Abstract
Molecular differentiation in 24 accessions representing 19 taxa of Indian Citrus has been examined through sequence analysis of Internal Transcribed Spacer (ITS) region of nrDNA. Sequence length in the 24 accessions of Citrus taxa ranged from 512 to 665 bp (ITS1 & ITS2 partial and 5.8S complete sequence). The ITS sequences were very rich in G+C content ranging from 61.40 to 66.60% with an average of 64.2%. Genetic distance within Citrus group ranged from 0 to 13.4% with an average of 4.6%, showing moderate rate of nucleotide divergence. The phylogeny was inferred using the Maximum parsimony (MP) and Neighbor-Joining (NJ) methods. Both MP and NJ trees separated all the 24 accessions of Citrus into six distinct clusters. The disposition of all the accessions of Citrus in separate clusters in ITS-derived dendrograms was partly in accordance with the morpho-taxonomic affinities of the target taxa. This study supports the concept of Citrus medica (citron), C. reticulata (mandarin), and C. maxima (pummelo) as the basic species of the genus. However, ITS marker could not find any clear cut differentiation between subgenera Citrus and Papeda as proposed in Swingle’s Citrus classification system. The present study also supports the distinctiveness of C. indica (Indian wild orange), C. latipes (Khasi papeda) and C. hystrix (Melanesian papeda) as true species, besides elucidating the probable hybrid origin and relationships among the cultivated species/biotypes, such as Citrus ×aurantiifolia (sour lime) C. ×limon (lemon), C. ×taitensis (Indian rough lemon), C. limettioides (sweet lime), C. ×aurantium (including sour and sweet oranges and grapefruit), and other indigenous varieties of Indian origin: C. megaloxycarpa (sour pummelo), C. karna (karna orange), C. pseudolimon (Hill lemon), ‘Memang athur’, ‘Pummelo-lemon’ and ‘Kathairi nimbu’.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Citrus L. belongs to the subfamily Aurantioideae of the family Rutaceae (Swingle and Reece 1967). It includes some of the major fruit crops of the world, such as the citrons, lemons, limes, mandarins, sour oranges, sweet oranges, pummelos, grapefruits, kumquats, etc. (Mabberley 2008). Citrus fruits are well known for their dietary, nutritional, medicinal and cosmetic properties and are also good sources of citric acid, flavonoids, phenolics, pectins, limonoids, etc. (Dugo and Di Giacomo 2002). Recent studies support the traditional uses of citrus fruits in several diseases like scurvy, cancer, HIV/AIDS, contraception, cough, and reducing blood pressure (Mabberley 2004).
Citrus is believed to have its primary centre of origin in south and south-east Asia, particularly in the region extending from northeast India, eastward through the Malayan Archipelago to China and Japan, and southward to Australia (Nicolosi 2007; Pfeil and Crisp 2008). Citrus fruits are widely cultivated throughout the tropical and subtropical regions of the world (Webber 1943). Citrus is the third most important fruit crop grown in India after mango and banana with an estimated production of 8,608,000 MT in a total area of 9,23,000 HA (Anonymous 2010).
Despite its manifold economic importance and increasing demands in the global citrus industry, the taxonomy of Citrus is still controversial, mainly due to the sexual compatibility between Citrus and its related genera, apomixis (adventive nucellar polyembryony), high frequency of bud mutations, long history of cultivation, and wide dispersion (Moore 2001). Consequently, there has been no consensus among the taxonomists as to the actual number of species that constitute the genus Citrus. Among the two principal Citrus classification systems in current practice, Swingle (1943; revised by Swingle and Reece 1967) included 16 species (10 spp. in subgenus Citrus and 6 spp. in subgenus Papeda) while that of Tanaka (1954, 1977) recognized up to 162 species in two subgenera: Archicitrus and Metacitrus. Advanced studies, based on the biochemical and morphological characterization, suggest that there are only three basic species, i.e. citron (C. medica L.), mandarin (C. reticulata Blanco), and pummelo [C. maxima (Burm.) Merr.] within the subgenus Citrus whereas the other edible citrus (e.g. lemon, lime, sour orange, sweet orange, grapefruit, etc.) have been considered as apomictically perpetuated biotypes of probable hybrid origin (Barrett and Rhodes 1976; Scora 1988). The concept of basic species was well- supported by Moore (2001) and Mabberley (1997, 2004).
Taxonomic characterization leading to unambiguous identification of Citrus species and their genetic resources are essential requisites for citrus breeding, citriculture and citrus industry. Since morphological characters are only of limited use, alternate approaches, including application of appropriate molecular markers, have now been increasingly adopted to address the problems in Citrus taxonomy. Several workers have revisited the taxonomy and phylogeny of Citrus and related genera using molecular markers such as isozymes (Herrero et al. 1996), RAPD & PCR–RFLP (Federici et al. 1998; Abkenar et al. 2004), RAPD, SCAR & PCR–RFLP (Nicolosi et al. 2000; Jena et al. 2009), AFLP (Liang et al. 2007; Pang et al. 2007), SSR (Barkley et al. 2006), ISSR (Fang et al. 1998; Shahsavar et al. 2007) and sequence data analysis of ITS region of nrDNA (Xu et al. 2006; Kyndt et al. 2010; Pessina et al. 2011) and non-coding chloroplast DNA (cpDNA) regions (Chase et al. 1999; Araujo et al. 2003; Morton et al. 2003; Lu et al. 2011). Bayer et al. (2009), in a recent molecular analysis based on nine cpDNA sequences, broadened the circumscription of Citrus to include seven other closely related genera of the orange subfamily, such as Clymenia Swingle, Fortunella Swingle, Poncirus Raf., Microcitrus Swingle, Eremocitrus Swingle, Oxanthera Montrouz., and Feroniella Swingle.
India is rich in Citrus genetic resources, both in cultivation and wild. In a systematic account on Indian Citrus, Nair and Nayar (1997) followed primarily Swingle and Reece (1967) and partly Tanaka (1977) and included 18 taxa, comprising of eight species under subgenus Citrus, three under subgenus Papeda, and seven other indigenous Citrus varieties with a suspected hybrid origin and uncertain taxonomic affinities. In an earlier study (Jena et al. 2009), the present authors examined the molecular phylogeny of Indian Citrus using PCR–RFLP of the trnD-trnT and rbcL-ORF 106 regions as well as sequence data analysis of the trnL-trnF intergenic spacer region of cpDNA. In the present study, we have revisited the phylogenetic relationships among the Indian Citrus species/varieties using sequence variation in ITS (internal transcribed spacer) region of nuclear ribosomal DNA.
Materials and methods
Plant samples
Fifty accessions of 19 Citrus taxa or biotypes (species/cultivars/hybrids) and one out- group taxon [Atalantia monophylla (L.) DC.] were collected from wild as well as domesticated stocks from different parts of India. Young fresh leaf tissues from all the sample materials were collected and stored in silica gel (20–60 mesh) and were used subsequently for genomic DNA isolation. Details of accessions used for morpho-metric and ITS sequence analyses are given in Table 1. Voucher specimens of all the accessions were deposited in the Herbarium of the National Botanical Research Institute (LWG), Lucknow, India.
Morphological characterization
Fifty accessions of Citrus (Table 1) were used for morpho-metric analysis. Seventy-six discrete morphological characters (33 quantitative and 43 qualitative characters) were selected from taxonomic literature (Barrett and Rhodes 1976; IPGRI 1999; Nair and Nayar 1997) and by examination of living plants and herbarium collections of Indian Citrus (ESM_1.pdf). The characters were converted into bi-states and multi-states (interval) code (ESM_2.pdf). Standardization of morphological data was done based on YBAR option with the software NTSYS ver. 2.10e (Rohlf 2000). A pair wise similarity matrix was generated using Simple Matching coefficient and a dendrogram was constructed based on UPGMA (Unweighted Pair Group Method by Arithmetic averages) with the same software. Principal Coordinate Analysis (PCOA) was performed to analyse non-hierarchical relationship among the accessions (Gower 1966). This analysis was executed by calculating the eigenvectors and eigenvalues from Eigen programme in the NTSYS software, which resulted in a two -dimensional plot.
DNA extraction
Total genomic DNA was isolated from a final set of 25 representative accessions through Cetyl Trimethyl Ammonium Bromide (CTAB) method (Rogstad 1993). Quantitation of isolated DNA was done spectrophotometrically and its quality checked by electrophoresis on 0.8% agarose gel.
ITS-PCR amplification, Sequencing and Sequence analysis
Entire ITS region (ITS1, 5.8S, ITS2) of Citrus and outgroup accessions was amplified using a pair of universal primers, i.e. ITSP4—TCCTCCGCTTATTGATATGC (White et al. 1990) and ITSP5—AAGTCGTAACAAGGTTTCCGTAG (Kollipara et al. 1997). The concentration of PCR components was optimized for amplification of ITS region: 10 mM Tris (pH 8.9), 50 mM KCl, 0.2 mM dNTP each, 1.5 mM MgCl2, 1 U Taq DNA polymerase, 10 pmol primer each (ITSP4 and ITSP5) and 50 ng genomic DNA in 50 μl final reaction volume. PCR was programmed as pre-denaturation at 94°C for 3 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, extension at 72°C for 1.5 min and final extension at 72°C for 5 min.
Total ITS-PCR products were electrophoresed in 0.8% low melting agarose gel (Bangalore Genei) at 80 V for 3 h, and bands were visualized in UVITec Gel Documentation System. The bands were excised and purified using Clean Genei kit (Bangalore Genei). The yield of purified DNA was quantified using UV spectrophotometer. Eluted PCR products were sequenced using an Applied Biosystems Automated Sequencer (Model 3730, version 3.1) using both forward and reverse primer. Sequences of 25 accessions of Citrus including one outgroup were annotated and submitted to the NCBI GenBank (accessions nos. GQ225843—GQ225867).
The identity of sequences was confirmed through a BLASTn search in NCBI data base (Altschul et al. 1997) for determining their homology with sequences of related taxa available in EMBL/GenBank Data bases. The sequences were aligned using Clustal-W program (Higgins et al. 1994) with the default settings. Phylogenetic analysis was carried out in MEGA 4 software (Tamura et al. 2007). Pair-wise sequence divergence rates between accessions were calculated using Maximum Composite Likelihood method (Tamura et al. 2004). Phylogeny reconstruction was carried out using Maximum Parsimony (MP) and Neighbor Joining (NJ) methods. MP tree was constructed using the Close-Neighbor-Interchange algorithm with search level 3 in which the initial trees were obtained with the random addition of sequences (10 replicates), while NJ tree was obtained using the Maximum Composite Likelihood criterion. In MP analysis all the characters were assigned equal weights at all nucleotide positions (Fietch 1971). In the MP and NJ analyses, all positions containing gaps and missing data were eliminated from the dataset (Complete Deletion option). Support values of the internal branches of MP and NJ trees were evaluated through boot strap method (500 replicates) (Felsenstein 1985).
Results
Morpho-metric analysis
Similarity values of all the fifty accessions of Citrus ranged from 0.18 to 1.00 with an average of 0.39 (ESM_3.pdf). C. megaloxycarpa Lush. (CMEG-S1) showed minimum similarity value of 0.18 with all accessions of C. reticulata (CRET-B1, CRET-S2 and CRET-L3). Maximum similarity at interspecific level was observed between C. pseudolimon Tanaka (CPSE-L4) and C. ×limon (L.) Burm. f. (CLIM-L1 and CLIM-L2).
The UPGMA dendrogram (Fig. 1) resolved four main clusters and eight groups as shown below:
-
Cluster I: Group I: C. megaloxycarpa Lush. (Sour Pommelo) and C. sp. (Pummelo-lemon); Group II: C. maxima (Burm.) Merr. (Sweet pummelo), C. sp. (Kathairi nimbu) and C. ×aurantium L. (Grapefruit),
-
Cluster II: Group I: C. ×aurantium L., (Sour orange), C. ×aurantium L. (Sweet orange); Group II: C. latipes (Swingle) Tanaka (Khasi papeda), C. hystrix DC. (Melanesian papeda),
-
Cluster III: Group I: C. ×aurantiifolia (Christm.) Swingle (Sour lime), C. limettioides Tanaka (Sweet lime); Group II: C. medica L. (Citron), C. ×limon (L.) Burm. f. (Lemon), C. pseudolimon Tanaka (Hill lemon), C. karna Raf. (Karna orange), C. ×taitensis Lush. (Indian rough lemon),
-
Cluster IV: Group I: C. reticulata Blanco (Mandarin); Group II: C. indica Tanaka (Indian wild orange), and C. sp. (Memang athur).
Mantel test was carried out for comparing the UPGMA cluster analysis and similarity matrix. A correlation value (r = 0.93) showed very good fit of UPGMA clustering pattern to the data. PCOA was used for identifying multi-dimensional relationships among characters for the definition of groups. In this analysis, 1st and 2nd principal co-ordinates accounted for 28.01 and 19.13% of the total variation, respectively (Table 2). 2-D plot (Fig. 2) generated through PCOA also showed the same grouping pattern as the UPGMA dendrogram.
2-D plot of the first and second co-ordinate axes, derived from principal coordinate analysis of 50 accessions of Citrus using morpho-metric data. The 1st and 2nd co-ordinates are 28.01 and 19.13% respectively (Note: Numbers are equivalent to those listed in Table 1)
ITS sequence data
The BLASTn search helped determine that the new sequences were from ITS region and maximum homology was obtained from ITS sequences of Citrus and related taxa of Rutaceae. On the basis of the angiosperm consensus motif determined by Jobes and Thien (1997), the putative start and end points of 5.8 S regions in the aligned sequences were identified. The aligned ITS sequences are shown in Fig. 3. Sequence length in the 24 Citrus accessions ranged from 512 to 665 bp (ITS1 & ITS2 partial and 5.8S complete sequence) and 564 bp in A. monophylla. In Citrus, the length of ITS1 ranged from 118 to 269, ITS2 from 150 to 276 and 5.8S from 162 to 165. The data set including alignment gaps and missing data comprised 741 bp aligned nucleotide positions, which included 388 conserved sites, 320 variable sites and 144 parsimony informative sites. When missing data were excluded, the sequence length was 463 bp, including 293 conserved, 159 variable and 82 parsimony informative sites. The ITS sequences were very rich in G+C content ranging from 61.40% (C. ×aurantiifolia) to 66.60% (C. maxima) with an average of 64.2%. The nucleotide frequencies were found as 0.208 (A), 0.316 (C), 0.304 (G), and 0.171 (T). The transition/transversion rate ratios were k1 = 1.167 (purines) and k2 = 2.796 (pyrimidines). Transition/transversion bias (R) was 1.158. Summary of ITS sequence data is given in Table 3 and 4.
ITS sequence analysis showed moderate rate of nucleotide divergence within and among the Citrus taxa and A. monophylla (Table 5). Genetic divergence within Citrus group ranged from 0 to 13.4% with an average of 4.6%. C. megaloxycarpa showed 0% nucleotide divergence with C. ×limon, while maximum distance (13.4%) was found between C. indica (CIND-D08) and C. ×aurantiifolia. Sequence divergence within C. indica accessions ranged from 0.8% (CIND-N25 and CIND-K26) to 4.1% (CIND-D08 and CIND-D18) with an average of 2.5%. Nucleotide divergence between the basic species of Citrus was 3.6% (C. medica and C. maxima), 2.1% (C. maxima and C. reticulata), and 2.5% (C. medica and C. reticulata).
In the aligned sequence, three substitutions were recorded only in C. indica and ‘Memang athur’ at coordinate 265 (C→T), 316 (C→T) and 459 (A→T) (Fig. 3). One substitution (coordinate 26, G→A) was observed only in citron group (C. medica, C. ×taitensis, C. ×limon, C. ×aurantiifolia, C. limettioides) and ‘Memang athur’. Similarly, one substitution occurred at coordinate 390 (A→G) only in C. ×taitensis and ‘Memang athur’. Two substitutions occurred at coordinate 10 (C→T) and 353 (C→T) in C. maxima, C. ×aurantium (Sour orange) and C. ×aurantium (Grapefruit).
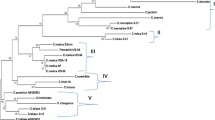
Maximum Parsimony (MP) analysis resulted in 63 most parsimonious trees (length = 154), out of which a single fully resolved consensus tree is shown in Fig. 4, with consistency index (CI—0.6804) and retention index (RI- 0.7350). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown at the nodes. There were a total of 380 positions in the final dataset, out of which 50 were parsimony informative. Maximum parsimony tree (Fig. 4 and 5) placed all accessions in the following six clusters:
-
Cluster I: C. pseudolimon and ‘Pummelo-lemon’; ‘Kathairi nimbu’ and C. ×aurantiifolia;
-
Cluster II: C. medica –W and C. medica-D;
-
Cluster III: C. reticulata, C. ×aurantium (Grapefruit), and C. ×aurantium (Sour orange);
-
Cluster IV: C. ×taitensis, C. limettioides, C. ×limon, C. megaloxycarpa, and C. karna;
-
Cluster V: C. latipes, C. hystrix, C. maxima and C. ×aurantium (Sweet orange);
-
Cluster VI: C. indica, and ‘Memang athur’.
Atalantia monophylla was separately attached at the base of tree as the diverging Citrus relative’s lineage. The phylogeny was also inferred using the Neighbor-Joining method, which resulted in an optimal tree with the SBL (sum of branch length) of 0.3911. There were a total of 380 positions in the final dataset. The optimal NJ tree (Fig. 6) separated all the 24 accessions in the six clusters as similar to MP tree.
Discussion
All major morphology-based classification of Citrus (Swingle 1943; Swingle and Reece 1967; Tanaka 1954, 1977; Bhattacharya and Dutta 1956; Hodgson 1965; Singh 1967; Singh and Nath 1969) relied on combination of a few special characters of the foliage (size and shape of the petiole wings and the ratio between length/breadth of petiole and lamina), flower (number of flowers per inflorescence, colour of buds and petals, cohesion of stamens), fruits (size, shape, texture and colour of epicarp, presence or absence of mamillate apex, thickness and texture of mesocarp, taste of juice) and seed (size, shape, colour of cotyledon and chalazal cap). Frequent hybridization, introgression, bud mutations and polyploidy have created innumerable hybrids and mutant varieties of Citrus throughout the Citrus belt of the world. This in turn created confusion among taxonomists in comprehending the species and generic limits within Citrus and closely related genera within the orange subfamily. Those who followed the biological species concept did not accord true species status to natural or human-made hybrids and bud sports of Citrus fruits (Swingle and Reece 1967; Mabberley 1998, 2004, 2008), whereas others like Tanaka (1977), who followed an horticultural concept, gave true species status to any notable variants of Citrus fruits, irrespective of their actual mode of origin and taxonomic affinities. While both the above approaches have their own merits and demerits, a proper circumscription and classification of Citrus at global, regional and national scale is still awaited. In the present study we have analysed the resolving power of morphological and molecular (nr DNA ITS) markers in discriminating the true basal species, probable hybrids and varieties of Indian Citrus. The main results of morpho-metric and ITS sequence analyses are discussed to elucidate the taxonomic identity, origin and phylogeny of 19 biotypes of Indian Citrus studied.
ITS sequence variations
ITS analysis carried out in Indian taxa of Citrus was useful in differentiating all the true species and species/varieties of probable hybrid origin in distinct clusters or groups. Range of ITS sequence length (512–665 bp) of Citrus was almost similar to that of ITS sequence length (565–634 bp) of Phebalium group (Rutaceae: Boronieae) (Mole et al. 2004). ITS1 sequence length in Citrus species ranged from 118 (including partial sequences) to 269 bp, while ITS2 region ranged from 150 (including partial sequences) to 276 bp. Similar length variation was also reported in several angiosperms (Baldwin 1993; Baldwin et al. 1995). GC content of ITS region was also found very high (61.40–66.60%) in Citrus, which is slightly higher (50–67%) than that reported in Cucurbita, Cucumis and other genera of Cucurbitaceae (Jobst et al. 1998). Similar case was also observed in the plants of Poaceae, in which higher GC content was reported in arid region than the plants from temperate region (Salinas et al. 1988).
In Citrus, 293 conserved, 159 variable and 82 parsimony informative sites were found in the aligned ITS sequences (463 bp, excluding missing data). Maximum conserved regions were observed in 5.8S and ITS2 regions. Maximum variable sites were found in ITS1 region. Similar results were also reported in Citrus by Xu et al. (2006), who observed that the ITS1 region in Citrus taxa showed maximum nucleotide variations due to high rate of point mutation. Inter-specific sequence divergence in Citrus ranged from 0 to 13.4%, which was comparatively lower than that of reported nucleotide sequence divergence in Coffea (Rubiaceae; 1.5–39%; Lashermes et al. 1997). However, low sequence divergence (0.0–4.86%) was also observed in Cistus (Cistaceae) (Guzmán and Vargas 2005).
Phylogenetic relationships among Indian Citrus
The UPGMA dendrogram based on morpho-metric analysis (Fig. 1) and the MP and NJ trees (Fig. 4, 5 and 6) generated through nr DNA ITS sequence analysis segregated C. maxima, C. medica and C. reticulata in separate clusters. The clear separation of these three species in three distinct clusters supports the concept of basic species within Citrus.
Citrus medica (citron) is believed to have acted as male parent in the origin of several hybrids/cultivars of Citrus (Federici et al. 1998; Nicolosi et al. 2000; Moore 2001; Mabberley 2004). In our morpho-metric study, all the accessions of C. ×limon, C. pseudolimon, C. karna, C. ×taitensis, C.×aurantiifolia and C. limettioides grouped along with C. medica in cluster III. Our ITS data recognized C. medica as a true basic species as both wild and domesticated accessions of the species grouped in the cluster II with a very high bootstrap value of 95% (NJ tree) and 86% (MP tree). One substitution (coordinate 26, G→A) was observed only in citron group (C. medica, C. ×taitensis, C. ×limon, C. ×aurantiifolia, C. limettioides) and ‘Memang athur’. This substitution indicates involvement of C. medica as one of parents in the hybrid origin of lemons and limes. C. ×taitensis, C. limettioides, C. megaloxycarpa, and C. karna grouped in the same cluster in both NJ and MP trees, with a robust bootstrap value of 85%, indicative of their common lineage. Based on morphology all these species (except C. limettioides) are characterized by having mammillate fruit apex, which is characteristic of C. medica. So the involvement of citron as one of the parents in the origin of C. ×taitensis, C. megaloxycarpa and C. karna could not be ruled out. This result also indicates that one of the species of citron group may be acted as putative parent in the origin of ‘Memang athur’. This inference further gained support by substitution occurred at coordinate 390 (A→G) only in C. ×taitensis and ‘Memang athur’, as both are morphologically most similar in fruit characters.
Citrus maxima and C. reticulata are believed to have contributed to the development of several commercial Citrus fruits, such as sour orange (C. ×aurantium, a cross between mandarin and pummelo), sweet orange (C. ×aurantium; syn. C. sinensis (L.) Osbeck (a backcross between pummelo and mandarin), grapefruit (C. ×aurantium; syn. C. paradisi (a backcross between pummelo and sweet orange) (Moore 2001; Mabberley 2004). In UPGMA tree, the sour and sweet oranges were grouped together in a separate cluster along with the Khasi papeda and Melanesian papeda, while in the MP and NJ trees the grapefruit and sour orange formed a separate cluster along with C. reticulata, and the sweet orange grouped with C. maxima along with the Khasi papeda and Melanesian papeda. Our previous study, based on cpDNA data, also elucidated the involvement of C. reticulata as a maternal parent in the origin of sweet orange (Jena et al. 2009). The consistent grouping of sweet orange with C. maxima in the ITS-derived trees indicates the role of C. maxima as a male parent in the origin of sweet oranges. Moreover, two substitutions at coordinate 10 (C→T) and 353 (C→T) in C. maxima, C. ×aurantium (sour orange) and C. ×aurantium (grapefruit) support a common genetic lineage of the three species. These results support that involvement of both C. maxima and C. reticulata in the hybrid origin of the sour orange, sweet orange and grapefruit.
Swingle and Reece (1967) divided Citrus into two subgenera: Citrus and Papeda. The wild species including C. latipes and C. hystrix were classified under subgenus Papeda of the genus Citrus by Swingle (1943). Tanaka (1954) also classified the wild species of Citrus under Archicitrus, which also included other cultivated species like citrons, lemons, limes, oranges and pummelos. Our ITS data analysis, however, could not find any clear cut differentiation between subgenera Citrus and Papeda according to Swingle’s system. This supports the earlier findings of Nicolosi et al. (2000) and Pang et al. (2007). In NJ and MP trees, based on ITS sequence data, C. latipes and C. hystrix were grouped with C. maxima and C. ×aurantium (sweet orange) in the same cluster with a robust bootstrap value (85%). This grouping showed close genetic affinity of the Papedas with the Pummelos as supported by earlier studies based on RAPD and RFLP (Federici et al. 1998), cpDNA (Nicolosi et al. 2000), and AFLP (Pang et al. 2007).
Citrus indica is a true wild species endemic to the Garo Hills in Meghalaya. Tanaka (1928) was the first to describe it as a new species. He (Tanaka 1977) placed C. indica in section Acrumen of the Subgenus Metacitrus. Cluster IV in our UPGMA tree included C. indica, ‘Memang athur’ and C. reticulata, which diverged from other clusters with similarity value of 0.34. ‘Memang athur’ consistently grouped with C. indica with maximum similarity (0.56), indicating closer relationship among the two. All accessions of C. indica and ‘Memang athur’ diverged from C. reticulata group with similarity value of 0.43, showing maximum similarity with C. reticulata based on morphology. This grouping supports Tanaka’s placement of C. indica with the mandarins. Swingle and Reece (1967) suspected C. indica to be of hybrid origin involving a wild species of Citrus (C. latipes?) and one of the cultivated species of Citrus as putative parents. Mabberley (2004) also subscribed Swingle’s view in treating C. indica as a species of suspected hybrid origin. Based on RAPD and PCR–RFLP data, Federici et al. (1998) argued against the hybrid origin of C. indica. In our ITS analysis, C. indica was independently grouped with its close variant ‘Memang athur’ in the cluster VI in both MP and NJ trees. This result, therefore, does not support the hybrid origin of C. indica as it consistently separated out as a distinct cluster with good boot strap support (67 and 77%). Similar result was also found in our previous study based on sequence analysis of trnL-trnF region cpDNA (Jena et al. 2009).
‘Memang athur’ is a hitherto unidentified Citrus fruit, which we could locate in one of the Garo tribal settlements in Daribokgre in NBR and its vicinity. Morphologically, ‘Memang athur’ looks more similar to C. indica in the leaf shape, small and scarlet red fruits, and medium to large sized plumpy seeds. Some characters, like petiole size, serrations on leaf margin, flowers with thick fleshy, 4 or 5 purplish tinged petals, mammiform fruit apex, longitudinal furrow and ridge on the surface, and reddish chalazal cap bring ‘Memang athur’ much closer to C. medica. Based on the morphological characters and partial seed sterility, ‘Memang athur’ appears to be a probable hybrid. Malik et al. (2006) suspected it to be a hybrid between C. indica and one of the cultivated Citrus species. In the aligned sequence, three substitutions at coordinate 265 (C→T), 316 (C→T) and 459 (A→T) in C. indica and ‘Memang athur’ and grouping together in the same cluster of MP and NJ trees support our previous study in Citrus based on ISSR markers (Kumar et al. 2010) that C. indica is perhaps one of the putative parents involved in the hybrid origin of Memang athur. This gained further support from phylogenetic trees that ‘Memang athur’ consistently grouped along with C. indica in all the trees. ‘Memang athur’ clearly separated from C. indica with a robust bootstrap value of 85% (NJ tree) and 67% (MP tree), within the cluster VI of ITS derived trees, which showed close similarity with C. indica. The results support that ‘Memang athur’ is closely related to C. indica genetically.
Citrus megaloxycarpa (sour pummelo) is suspected to be a probable hybrid between C. maxima and C. ×limon (Nair and Nayar 1997). It is morphologically close to C. maxima, except difference in some characters like presence of marginate or narrowly winged petiole, purplish tinged petal, sour juice and purple chalazal cap. Lushington (1910) established it as a valid species. Bhattacharya and Dutta (1956) and Tanaka (1977) also accepted it as a valid species, while Swingle and Reece (1967) and Nair and Nayar (1997) considered it as a probable hybrid. The present morpho-metric data support that C. megaloxycarpa is very close to C. maxima, while our ITS data show the placement of C. megaloxycarpa in the C. ×limon cluster in both MP and NJ trees. The genetic identity (100% similarity) between C. megaloxycarpa and C. ×limon as shown in the nucleotide divergence rate (0%; Table 5) is indicative of the probable hybrid origin and close phylogenetic affinity of sour pummelo with the lemon group.
C. pseudolimon (the ‘Hill-lemon or ‘Gulgal’ or ‘Kumaon lemon’) has been considered as a variety of lemon. Morpho-metric data placed the ‘Hill lemon’ in the citron cluster along with lemon and lime groups. ITS analysis, on the other hand supported its placement in the sour lime group along with ‘pummelo-lemon’ and ‘Kathairi nimbu’. ‘Kathairi nimbu’ is an unusually interesting citrus fruit characterized by prominently bumpy-warty epicarp, broadly mammillae or depressed apex and a prominently collared neck at base. It is a seedless form, clonally propagated through suckers or stem cuttings. Morpho-metric results showed close morphological affinity of ‘Kathairi nimbu’ to C. maxima, whereas in ITS analysis it consistently grouped with the sour lime (C. ×aurantiifolia) cluster. ‘Pummelo-lemon’ is another interesting variety/hybrid, showing intermediate characters of pummelo and lemon. Based on ITS data, it clustered along with Hill lemon and lime group while morphological data supported the close relationship of Pummelo- lemon with pummelo (C. maxima).
Conclusions
ITS analysis carried out in Indian taxa of Citrus was useful in differentiating all the true species and species/varieties of probable hybrid origin in distinct clusters or groups. The disposition of all the accessions of Citrus in distinct clusters based on ITS sequence data was partly in accordance with the morpho-taxonomic affinities of different taxa. The separation of Citrus maxima, C. medica and C. reticulata in distinct clusters or subclusters supports their distinctiveness as the basic species of edible Citrus. ITS sequence analysis could not find any clear cut differentiation between subgenera Citrus and Papeda according to Swingle’s system. The study also supported the distinctiveness of C. indica, C. latipes and C. hystrix as true species, besides elucidating the hybrid origin and relationships among the cultivated species/biotypes, such as C. ×aurantiifolia, C. ×limon, C. ×taitensis, C. limettioides, C. ×aurantium (including sour and sweet oranges and grapefruit), C. megaloxycarpa, C. karna, C. pseudolimon, ‘Memang athur’, ‘Pummelo-lemon’ and ‘Kathairi nimbu’. The outcomes of this study will be useful for correct taxonomic identification, documentation, characterization and evaluation of Indian Citrus and its genetic resources.
References
Abkenar AA, Isshiki S, Tashino Y (2004) Phylogenetic relationships in the true Citrus fruit trees revealed by PCR-RFLP analysis of cp DNA. Sci Hortic 102:233–242
Altschul SF, Thomas LM, Alejandro AS et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anonymous (2010) Indian horticulture database. National Horticulture Board, Ministry of Agriculture, Gurgaon
Araujo EF, Queiroz LP, Machado MA (2003) What is Citrus? Taxonomic implications from a study of cp-DNA evolution in the tribe Citreae (Rutaceae subfamily Aurantioideae). Org Divers Evol 3:55–62
Baldwin BG (1993) Molecular phylogenetics of calycadenia (Compositae) based on ITS sequences of nuclear ribosomal DNA: chromosomal and morphological evolution reexamined. Am J Bot 80:222–238
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard 82:247–277
Barkley NA, Roose ML, Krueger RR, Federici CT (2006) Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor Appl Genet 112:1519–1531
Barrett HC, Rhodes AM (1976) A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Syst Bot 1:105–136
Bayer RJ, Mabberley DJ, Morton CM, Cathy H, Sharma IK, Pfeil BE, Rich S, Hitchcock R, Sykes S (2009) A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. Am J Bot 96:668–685
Bhattacharya SC, Dutta S (1956) Classification of Citrus fruits of Assam. Sci Monogr 20. ICAR, New Delhi, p 110
Chase W, Morton CM, Kallunki JA (1999) Phylogenetic relationships of Rutaceae: a cladistic analysis of the subfamilies using evidence from rbcL and atpB sequence variation. Am J Bot 8:1191–1199
Dugo G, Di Giacomo A (2002) Citrus: the genus Citrus, medicinal and aromatic plants—industrial profiles. Taylor & Francis group, London
Fang DQ, Krueger RR, Roose ML (1998) Phylogenetic relationships among selected Citrus germplasm accessions revealed by inter-simple sequence repeat (ISSR) markers. J Amer Soc Hort Sci 123:612–617
Federici CT, Fang DQ, Scora RW et al (1998) Phylogenetic relationship within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theor Appl Genet 96:812–822
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fietch WM (1971) Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Guzmán B, Vargas P (2005) Systematics, character evolution, and biogeography of Cistus L. (Cistaceae) based on ITS, trnL-trnF, and matK sequences. Mol Phylogenet Evol 37:644–660
Herrero R, Asins MJ, Carbonell EA, Navarro L (1996) Genetic diversity in the orange subfamily Aurantoideae. I. Interspecies and intragenus genetic variability. Theor Appl Genet 92:599–609
Higgins D, Thompson J, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Hodgson RW (1965) Taxonomy and nomenclature in citrus fruits. In: Krishnamurthi S (ed) Advances in agricultural sciences and their applications. Madras Agric J, Madras, pp 317–331
IPGRI (1999) Descriptors for Citrus. International Plant Genetic Resources Institute, Rome
Jena SN, Kumar S, Nair KN (2009) Molecular phylogeny in Indian Citrus L. (Rutaceae) inferred through PCR-RFLP and trnL-trnF sequence data of chloroplast DNA. Sci Horti 199:403–416
Jobes DV, Thien LB (1997) A conserved motif in the 5.8S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences. Plant Mol Biol Rep 15:326–334
Jobst J, King K, Hemleben V (1998) Molecular evolution of the internal transcribed spacers (ITS1 and ITS2) and phylogenetic relationships among species of the family Cucurbitaceae. Mol Phylogenet Evol 9:204–219
Kollipara KP, Singh RJ, Hymowitz T (1997) Phylogenetic and genomic relationships in the genus Glycine Willd. Based on sequences from the ITS region of nuclear rDNA. Genome 40:57–68
Kumar S, Jena SN, Nair KN (2010) ISSR polymorphism in Indian Wild Orange, C. indica Tanaka (Rutaceae) and related wild species in North-east India. Sci Horti 123:350–359
Kyndt T, Dung TN, Goetghebeur P, Toan HT, Gheysen G (2010) Analysis of ITS of the rDNA to infer phylogenetic relationship among Veitnamese Citrus accessions. Genet Res Crop Evol 57:183–192
Lashermes P, Combes MC, Trouslot P, Charrier A (1997) Phylogenetic relationships of coffee-tree species (Coffea L.) as inferred from ITS sequences of nuclear ribosomal DNA. Theor Appl Genet 94:947–955
Liang G, Xiong G, Guo Q, He Q, Li X (2007) AFLP analysis and the taxonomy of Citrus. Acta Hortic (ISHS) 760:137–142
Lu Z, Zhou Z, Xie R (2011) Molecular phylogeny of the true Citrus fruit trees group (Aurantioideae, Rutaceae) as inferred from chloroplast DNA sequences. Agri Sci China 10:49–57
Lushington AW (1910) The genus Citrus. Indian Forester 36:323–353
Mabberley DJ (1997) A classification of edible Citrus (Rutaceae). Telopea 7:167–172
Mabberley DJ (1998) Australian Citreae with notes on other Aurantioideae (Rutaceae). Telopea 7:333–344
Mabberley DJ (2004) Citrus (Rutaceae): a review of recent advances in etymology, systematics and medical applications. Blumea 49:481–498
Mabberley DJ (2008) Mabberley’s plant-book: a portable dictionary of plants, 3rd edn. Cambridge University Press, Avon
Malik SK, Chaudhury R, Dhariwal OP, Kalia RK (2006) Collection and characterization of Citrus indica Tanaka and C. macroptera Montr.: wild endangered species of northeastern India. Genet Resour Crop Evol 53:1485–1493
Mole BJ, Udovicic F, Ladiges PY, Duretto MF (2004) Molecular phylogeny of Phebalium (Rutaceae: Boronieae) and related genera based on the nrDNA regions ITS 1 + 2. Pl Syst Evol 249:197–212
Moore GA (2001) Oranges and lemons: clues to the taxonomy of Citrus from molecular markers. Trends Genet 17:536–540
Morton CM, Grant M, Blackmore S (2003) Phylogenetic relationships of the Aurantioideae inferred from cpDNA sequenced data. Am J Bot 90:1463–1469
Nair KN, Nayar MP (1997) Rutaceae. In: Hajra PK, Nair VJ, Daniel P (eds) Flora of India, vol IV. Botanical Survey of India, Calcutta, pp 229–407
Nicolosi E, Deng ZN, Genetile A et al (2000) Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet 100:1155–1166
Nicolosi E (2007) Origin and taxonomy. In: Khan I (ed) Citrus: genetics. Breeding and Biotechnology, CAB International, UK, pp 19–43
Pang XM, Hu CG, Deng XX (2007) Phylogenetic relationships within Citrus and its related genera as inferred from AFLP markers. Genet Resour Crop Evol 54:429–436
Pessina D, Gentili R, Barcacciab G, Nicolèb S, Rossic G, Barbestia S, Sgorbatia S (2011) DNA content, morphometric and molecular marker analyses of Citrus limonimedica, C. limon and C. medica for the determination of their variability and genetic relationships within the genus Citrus. Sci Horti 129:663–673
Pfeil BE, Crisp MD (2008) The age and biogeography of Citrus and the orange subfamily (Rutaceae:Aurantioideae) in Australasia and New Caledonia. Am J Bot 95:1621–1631
Rogstad SH (1993) Saturated NaCl-CTAB solution as a means of field preservation leaves for DNA analysis. Taxon 41:701–708
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system, ver. 2.10e, Exeter Ltd., Setauket, NY
Salinas J, Matassi G, Montero LM, Bernardi G (1988) Compositional compartmentalization and compositional patterns in the nuclear genomes of plants. Nucleic Acids Res 16:4269–4285
Scora RW (1988) Biochemistry, taxonomy and evolution of modern cultivated Citrus. In: Goren RK, Mendel K (eds) Proceedings of 6th international Citrus congress, Margraf Scientific Books, Weikesheim, pp 277–289
Shahsavar AR, Izadpanah K, Tafazoli E, Tabatabaei BES (2007) Characterization of Citrus germplasm including unknown variants by inter-simple sequence repeat (ISSR) markers. Sci Horti 112:310–314
Singh R (1967) A key of the Citrus fruits. Ind J Hortic 4:71–83
Singh R, Nath N (1969) Practical approach to the classification of Citrus. In: Chapman HD (ed) Proceedings of International Citrus Symposium, vol 1, pp 435–440
Swingle WT (1943) The botany of Citrus and its wild relatives. In: Webber HJ, Batchelor DL (eds) The Citrus industry, vol 1. University of California, Berkeley, pp 128–474
Swingle WT, Reece PC (1967) The botany of Citrus and its wild relatives in the orange subfamily. In: Reuther W, Webber HJ, Batchelor DL (eds) The Citrus industry, vol 1. University of California, Berkeley, pp 190–340
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenetics by using the NJ methods. Proc Nat Acad Sci (USA) 101:11030–11035
Tamura K, Dudley J, Nei M, et al. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. doi:10.1093/molbev/msm092
Tanaka T (1928) On certain new species of Citrus. Studia Citrologia 2:155–164. (Japanese with English & Latin resume)
Tanaka T (1954) Species problem in Citrus. Japanese Society for promotion of Science, Ueno, p 152
Tanaka T (1977) Fundamental discussion of Citrus classification. Stud Citrol 14:1–6
Webber HJ (1943) Cultivated varieties of Citrus. In: Webber HJ, Batchelor DL (eds) The Citrus industry, vol I. University of California, Berkeley, pp 475–668
White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis N, Gelfand D, Sninsky J, White J (eds) PCR—protocols and applications—a laboratory manual. Academic Press, New York, pp 315–322
Xu CJ, Bau L, Zhang B, Bei ZM, Ye XY, Zhang SL, Chen KS (2006) Parentage analysis of huyou (Citrus changshanensis) based on internal transcribed spacer sequences. Pl Breed 125:519–522
Acknowledgments
The authors wish to thank the Director, CSIR-National Botanical Research Institute (NBRI) for providing facilities to carry out this work, and the officers & staff of Nokrek Biosphere Reserve (NBR), Tura, Meghalaya for their help and support during exploration trips to NBR. The financial support received from the Department of Biotechnology, New Delhi and the Council of Scientific and Industrial Research (CSIR), New Delhi to carry out this work is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, S., Nair, K.N. & Jena, S.N. Molecular differentiation in Indian Citrus L. (Rutaceae) inferred from nrDNA ITS sequence analysis. Genet Resour Crop Evol 60, 59–75 (2013). https://doi.org/10.1007/s10722-012-9814-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-012-9814-x