Abstract
Flax (Linum usitatissimum L.) is the earliest oil and fibre crop, but little is known about its domestication process. Attempt was made here to assess genetic relationships of 63 Linum accessions representing seven typical groups of cultivated flax and its wild progenitor, pale flax (Linum bienne Mill.), by using 49 informative expressed sequence tag-derived simple sequence repeat (EST-SSR) primer pairs. The seven groups were pale flax from Turkey, pale flax from other countries, and five groups of cultivated flax (landrace, fibre, oil, winter, and dehiscent). From these 63 samples, 366 polymorphic bands were detected, which likely represented 79 loci. These polymorphic bands had frequencies that ranged from 0.016 to 0.984 and averaged 0.284. Group-specific EST-SSR variation (Fst values) ranged from 0.339 to 0.373 and averaged 0.349 and pairwise group EST-SSR variation ranged from 0.067 to 0.507. A neighbor-joining clustering of these seven groups revealed that dehiscent flax clustered most closely to its wild progenitor, pale flax, followed by oil flax and fibre flax. Winter flax clustered most closely to oil flax and less to pale flax. These clustering patterns were essentially the same when individual samples were analyzed via neighbor-joining. These findings strongly suggest that capsular dehiscence was among the first flax traits modified by human after initial domestication, reflecting the importance that reducing capsular dehiscence likely played in early flax domestication.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The last two decades have seen an increase in genetic studies of crop domestication, thanks to the development of many informative molecular markers (Zeder et al. 2006; Purugganan and Fuller 2009). Significant efforts have been made to determine the genetic basis of plant traits associated with domestication such as plant branching and seed shattering in model crop species (Doebley et al. 2006; Li et al. 2006; Burke et al. 2007). Considerable research has also been conducted to identify wild progenitors of domesticated crops (Fu et al. 2002; Matsuoka 2005) and investigate domestication events (Heun et al. 1997; Matsuoka et al. 2002; Morrell and Clegg 2007). However, less effort has been made to trace domesticate dispersals and infer temporal domestication sequences (Sweeney et al. 2007; Konishi et al. 2008). Consequently, genetic evidence is still largely lacking for understanding the domestication processes of many agricultural crops (Allaby 2010).
Flax (Linum usitatissimum L.) is one of the founding crops in the Near Eastern agriculture, was a principal source of oil and fibre from prehistoric times until the early twentieth century, and still remains a crop of considerable economic importance (Muir and Westcott 2003). It was domesticated for both oil and fibre use more than 8,000 years ago in the Near East, as revealed by many archaeological finds (Helbaek 1959; van Zeist and Bakker-Heeres 1975). Its wild progenitor is pale flax (Linum bienne Mill.), as confirmed with morphological, cytological and molecular characterizations (Tammes 1928; Gill 1966, 1987; Diederichsen and Hammer 1995; Fu et al. 2002; Allaby et al. 2005). Recent molecular evidence suggests that cultivated flax is probably descended from a single domestication of pale flax, apparently for its oil, rather than fibre, use (Allaby et al. 2005). However, the rest of the early history of flax domestication remains unclear (Zohary and Hopf 2000; Allaby et al. 2005).
The archaeological finds of pale flax came first from Tell Abu Hureyra in northern Syria (11,200–10,500 years ago) (Hillman 1975) and occurred throughout the Near East by the 8th millennium BC (Zohary and Hopf 2000). The first occurrence of cultivated forms of flax with an increase in seed size is evidenced in archaeological records from Tell Ramad in Syria 9,000 years ago (van Zeist and Bakker-Heeres 1975). Flax then spread from the Near East to Europe and the Nile Valley. The flax varieties that spread into the Danube valley were winter oil varieties. However, summer fibre varieties developed in eastern Europe also spread into central Europe and replaced the original varieties (Helbaek 1959; Diederichsen and Hammer 1995). It is uncertain whether fibre flax from eastern Europe resulted from a domestication event independent to that of fibre flax in the Near East, but all modern fibre varieties in use today are thought to have originated from eastern Europe (Helbaek 1959).
Earlier genetic studies of flax domestication have identified its wild progenitor (Tammes 1928; Gill 1987; Fu et al. 2002; Fu and Allaby 2010) and inferred the domestication event and purpose (Allaby et al. 2005; Uysal et al. 2010). These studies, although useful, are largely limited in scope due to the lack of informative molecular markers and insufficient sampling of pale flax. The recent development of flax expressed sequence tag-derived simple sequence repeat (EST-SSR) markers (Cloutier et al. 2009; Fu and Peterson 2010) and recent collections of Turkish pale flax germplasm (Uysal et al. 2010) made further genetic inferences of flax domestication processes possible. For example, assessments of genetic relationships among various groups of cultivated flax with unique domestication traits may shed some insight into the path of flax domestication, as domestication processes influenced flax traits and specific flax trait groups may carry genetic signatures of such domestication processes over time.
Pale flax is a winter annual or perennial plant with narrow leaves and dehiscent capsules, and usually displays large variation in the vegetative plant parts and variable growth habit (Diederichsen and Hammer 1995; Uysal et al. 2011). In contrast, cultivated flax has variable seed dormancy, grows fast with large variation in the generative plant parts, and has early flowering, almost indehiscent capsules and large seeds. Interestingly, some of these domestication syndromes (Hammer 1984) have been used to group cultivated flax such as dehiscent cultivated flax with spontaneously opening capsules and winter flax with a vernalization requirement (Elladi 1940; Dillman 1953; Kulpa and Danert 1962; Diederichsen and Fu 2006). These intraspecific classifications should not only facilitate flax germplasm management and utilization, but also enhance genetic studies of flax domestication history.
The objective of this study was to assess the genetic relationships of 63 Linum accessions representing seven typical groups of cultivated flax and its wild progenitor pale flax, by using 49 informative EST-SSR primer pairs. The seven groups are pale flax from Turkey, pale flax from other countries, and five groups of cultivated flax (landrace, fibre, oil, winter, and dehiscent). The last four groups represent cultivated flax with domestication-related traits and may carry genetic signatures of related domestication processes.
Materials and methods
Sixty-three Linum accessions (Table 1) were selected for this study and they represent three closely related Linum species (L. usitatissimum, L. bienne, and Linum decumbens Desf.) and five major groups of cultivated flax (landrace, fibre, oil, winter and dehiscent). The landrace group represents a collection of local oil and/or fibre varieties from different countries. The accession selection process also took into account the country of origin to widen genetic diversity for this study. About 10 seeds of the selected accessions were obtained from the world flax collection at Plant Gene Resources of Canada and planted in greenhouse at the Saskatoon Research Centre, Agriculture and Agri-Food Canada. Young leaves were individually collected, freeze-dried (in a Labconco Freeze Dry System for 1–3 days), and stored at −20°C. A freeze-dried leaf sample of one individual plant from each accession was selected, and its genomic DNA was extracted with the DNEasy Plant Mini kit (Qiagen, Mississauga, ON, Canada). Extracted DNA was quantified with a Thermo Scientific NanoDrop 8000 spectrometer (Fisher Scientific Canada, Toronto, Ontario, Canada) by following the instrument’s standard protocols for double-stranded DNA and then was diluted to 25 ng μL−1 for EST-SSR analysis.
Based on the characterization effort previously made in Linum species (Fu and Peterson 2010), 49 of the most informative EST-SSR primer pairs developed by Cloutier et al. (2009) were selected and applied to screen 63 samples. The PCR conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 10 s, 50°C for 20 s, 72°C for 1 min, and a final elongation step of 72°C for 5 min on either a DYAD or PTC-200 thermocycler (Bio Rad, Mississauga, ON, Canada). Each reaction consisted of 1X Standard Buffer with 1.5 mM MgCl2 (New England BioLabs, Pickering, ON, Canada), 0.2 mM dNTP (Promega/Fisher Scientific, Nepean, ON, Canada), 5 pmol each forward and reverse primer, 0.5 U of Taq polymerase (New England Biolabs, Pickering, ON, Canada) and 25 ng of genomic DNA in a final volume of 25 μL. Amplification products were separated with the Mega Gel system (CBS Scientific, Del Mar, CA, USA) on a non-denaturing, 5% 19:1 polyacrylamide gel (Wang et al. 2003) for up to 2.5 h at 300 V. Fragments were stained with 0.5 μg L−1 ethidium bromide in the running buffer, and recorded on a digital gel-documentation system.
To generate a dataset of EST-SSR allele counts for each sample, DNA fragments amplified by EST-SSR primer pairs were identified based on their sizes in base pairs measured with a 50 bp DNA ladder (New England BioLabs, Pickering, ON, Canada) and compared with fragment sizes reported in the literature (Cloutier et al. 2009; Fu and Peterson 2010). Multiple loci per primer pair were inferred as likely multiple loci, based on the number of bands observed for individual cultivated flax samples and the band pattern(s) across the 63 samples. Multiple alleles at a locus may exist at the species level, but only two bands per locus are expected for a diploid sample like those of cultivated flax. The DNA fragments were manually scored as 1 for presence or 0 for absence. Levels of polymorphism were analyzed with respect to primer and locus by counting the number of polymorphic bands and generating summary statistics on band frequencies. Shannon entropy was calculated following Russell et al. (1993) to estimate the diversity content per locus, as this estimate does not require strict genetic assumptions. Essentially, the entropy-based diversity content provides a measure of the effective number of alleles per marker locus (Reyes-Valdes and Williams 2005). These analyses were performed by using a SAS program written in SAS IML (SAS Institute Inc. 2004).
Genetic relationships of the 63 individual samples were inferred by using PAUP* (Swofford 2002) with a neighbor-joining method and a radiation tree was displayed by using MEGA 4.01 (Tamura et al. 2007). An analysis of molecular variance (AMOVA) was also performed using Arlequin version 3.01 (Excoffier et al. 2005) to assess genetic variation among various groups of Linum accessions. Four models of genetic structuring were considered: pale vs cultivated flax, two originating groups of pale flax, five groups of cultivated flax, and seven groups of pale and cultivated flax. The significance of variance components and inter-group genetic distances for each model was tested with 10,010 random permutations. The last model also generated genetic distances as the proportional EST-SSR variations among seven groups of Linum accessions. Based on these groupwise genetic distances, a neighbor-joining clustering of the seven groups was also made with NTSYS-pc 2.3 (Rohlf 1997).
Results
Screening 49 EST-SSR primer pairs detected a total of 366 polymorphic bands across the 63 samples (Table 2). Based on reported fragment sizes and band patterns, 79 likely loci were inferred. Nine primer pairs each amplified a pattern of three likely loci; 12 primer pairs each detected two likely loci; and the remaining primer pairs each generated a single locus (Table 2). The number of bands detected per locus ranged from one to 29 (for the locus Lu151a) with an average of 4.6 bands per locus. Band frequencies across the 63 samples ranged from 0.016 to 0.984 and averaged 0.283. There were 84 bands that occurred in three or fewer samples (i.e., with a frequency 0.048 or smaller). An assessment of diversity content per locus revealed 13 loci with a Shannon entropy ≥ 1.5 and 5 loci with a Shannon entropy ≥ 2 (Table 2). A linear regression analysis revealed that these diversity contents were significantly (P < 0.0001) associated with the numbers of bands detected for these loci. As these loci were not mapped, the exact genome coverage of these markers remains unknown, but they should sample many of the transcribed chromosomal segments of flax genome.
Patterns of genetic structure within and among these flax accessions were assessed with four genetic-structure models (Table 3). Overall, 28.2% of the total EST-SSR variation could be attributed to difference between samples of pale and cultivated flax and 71.8% occurred within samples of each species. More variation was observed within cultivated flax than within pale flax. Considerable variation (16.6%) was noted between pale flax samples from Turkey and those from other countries, and slightly more variation was observed within the Turkish than within other countries’ pale flax samples. For cultivated flax, 28.5% of the total variation resided among five cultivated flax groups. The oil flax samples displayed the most within-group variation, followed by the winter, landrace, fibre, and dehiscent flax, respectively.
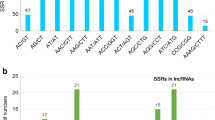
Considering seven groups of pale and cultivated flax together, 34.8% of the total variation differentiated the seven groups, and 65.2% was found within groups (Table 3). The groups of oil flax and pale flax from other countries displayed slightly more within-group variation than did the other groups (Table 4). Assessments of pairwise group variation measured with the proportional EST-SSR variation (Table 4) showed that dehiscent flax samples were the most distant from Turkish pale flax samples (0.507), followed by fibre (0.445, winter (0.384), landrace (0.372), and oil flax samples (0.369).
Neighbor-joining clustering of these seven groups based on proportional EST-SSR variation revealed that dehiscent flax samples clustered most closely to pale flax samples, followed by oil flax (and/or winter flax) and fibre flax (and/or landrace flax) samples (Fig. 1). This clustering pattern remained unchanged either when the landrace and winter flax samples were excluded or when pale flax accessions from Turkey and other countries were formed as a single group. Neighbor-joining analysis of the 63 individual samples revealed the same pattern for the seven groups of pale and cultivated flax (Fig. 2). For example, dehiscent flax samples still clustered most closely to pale flax samples, followed by fibre flax group mixed with landrace flax. Winter flax samples clustered most closely to oil flax samples and were less like its wild progenitor. Only one winter flax sample was clustered with the fibre flax group. Four of the eight landrace samples were clustered with the fibre flax samples. One fibre flax sample (Uf7) was clustered with oil flax group and (Fig. 2). The four Greek pale flax samples grouped together, but the Turkish pale flax samples were more divergent. The other closely related wild Linum species, L. decumbens, separated clearly from both pale and cultivated flax (Fig. 2).
Genetic relationships of seven groups of pale and cultivated flax accessions based on genetic distances (as the proportional EST-SSR variation) obtained from the analysis of molecular variance for 366 polymorphic EST-SSR bands. The seven groups are pale flax from Turkey, pale flax from other countries, and five groups of cultivated flax (landrace, fibre, oil, winter, and dehiscent)
Clustering of 63 Linum accessions representing three species and five groups of cultivated flax obtained from the neighbor-joining analysis of 366 polymorphic SSR bands. Each accession is labeled as in Table 1 with the first letter for species (B = L. bienne; D = L. decumbens, and U = L. usitatissimum), the second letter (if any) for the country of L. bienne accessions (t Turkey, g Greece, m multiple countries) and for the group of cultivated flax (n landrace, f fibre, o oil, w winter, d dehiscent), and the numbering for multiple accessions of the same group. The distance scale reflects the proportion of EST-SSR variation
Discussion
This EST-SSR analysis demonstrated that dehiscent flax most closely resembled its wild progenitor, pale flax, followed by oil flax and fibre flax. It also showed that winter flax closely resembled oil flax and was less like pale flax. These results were consistent with clustering pattern obtained with ISSR markers, indicating that Turkish pale flax clustered most closely to dehiscent flax (Uysal et al. 2010). These findings together provide the first genetic evidence that capsular dehiscence was among the flax traits modified after initial human selection and indicate that reducing capsular dehiscence was an important step in early flax domestication.
The importance of reducing capsular dehiscence and minimizing vernalization requirement in early domestication of many founding crops, although with little empirical support available, has been well recognized (Hammer 1984; Zohary and Hopf 2000; Zeder et al. 2006; Li et al. 2006), as their wild progenitors usually are winter annual or perennial plant with dehiscent capsules and modern cultivars largely have indehiscent capsules and little vernalization requirement. However, the importance of minimizing flax vernalization requirement in early flax domestication does not seem to be as clear-cut as for capsular dehiscence. First, winter flax is genetically so distant from pale flax while dehiscent flax (a summer annual) is much less so (Figs. 1 and 2). Second, winter flax is genetically intermingled with oil flax while dehiscent flax formed its own cluster (Fig. 2). Based on these findings, it is difficult to establish the case for early flax domestication with winter hardiness.
It is possible that capsular dehiscence in flax has a simple genetic control like those in other crops do (e.g., Bailey et al. 1997; Li et al. 2006; Kaga et al. 2008), mass selection for capsular indehiscence was successful at the time of initial flax domestication, and further selection was not required at later stages of domestication for other genetically complex traits such as oil or fibre. Thus, the unique genetic background of dehiscent flax was able to be maintained. It was also unlikely that dehiscent flax was a result of more recent hybridization between highly selected flax and pale flax, as large EST-SSR variation was observed among dehiscent flax accessions (Table 4). In contrast, winter hardiness is a complex trait controlled by many genes across many chromosomes (e.g., Pan et al. 1994; Kahraman et al. 2004), and may have been long selected along with oil improvement in diverse environment remote from the locus of domestication. This seems to accord well with the adaptation of cultivated flax to cold environments in northern Europe when oil flax spread into the Danube valley (Helbaek 1959). Thus, most winter flax samples assayed were genetically intermingled with oil flax accessions and divergent from pale flax. It also is possible that the winter flax accessions assayed here may not adequately sample the gene pool of flax germplasm with vernalization requirement, as the use of winter flax for fibre use was also reported (Hegi 1925), and bias may exist in inferences of its departure from pale flax and its genetic similarity with oil flax.
The finding of fibre flax clustered more closely to pale flax (Fig. 2) implies that most of the fibre flax still maintained its unique genetic background, more similar than the oil flax to the pale flax. However, this finding seems to contradict with the early argument that flax was first domesticated for oil use (Allaby et al. 2005). Also, the assayed flax landrace samples do not appear to generate any new information for understanding flax domestication processes. This may reflect the nature of the landrace group with mixed members from other four forms of cultivated flax. Moreover, the inferred clusters may reflect roughly the genome similarities, not the precise inferences of evolutionary relationship, between cultivated and pale flax, as the assayed SSR alleles of same size may not always be identical by descent between two species.
In spite of these limitations, the inferred genetic relationships among various groups of pale and cultivated flax are significant for understanding flax domestication. They may reflect the path of flax domestication over the past 9,000 years (Helbaek 1959) from domesticating pale flax to reduced capsular dehiscence, increased fibre length and seed size for oil content, along with improved winter hardiness, and eventually to modern cultivars. Of course, the exact events and sequence of these modifications remain to be determined. Next-generation DNA sequence studies (Nordborg and Weigel 2008), together with phylogenetic analyses including appropriate outgroups to establish polarity of characters, should help to clarify relationships between pale flax and the various forms of cultivated flax. In addition, specific domestication-associated trait groups in flax may carry abundant genetic signatures of past domestication events (Allaby 2010), which should spur future research.
References
Allaby R (2010) Integrating the processes in the evolutionary systems of domestication. J Exp Bot 61:935–944
Allaby RG, Peterson GW, Merriwether A, Fu YB (2005) Evidence of the domestication history of flax (Linum usitatissimum) from genetic diversity of the sad2 locus. Theor Appl Genet 112:58–65
Bailey MA, Mian MAR, Carter TE Jr, Askley DA, Boerma HR (1997) Pod dehiscence of soybean: identification of quantitative trait loci. J Hered 88:152–154
Burke JM, Burger JC, Chapman MA (2007) Crop evolution: from genetics to genomics. Curr Opin Genet Dev 17:525–532
Cloutier S, Niu X, Datla R, Duguid S (2009) Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor Appl Genet 119:53–63
Diederichsen A, Fu YB (2006) Phenotypic and molecular (RAPD) differentiation of four infraspecific groups of cultivated flax (Linum usitatissimum L. subsp. usitatissimum). Genet Resour Crop Evol 53:77–90
Diederichsen A, Hammer K (1995) Variation of cultivated flax (Linum usitatissimum L. subsp. usitatissimum) and its wild progenitor pale flax (subsp. angustifolium (Huds.) Thell.). Genet Resour Crop Evol 42:263–272
Dillman AC (1953) Classification of flax varieties, 1946. USDA Technical Bulletin No. 1054. United States Department of Agriculture, Washington, DC, p 56
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127:1309–1321
Elladi VN (1940) Linum usitatissimum (L.) Vav. consp. nov.—Len. (Russ.). In: Vul’f EV, Vavilov NI (eds) Kul’turnaja flora SSSR, prjadil’nye [Flora of cultivated plants of the USSR, fibre plants]. Sel’chozgiz, Moscow, Leningrad, vol 5, Part 1, pp 109–207
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.01: an integrated software package for population genetics data analysis. Evol Bioinfo Online 1:47–50
Fu YB, Allaby RG (2010) Phylogenetic network of Linum species as revealed by non-coding chloroplast DNA sequences. Genet Resour Crop Evol 57:667–677
Fu YB, Peterson GW (2010) Characterization of expressed sequence tag derived simple sequence repeat markers for 17 Linum species. Botany 88:537–543
Fu YB, Peterson G, Diederichsen A, Richards KW (2002) RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet Resour Crop Evol 49:253–259
Gill KS (1966) Evolutionary relationships among Linum species. Ph.D. Thesis, University of California, Riverside, CA, USA
Gill KS (1987) Linseed. Indian Council of Agricultural Research, New Delhi
Hammer K (1984) Das Domestikationssyndrom. Kulturpflanze 32:11–34
Hegi G (1925) Illustrierte Flora von Mitteleuropa. Band V, Teil 3. Parey, Berlin-Hamburg
Helbaek H (1959) Domestication of food plants in the Old World. Science 130:365–372
Heun M, Schäfer-Pregl R, Klawan D, Castagna R, Accerbi M, Borghi B, Salamini F (1997) Site of einkorn wheat domestication identified by DNA fingerprinting. Science 278:1312–1314
Hillman G (1975) The plant remains from Tell Abu Hureyra: a preliminary report. Proc Prehist Soc 41:70–73
Kaga A, Isemura T, Tomooka N, Vaughan DA (2008) The genetics of domestication of the azuki bean (Vigna angularis). Genetics 178:1013–1036
Kahraman A, Kusmenoglu I, Aydin N, Aydogan A, Erskine W, Muehlbauer FJ (2004) Genetics of winter hardiness in 10 lentil recombinant inbred line populations. Crop Sci 44:5–12
Konishi S, Ebana K, Izawa T (2008) Inference of the japonica rice domestication process from the distribution of six functional nucleotide polymorphisms of domestication-related genes in various landraces and modern cultivars. Plant Cell Phyisol 49:1283–1293
Kulpa W, Danert S (1962) Zur Systematik von Linum usitatissimum L. Kulturpflanze (Beiheft 3):341–388
Li CB, Zhou AL, Sang T (2006) Rice domestication by reducing shattering. Science 311:1936–1939
Matsuoka Y (2005) Origin matters: lessons from the search for the wild ancestor of maize. Breed Sci 55:383–390
Matsuoka Y, Vigouroux Y, Goodman MM, Jesus Sanchez G, Buckler E, Doebley J (2002) A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99:6080–6084
Morrell PL, Clegg MT (2007) Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the fertile crescent. Proc Natl Acad Sci USA 104:3289–3294
Muir A, Westcott N (2003) Flax, the genus Linum. Taylor and Francis, London
Nordborg M, Weigel D (2008) Next-generation genetics in plants. Nature 456:720–723
Pan A, Hayes PM, Chen F, Chen THH, Blake T, Wright S, Karsai I, Bedo Z (1994) Genetic analysis of the components of winter hardiness in barley (Hordeum vulgare L.). Theor Appl Genet 89:900–910
Purugganan MD, Fuller DQ (2009) The nature of selection during plant domestication. Nature 457:843–848
Reyes-Valdes MH, Williams CG (2005) An entropy-based measure of founder informativeness. Genet Res 85:81–88
Rohlf FJ (1997) NTSYS-pc 2.1. Numerical taxonomy and multivariate analysis system. Exeter Software, Setauket
Russell JR, Hosein F, Johnson E, Waugh R, Powell W (1993) Genetic differentiation of cocoa (Theobroma cacao L.) populations revealed by RAPD analysis. Mol Ecol 2:9–97
SAS Institute Inc (2004) The SAS system for windows V8.02. SAS Institute Inc., Cary, NC
Sweeney MT, Thomson MJ, Cho YG, Park YJ, Williamson SH, Bustamante CD, McCouch SR (2007) Global dissemination of a single mutation conferring white pericarp in rice. PLOS Genet 3:11418–11424
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland
Tammes T (1928) The genetics of the genus Linum. Bibliogr Genet 4:1–36
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Uysal H, Fu YB, Kurt O, Peterson GW, Diederichsen A, Kusters P (2010) Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne Mill.) as revealed by ISSR markers. Genet Resour Crop Evol 57:1109–1119
Uysal H, Kurt O, Fu YB, Diederichsen A, Kusters P (2011) Variation in morphological and phenotypic characters of pale flax (Linum bienne Mill.) from Turkey. Genet Resour Crop Evol (accepted)
van Zeist W, Bakker-Heeres JAH (1975) Evidence for linseed cultivation before 6000 BC. J Archaeol Sci 2:215–219
Wang D, Shi J, Carlson SR, Cregan PB, Ward RW, Diers BW (2003) A low-cost, high-throughput polyacrylamide gel electrophoresis system for genotyping with microsatellite DNA markers. Crop Sci 43:1828–1832
Zeder MA, Bradley DG, Emshwiller E, Smith BD (2006) Documenting domestication: new genetic and archaeological paradigms. University of California Press, Berkeley and Los Angeles
Zohary D, Hopf M (2000) Domestication of plants in the Old World, 3rd edn. Oxford University Press, Oxford. pp 125–132
Acknowledgments
The author would like to thank Dallas Kessler, Greg Peterson and Kimberly MacKay for their technical support in sampling and planting seeds, EST-SSR screening and scoring of Linum samples; Axel Diederichsen for his stimulating discussion on this project; and two anonymous reviewers for their helpful comments on the early version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fu, YB. Genetic evidence for early flax domestication with capsular dehiscence. Genet Resour Crop Evol 58, 1119–1128 (2011). https://doi.org/10.1007/s10722-010-9650-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-010-9650-9