Abstract
Genetic diversity among 19 Triticum aestivum accessions and 73 accessions of closely related species was analyzed using simple sequence repeat (SSR) markers. Forty-four out of 497 SSR markers were polymorphic. In total 274 alleles were detected (mean 6.32 alleles per locus). The polymorphic information content (PIC) of the loci ranged from 0.3589 to 0.8854 (mean 0.7538). The D genome contained the highest mean number of alleles (6.32) followed by the A and B genomes (6.13 and 5.94, respectively). The correlation between PIC and allele number was significant in all genome groups (0.7540, 0.7361 and 0.7482 for A, B and D genomes, respectively). Among the seven homologous chromosome groups, genetic diversity was lowest in group 7 and highest in group 5. In cluster and principal component analyses, all accessions grouped according to their genomes were consistent with their taxonomic classification. Accessions with the A and D genomes were clustered into two distinct groups, and AABB accessions showed abundant genetic diversity and a close relationship. Triticum durum and T. turgidum were clustered together, consistent with their morphological similarity. Cluster analysis indicated emmer is closely related to hexaploid wheat. Compared with common wheat, higher genetic variation was detected in spelt, T. aestivum subsp. yunnanense and subsp. tibetanum. In addition, a close genetic relationship between T. polonicum and T. macha was observed. The results of the clustering and principal component analyses were essentially consistent, but the latter method more explicitly displayed the relationships among wheat and closely related species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As population growth and conflicting resource requirements continue to increase, coupled with a significant reduction in the area of arable land, environmental changes and loss of genetic diversity, improving wheat production is becoming increasingly challenging. The loss of genetic diversity not only limits further improvement of yield and quality, but also increases the vulnerability of wheat to biotic and abiotic stresses. On the other hand, numerous wild species closely related to common wheat are a particularly valuable source of genes for disease, cold, insect and salinity resistance and high grain quality (Orth and Bushuk 1973; Limin and Fowler 1981; Schachtman et al. 1992). Therefore wheat-related species are an excellent potential source of genes for wheat improvement (Knaggs et al. 2000).
Molecular markers have been widely used in wheat genetics research and breeding over the last 20 years. Studies of genetic diversity in wheat have used different types of molecular markers such as randomly amplified polymorphic DNA (Joshi and Nguyen 1993), restriction fragment length polymorphisms (Kim and Ward 2000), and amplified fragment length polymorphisms (Shoaib and Arabi 2006). Most of these molecular markers, however, show low level of polymorphisms in wheat. Another PCR-based marker type, SSRs, which are also known as microsatellites, offers a number of advantages, such as a high level of polymorphism, locus specificity, codominance, reproducibility, convenience, and random and uniform distribution throughout the genome (Röder et al. 2002). SSR markers are proven to be an efficient tool for estimation of genetic variation in wheat (Landjeva et al. 2006).
Among wheat and its close relatives, six species have the same chromosomal composition (AABBDD) as common wheat (Triticum aestivum L. s.str.), namely T. macha Dekapr. et Menabde, T. spelta L., T. vavilovii (Thum.) Jakubz., T. compactum Host, T. petropavlovskyi Udacz. et Migusch., and T. sphaerococcum Percival (Hammer et al. 2011). In China, several species have the AABBDD genomic composition, such as Xinjiang rice wheat (T. petropavlovskyi), T. aestivum subsp. tibetanum (Tibetan wheat) and T. aestivum subsp. yunnanense (Yunnan wheat) (Ward et al. 1998). These species are an important germplasm resource for improvement of common wheat and contain many desirable genes. Investigation of the genetic diversity of wheat germplasm is crucial in order to broaden the gene pool available for wheat breeding. Although several studies on genetic diversity of wheat and closely related species have been reported, most of these studies provided limited information because they evaluated a narrowly diverse germplasm selection (Maher et al. 2005; Naghavi et al. 2009a, b; Achtar et al. 2010).
In the present study, SSR markers were used to investigate the genetic diversity of 92 accessions of wheat and closely related species. This information will be useful to facilitate the efficient utilization of wheat-related species in the breeding of novel wheat cultivars.
Materials and methods
Experimental materials
We examined genetic diversity among 19 Triticum aestivum accessions and 74 accessions of closely related species, comprising seven accessions of Aegilops tauschii Coss., seven accessions of T. boeoticum Boiss., 13 accessions of T. durum Desf., 13 accessions of T. turgidum L., three accessions of T. dicoccoides (Körn. ex Asch. et Graebn.) Schweinf., seven accessions of T. dicoccon Schrank, six accessions of T. polonicum L., two accessions of T. carthlicum Nevski for T. persicum, two accessions of T. macha Dek. et Men., two accessions of T. araraticum Jakubz., two accessions of T. turanicum Jacubc. for T. orientale, nine accessions of T. spelta L., two accessions of T. aestivum L. subsp. yunnanense King (Yunnan wheat), nine accessions of T. aestivum subsp. tibetanicum Shao (Tibetan wheat), and eight accessions of T. aestivum L. The accessions were provided by Dr. Lihui Li from the Chinese Academy of Agriculture Sciences, Beijing, China. Detailed information for these accessions is listed in Table 1.
Extraction of genomic DNA
Genomic DNA was extracted using the CTAB method (Saghai-Maroof et al. 1984). The quality and quantity of the extracted DNA was measured by agarose gel electrophoresis and UV spectrophotometry, respectively. The final DNA concentration was adjusted to 50 ng/l.
Selection of SSR markers and detection of PCR products
A total of 497 SSR markers were selected from the following primer sets: GWM (Röder et al. 1998), WMC (Somers et al. 2004) and BARC (Song et al. 2005). The sequences of the SSR primers were obtained from the GrainGenes Database (http://wheat.pw.usda.gov/GG2/index.shtm).
PCR amplifications were performed in 20 μl volumes containing 0.31 μM of each primer, 0.9 mM dNTP mix, 2.5 mM MgCl2, 1U Taq polymerase, 1×PCR buffer and 50 ng DNA template. The amplification protocol consisted of 4 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at an annealing temperature of 50–60 °C, and 1 min at 72 °C, and final extension for 8 min at 72 °C. The amplification products from the markers were separated in an 8 % polyacrylamide gel, visualized with silver and photographed.
Data analyses
The presence of a band in the same migration site of the gel produced in two independent experiments was recorded as 1 and the absence of the band was recorded as 0. Accordingly, a binary (0, 1) data matrix was constructed (Nei and Li 1979). The average polymorphic information content (PIC) was calculated for each marker in accordance with the method of Maher et al. (2005). Dice’s coefficient was calculated with the similarity analysis module (Qualitative) in the NTSYSpc2.1 software (Rohlf 2000). Cluster analysis was performed with the SAHN module of NTSYSpc2.1 using the unweighted pair-group method with arithmetic averages (UPGMA) and a dendrogram was generated accordingly. Two dimensional (2D) analyses of the Dice coefficient matrix were performed with Decneter in the Output & transf module and Eigen in the Ordination module (principal component analysis) of NTSYSpc2.1.
Results
Polymorphism of SSR loci
Forty-four out of the 497 SSR markers were polymorphic and were used to characterize and estimate the genetic diversity of the 92 genotypes. The total number of detected alleles was 274 with an average allele number of 6.32 per locus. The number of alleles per locus ranged from two for the Xwmc296 locus in the A genome to 13 for the Xwmc506 locus in the D genome. The average PIC values reflecting the genetic diversity among the 44 microsatellite loci was 0.7482. The PIC values ranged from 0.3589 for locus Xgwm295-7D to 0.8854 for locus Xwmc707-4A (Table 2).
Genetic diversity among the wheat genomes and chromosome groups
The 44 loci were evenly distributed in the A, B, and D genomes (16, 18, and 10, respectively). The highest mean number of alleles was detected in the D genome (6.90) followed by the A genome (6.13), and the lowest number was detected in the B genome (5.94). The highest mean PIC was recorded in the A genome (0.7540), followed by the D genome (0.7482) and the B genome (0.7361) (Fig. 1).
The highest PIC value of 0.1585 was observed for the fifth homologous chromosome group markers, and the lowest value of 0.0882 was observed for the seventh homologous group markers. Among the seven homologous chromosome groups in wheat, the ranking for average genetic richness was 4 (7.83) > 5 (7.00) = 6 (7.00) > 7 (6.25) > 1 (6.00) > 3 (5.80) > 2 (4.00), whereas the ranking for the genetic diversity index was 5 (0.1585) > 3 (0.1499) > 4 (0.1383) > 6 (0.1293) > 2 (0.1264) > 1 (0.0935) > 7 (0.0882). Overall, we detected considerable genetic diversity as measured by the average genetic richness and genetic diversity index (Fig. 2).
Cluster analysis of the 92 accessions
In order to explore relationships among the 92 genotypes, the Dice genetic similarity (GS) coefficient was calculated according to the binary (1, 0) data matrix of the amplified bands for the 92 wheat samples. The results showed that the GS coefficient varied from 0.57 to 0.94 among the samples with an average value of 0.7638 (Fig. 3a). The highest GS coefficient was observed between T. turgidum L. (no. 31) and T. turgidum L. (no. 32), indicating that these two accessions showed the highest genetic similarity and the closest genetic relationship.
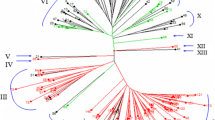
Dendrogram generated from an UPGMA cluster analysis (a) and two-dimensional principal component analysis scatterplot (b) derived from data for 44 SSR markers for 92 accessions of wheat and closely related species. The number beside each accession name in (a) and each plotted point in (b) corresponds to the respective accession numbers listed in Table 1. L was used as the cutoff for the average genetic similarity coefficient
UPGMA cluster analysis based on the GS matrix was used to analyze the 274 polymorphic bands amplified by the SSR primers. Two main clusters were resolved at a similarity of 0.57(L). Four groups were separated at 0.59(L) (Fig. 3a); the first group contained 17 genotypes, including all accessions of the diploid species Ae. tauschii Coss. and T. boeoticum Boiss.; the second group contained 39 genotypes, including all of the tetraploid wheat accessions; the third group contained 33 accessions, including all hexaploid wheat genotypes; and the fourth group consisted of a single T. spelta accession (no. 66). Accessions that shared the same genome were clustered in the same group, which is consistent with the genomic and taxonomic classification. However, some accessions of the same species were clustered into two groups, such as accessions nos. 57 and 58 of T. carthlicum, which were separated into groups I and II, respectively, and thus indicated that a wealth of genetic diversity exists in this species. Some conspecific accessions were placed in the same cluster and were indicated to have a close relationship, such as accessions nos. 31 and 32 of T. turgidum. Further research is needed to clarify the relationships of these accessions.
The principal coordinate analysis divided the 92 accessions into three groups (Fig. 3b). These results were highly consistent with those obtained with UPGMA clustering. However, principal component analysis can more explicitly display the relationships between accessions than other methods. T. durum (nos. 15–27) and T. turgidum (nos. 28–40, except no 38) were clustered in the same group as common wheat accessions, which indicated that these two species shared a close genetic relationship. Abundant genetic variation in the genomes of Yunnan wheat and Tibetan wheat was indicated, which could be utilized to broaden the gene pool of common wheat. In addition, a close genetic relationship was observed between T. polonicum and T. macha, which were clustered in the same group.
Cluster analysis of wheat genomes among the accessions
The relationship between the A and D genomes among the wheat genotypes was examined by means of a cluster analysis. UPGMA cluster analysis of the GS matrix was performed to analyze the 199 polymorphic bands amplified with the SSR primers among the 14 accessions with A or D genomic constitutions. The accessions were divided into two distinct clusters at a similarity coefficient of 0.66(L) and were separated from all other accessions, with only one accession with an A genomic constitution (T. boeoticum no 12, Hungary) placed independently.
The average GS coefficient among accessions with either A or D genomic constitutions was 0.6612 and ranged from 0.45 to 0.87 (Fig. 4). The average GS coefficient among accessions with an A genomic constitution (0.7682) was higher than that for accessions with a D genomic constitution (0.6872), which indicated that the latter showed more abundant genetic diversity than those with the A genome.
Dendrogram generated from an UPGMA cluster analysis (a) and two-dimensional principal component analysis scatterplot (b) derived from data for 44 SSR markers for accessions of wheat and closely related species carrying a single genome. The number beside each accession name in (a) and each plotted point in (b) corresponds to the respective accession numbers listed in Table 1
Accessions with an AABB genomic constitution showed abundant genetic diversity and a close relationship. UPGMA cluster analysis of the GS matrix was performed to analyze the 268 polymorphic bands amplified with the SSR primers among the 48 accessions with AABB genomic constitutions, basically the same species of materials in a group. For most species, such as T. durum Desf. and T. turgidum L., multiple accessions were clustered in the same group (Fig. 5). However, some accessions of the same species were not clustered together, such as T. araraticum nos. 61 and 62, T. turanicum nos. 63 and 64, persicum in carthlicum, and the accessions T. durum no 15, T. polonicum no 51 and T. turgidum no 38 were not clustered with other conspecific accessions. The average GS coefficient among these accessions was 0.6948 and ranged from 0.57 to 0.94 (Fig. 5). The highest GS coefficient (0.94) was observed between T. turgidum L. accession nos. 31 and 32, which indicated that these two taxa showed the lowest genetic similarity. The average GS coefficients of species with an AABB genomic constitution were ranked as follows: T. turanicum for T. orientale (0.6506) < T. dicoccon Schrank (0.6548) < T. carthlicum (0.6615) < T. polonicum L. (0.6734) < T. araraticum Jakubz. (0.6874) < T. dicoccoides (0.6912) < T. durum Desf. (0.7505) < T. turgidum L. (0.7612).
Dendrogram generated from an UPGMA cluster analysis (a) and two-dimensional principal component analysis scatterplot (b) derived from data for 44 SSR markers for accessions of wheat and closely related species carrying two genomes. The number beside each accession name corresponds to the respective accession numbers listed in Table 1
Among genotypes with an AABBDD genomic constitution, UPGMA cluster analysis of the GS matrix was performed to analyze the 260 polymorphic bands amplified with the SSR primers among the 30 accessions with an AABBDD genomic constitution. Accessions of T. macha and T. spelta were clustered into separate groups. Accessions of T. aestivum subsp. yunnanense and subsp. tibetanum were clustered in the same group as common wheat accessions. The average GS coefficient was 0.7167 and ranged from 0.55 to 0.86 (Fig. 6). The lowest GS coefficient (0.552) was observed for T. spelta L.(no. 66). The highest GS coefficient (0.86) was between T. aestivum L. (no. 88) and T. aestivum L. (no. 89), which indicated that these two species showed the greatest genetic dissimilarity. The average GS coefficients of species with an AABBDD genomic constitution were ranked as follows: T. spelta L. (0.6812) < T. macha Dek. et Men. (0.7312) < T. aestivum subsp. tibetanum Shao (0.7602) < T. aestivum subsp. yunnanense King (0.7712) < T. aestivum L. (0.7852).
Dendrogram generated from an UPGMA cluster analysis (a) and two-dimensional principal component analysis scatterplot (b) derived from data for 44 primer pairs for accessions of wheat and closely related species carrying three genomes. The number beside each accession name corresponds to the respective accession numbers listed in Table l
Discussion
Most previous studies of wheat genetic diversity show that species closely related to wheat contain high genetic diversity. Prasad et al. (2000) analyzed data for 55 wheat genotypes sourced from six continents and reported that one to 13 alleles were amplified at each SSR locus. Chao et al. (2007) reported an average of 7.2 alleles per locus among US wheat accessions with a range of two to 24 alleles per locus. In contrast, some studies have observed lower allelic diversity in common wheat. For example, Chen et al. (2003) screened genetic diversity among Chinese winter wheat germplasm and reported an average of 3.5 alleles per locus. Zhan et al. (2006) used 79 pairs of SSR primers to analyze 129 wheat accessions cultivated in the Huang-huai District and detected a total of 335 alleles, with each primer detecting two to eight alleles (average 4.24). Ma and Zhang (2008) screened 57 accessions of Chinese wheat and detected an average of 3.09 amplified alleles per locus. In the present study, 92 accessions of wheat and closely related species were screened with 44 pairs of SSR primers. Between two and 13 alleles were amplified for each SSR locus with an average of 6.32 per locus.
Of the three wheat genomes, the highest genetic diversity was present in the D genome (0.0749), followed by the A genome (0.0471) and the B genome (0.0408). With regard to homologous chromosome groups, the highest PIC value of 0.1585 was observed for group 5 chromosome markers and the lowest value of 0.0882 was recorded for group 7 markers. These findings contrast with those of a previous study (Huang et al. 2002a, b) in which the highest PIC value was reported for the B genome followed by A and D genomes. Other studies report highest PIC values for homologous chromosome group 2 (Huang et al. 2002a, b; Röder et al. 2002), whereas in the present study the highest diversity was observed for homologous group 7 using a similar set of primers. This difference may reflect differences in the geographical provenances of the accessions analyzed in the present and previous studies. Chen and Li (2007) observed that the ranking of average locus diversity per genome was D > B > A in synthetic hexaploid wheat genotypes. Chao et al. (2007) reported a ranking of B > D > A among US wheat genotypes, and Dreisigacker et al. (2005) reported a ranking of B > A > D among 68 CIMMYT wheat lines.
In the present study, using cluster analysis and principal component analysis to analyze the genetic diversity among 92 accessions, all species were grouped according to their genomic constitution and consistent with their taxonomic classification. The average GS coefficient for each species differed among the 92 accessions analyzed and the following ranking of species was observed: T. turanicum (0.6506) < T. dicoccon (0.6548) < T. carthlicum (0.6615) < T. polonicum L. (0.6734) < T. spelta L. (0.6812) < Ae. tauschii Coss. (0.6872) < T. araraticum (0.6874) < T.dicoccoides (0.6912) < T. macha (0.7312) < T. durum Desf. (0.7505) < T. turgidum L. (0.7612) < Tibetan wheat (0.7602) < T. boeoticum Boiss. (0.7682) < Yunnan wheat (0.7712) < T. aestivum L. (0.7852). Yang et al. (2007), using 24 microsatellite molecular markers to investigate the genetic diversity of different wheat populations, reported a ranking of average genetic diversity as follows: Spelt wheat (0.5359) > wild emmer wheat (0.3513) > Ae. tauschii (0.3181) > T. aestivum subsp. tibetanum (0.3056) > T. compactum (0.2639) > common wheat (0.1363) > T. aestivum subsp. yunnanense and Xinjiang rice wheat (T. petropavlovskyi) (0.0952). In our study the average genetic distance of Tibetan wheat accessions was 0.7602, this results indicated that Tibetan wheat showed a certain level of genetic similarity and distance, the average genetic distance of Tibetan wheat observed in the present study was higher than that reported in other studies. Whether or not this reflects gene transfer among the Tibetan wheat accessions after long-term evolution and natural selection requires further investigation and showed some genetic diversity.
Allowing for the limited sampling of genotypes in the current study, the results indicate that considerable genetic diversity exists among the germplasm analyzed. The accessions with A and D genomic constitutions (Ae. tauschii and T. boeoticum were divided into two distinct classes, and were clearly resolved from accessions carrying AABB and AABBDD genomic constitutions. Accessions with the AABB genomic constitution contained abundant genetic diversity and showed a close relationship. T. durum and T. turgidum were clustered in the same group, which is consistent with the close similarity in their morphology. Compared with diploid wheat, the cluster analysis results suggested that emmer (T. dicoccoides) has a close ancestral relationship with hexaploid wheat. These results are consistent with previous research that suggested emmer is the donor of the AB genomes to hexaploid wheat (Yang et al. 2007). Compared with common wheat, Yunnan wheat and Tibetan wheat exhibited higher genetic diversity, which could be utilized to broaden the genetic base of common wheat. In addition, a close genetic relationship between T. polonicum and T. macha was observed, which were clustered in the same group. However, some accessions with the same genomic constitutions were clustered in different groups. The reason for this may reflect their different geographical provenances, such as T. turanicum nos. 63 and 64, which were clustered in groups III and II, respectively. However, the dendrograms generated in the cluster analyses were essentially consistent with groupings obtained in the principal component analysis scatterplots. Principal component analysis, however, can display more explicitly the relationships among wheat and closely related species.
Taken together, the present and previously published SSR data suggest that the level of genetic diversity contained in each wheat genome differs depending on the specific wheat genotypes analyzed. The present analysis describes the divergence of alleles specific for a particular geographical region. In a recent study (Landjeva et al. 2006), cultivars from different geographical regions of Bulgaria were clearly distinguished by SSR markers, which implied that different environments may favor different alleles. Differential distribution of alleles consistent with ecological divergence has also been reported by Stachel et al. (2000). However, in our paper, no relationship between the geographical environment and levels of genetic diversity were observed. The low number of alleles present in modern wheat germplasm provides a warning that there is a narrow genetic base for further varietal improvement.
In conclusion, a high level of genetic diversity exists among the wheat accessions analyzed. It is possible to distinguish closely related elite wheat germplasm, and to select lines and cultivars for maximum genetic diversity, using only a small number of microsatellite markers. A SSR marker system is a rapid and reliable method for cultivar identification that might also be used in quality control in certified seed production programs, to identify sources of seed contamination, and to maintain pure germplasm collections.
References
Achtar S, Moualla MY, Kalhout A, Röder MS, Miralid N (2010) Analysis of genetic diversity among syrian durum (Triticum ssp. durum) and bread wheat (Triticum aestivum L.) using SSR markers. Russ. J Genet 46(11):1320–1326
Chao SM, Zhang WJ, Dubcovsky J, Sorrells M (2007) Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci 47(3):1018–1030
Chen GY, Li LH (2007) Detection of genetic diversity in synthetic hexaploid wheats using microsatellite markers. Agric Sci China 6(12):1403–1410
Chen XM, He ZH, Shi JR, Xia LQ, Ward R, Zhou Y, Jiang GL (2003) Genetic diversity of high quality winter wheat varieties (lines) based on SSR markers. Acta Agron Sin 29(1):13–19
Dreisigacker S, Zhang P, Warburton ML, Skovmand B, Hoisington D, Melchinger AE (2005) Genetic Diversity among and within CIMMYT wheat landrace accessions investigated with SSRs and implications for plant genetic resources management. Crop Sci 45(2):653–661
Hammer K, Filatenko AA, Pistrick K (2011) Taxonomic remarks on Triticum L. and × Triticosecale Wittm. Genet Resour Crop Evol 58:3–10
Huang HL, Weng YJ, Zhang XZ, Lu P (2002a) Progress of genetic diversity study and protocols for in situ conservation on semi-wild Tibetan wheat. J Plant Genet Resour 3(2):28–33
Huang XQ, Börner A, Röder MS, Ganal MW (2002b) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor Appl Genet 105:699–707
Joshi CP, Nguyen HT (1993) RAPD (random amplified polymorphic DNA) analysis based intervarietal genetic relationships among hexaploid wheats. Plant Sci 93:95–103
Kim HS, Ward RW (2000) Patterns of RFLP-based genetic diversity in germplasm pools of common wheat with different geographical or breeding program origins. Euphytica 115:197–208
Knaggs P, Ambrose MJ, Reader SM, Miller TE (2000) Morphological characterization and evaluation of the subdivision of Aegilops tauschii Coss. Wheat Inf Serv 91:15–19
Landjeva S, Korzun V, Ganeva G (2006) Evaluation of genetic diversity among Bulgarian winter wheat (Triticum aestivum L.) varieties during the period 1925–2003 using microsatellites. Genet Resour Crop Evol 53:1605–1614
Limin AE, Fowler DB (1981) Cold hardiness of some relatives of hexaploid wheat. Can J Bot 59:572–573
Ma SQ, Zhang HQ (2008) Genetic diversity analysis on wheat and its related species using microsatellite fluorescence markers. J Henan Agric Sci 3:28–32
Maher M, Sonia H, Ahmed R, Michael B (2005) Analysis of genetic diversity in Tunisian durum wheat cultivars and related wild species by SSR and AFLP markers. Genet Resour Crop Evol 52(1):21–31
Naghavi MR, Ranjbar M, Zali A, Aghaei MJ, Mardi M, Pirseyedi SM (2009a) Genetic diversity of Aegilops crassa and its relationship with Aegilops tauschii and the D genome of wheat. Cereal Res Commun 37(2):159–167
Naghavi MR, Malaki M, Alizadeh H, Pirseiedi M, Mardi M (2009b) An Assessment of genetic diversity in wild diploid wheat Triticum boeoticum from west of Iran using RAPD, AFLP and SSR markers. J Agric Sci Tech 11(11):585–598
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Orth RA, Bushuk W (1973) Studies of glutenin. III. identification of subunits coded by the D-genome and their relation to breadmaking quality. Cereal Chem 50:680–687
Prasad M, Varshney RK, Roy JK, Balyan HS, Gupta PK (2000) The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. Theor Appl Genet 100:584–592
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Röder MS, Wendehake K, Korzun V, Bredemeijer G, Laborie D, Bertrand L, Isaac P, Rendell S, Jackson J, Cooke RJ, Vosmann B, Ganal MW (2002) Construction and analysis of a microsatellite-based database of European wheat cultivars. Theor Appl Genet 106:67–73
Rohlf JF (2000) NTSYSpc: numerical taxonomy and multi-variate analysis system. Version 2.1, users guide. Exeter Software, Setauket
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Schachtman DP, Lagudah ES, Munns R (1992) The expression of salt tolerance form Triticum tauschii in hexaploid wheat. Theor Appl Genet 84:714–719
Shoaib A, Arabi MIE (2006) Genetic diversity among Syrian cultivated and landraces wheat revealed by AFLP markers. Genet Resour Crop Evol 53:901–906
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Stachel M, Lelley T, Grausgruber H, Vollmann T (2000) Application of microsatellites in wheat (Triticum aestivum L.) for studying genetic differentiation caused by selection for adaptation and use. Theor Appl Genet 100:242–248
Ward RW, Yang ZL, Kim HS, Yen C (1998) RFLP diversity in landraces of Triticum aestivum and collections of T. tauschii from China and Southwest Asia. Theor Appl Genet 96:312–318
Yang XQ, Song X, Du JK, Ni ZF, Sun QX (2007) Genetic diversity revealed by chloroplast genomic microsatellite markers in hexaploid wheat, wild emmer and Aegilops tauschii. Sci Agric Sin 40(7):1324–1330
Zhan KH, Wang LH, Cheng XY, Xu HX, Dong ZD (2006) Genetic difference of some germplasm resources of wheat based on SSR markers in Huang-Huai area. J Agri Bio 14(4):578–584
Acknowledgments
The present study was supported by a grant from the National Science Technology Planning Project of China (No. 2011FY110200-3) and by the Zhongying Tang Breeding Fund of Northwest A&F University (2009–2011). We thank Dr Jose Costa (Department of Plant Science and Landscape Architecture, University of Maryland, College Park, MD, USA) for help and advice.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wang, Y., Wang, C., Zhang, H. et al. Genetic analysis of wheat (Triticum aestivum L.) and related species with SSR markers. Genet Resour Crop Evol 60, 1105–1117 (2013). https://doi.org/10.1007/s10722-012-9907-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-012-9907-6