adenolinum from the genus Linum based on chromosome banding, molecular markers and RAPD analysis. Genetica 135:245–255
Nirmala A, Rao PN (1996) Genetics of chromosome numerical mosaism in higher plants. Nucleus 39:151–175
Ossipov VI, Polyakov NA, Sidelnikov AN, Hazieva FM (2017) Proanthocyanidins in the roots and rhizomes of Potentilla alba (Rosaceae). Rastitelnye Resursy 53:114–125
Paule J, Sharbal TF, Dobes C (2011) Apomoctic and sexual lineages of the Potentilla argentea L. group (Rosaceae): cytotype and molecular genetic differentiation. Taxon 60:721–732
Pedersen C, Linde Laursen I (1994) Chromosomal locations of four minor rDNA loci and a marker microsatellite sequence in barley. Chromosome Res 2:65–71
Pinto-Maglio CAF (2006) Cytogenetics of coffee. Braz J Plant Physiol 18:37–44
Rani S, Kumar S, Jeelani SM, Gupta RC, Kumari S (2012) Additions to the cytologically investigated species of Potentilla L. (Rosaceae) from India. Plant Syst Evol 298:485–497
Samatadze TE, Zelenin AV, Amosova AV, Popov KV, Suslina SN, Zagumennikova TN, Tsytsylin AN, Bykov VA, Muravenko OV (2012) Comparative cytogenetic study of the forms of Macleaya cordata (Willd.) R. Br. from different localities. Russ J Genet 48:63–69
Samatadze TE, Amosova AV, Melnikova NV, Suslina SN, Zagumennikova TN, Zelenin AV, Bykov VA, Muravenko OV (2014) Comparative cytogenetic study of the tetraploid Matricaria chamomilla L. and Matricaria inodora L. Biol Bull 2:123–132
Sheidai M, Koobaz P, Zehzad B (2003) Meiotic studies of some Avena L. species and populations. Iran J Sci 14:121–131
Shimono Y, Watanabe M, Hirao AS, Wada N, Kudo G (2009) Morphological and genetic variations of Potentilla matsumurae (Rosaceae) between fellfield and snowbed populations. Am J Bot 96(4):728–737
Smyk GK, Krivenko VV (1975) White cinquefoil, an effective agent for treating thyroid gland diseases. Farm Zh 2:58–62
Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60:561–588
Stebbins GL (1950) Variation and evolution in plants. Columbia University Press, New York
Sybenga J (1996) Recombination and chiasmata: few but in triguing discrepancies. Genome 39:473–484
Tomasz I, Kołodziejek J (2008) Chromosome numbers of Potentilla subsect. Collinae (Rosaceae) from Poland. Caryologia 61:170–175
Turchaninova LI (2014) Experience of using phytopreparation Alba (root extract of the Potentilla alba) in complex treatment of thyroid pathology in children and adolescents. Likars’ka sprava 3:125–129
Wolf T (1908) Monographie der Gattung Potentilla L. Biblioth Bot 71:1–714
Yurkevich OYu, Naumenko-Svetlova AA, Bolsheva NL, Samatadze TE, Rachinskaya OA, Kudryavtseva AV, Zelenina DA, Volkov AA, Zelenin AV, Muravenko OV (2013) Investigation of genome polymorphism and seed coat anatomy of species of section Adenolinum from the genus Linum. Genet Resour Crop Evol 60:661–676
Yuzepchuk SV (1941) Genus Potentilla. In: Shishkin BK (ed) Flora SSSR (Flora of the Soviet Union), vol 10. Leningrad, Moscow, pp 78–223
Funding
This work was supported by the Program of Fundamental Research for State Academies (No. 01201363824) and Russian Foundation of Basic Research (No. 17-29-08-034; 18-016-00167).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests in this work.
Rights and permissions
About this article
Cite this article
Samatadze, T.E., Zoshchuk, S.A., Khomik, A.S. et al. Molecular cytogenetic characterization, leaf anatomy and ultrastructure of the medicinal plant Potentilla alba L.. Genet Resour Crop Evol 65, 1637–1647 (2018). https://doi.org/10.1007/s10722-018-0640-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-018-0640-7