Abstract
We present here the first comprehensive genetic characterization of melon landraces from the humid tropics of southern India. The genetic diversity among 50 melon landraces collected from 3 agro-ecological regions of southern India (6 agro-ecological sub-regions) was assessed by variation at 17 SSR loci, morphological traits of plant habit and fruit, 2 yield-associated traits, pest and disease resistance, biochemical composition (ascorbic acid, carotenoids, titrable acidity) and mineral content (P, K, Fe, Zn). Differences among accessions were observed in plant and fruit traits. Melon germplasm with high titrable acidity, higher than average amounts of mineral content and resistance to Cucumber mosaic virus, Zucchini yellow mosaic virus, powdery mildew (races 1, 2, 3, 5), Fusarium wilt (races 1, 2), Aphis gossypii and leafminer was recorded in the collection. A high level of genetic variability in melon germplasm was suggested by the SSR analysis. Comparative analysis using SSRs of the genetic variability between Indian melons from north, south, and east regions and reference accessions of melon from Spain, France, Japan, Korea, Iraq, Zambia showed regional differentiation between Indian melon accessions and that Indian germplasm was weakly related to the melon accessions from other parts of the world, suggesting that an important portion of the genetic variability found within this melon collection has not been used yet for the development of new cultivars. Additional collections of acidulus melon germplasm should be made in southern India and adequate management of this important genetic resource is clearly a necessity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Melon (Cucumis melo L.; 2n = 2x = 24) is a tropical old-world species and its geographical origin is still unclear. Africa is considered to be the centre of origin of melon, though the recent data supports the view that the origin of the genus Cucumis may be in Asia (Renner et al. 2007; Schaefer et al. 2009). Melon was first domesticated in Egypt and Iran during the second and third millennia BC, respectively (Pangalo 1929). The main center of diversity of melon is located in Asia, from the Mediterranean basin (Turkey) to Central Asia (Iran, Uzbekistan) to India to East Asia (China, Korea) (Robinson and Decker-Walters 1997).
Melon exhibits tremendous variation in fruit traits such as size, shape, colour, taste, texture, and biochemical composition. Information on the genetic variation in melon germplasm of the humid tropics of southern India is lacking in the literature. Global genebanks contain Indian melon accessions originating from the north (Rajasthan) and central (Madhya Pradesh) parts of India (McCreight et al. 1993; Staub and McCreight 2004) but not from southern India. McCreight et al. (2004) emphasised that the genetic variation in melon germplasm of north and central India might not represent the genetic diversity present in southern and eastern India. They recommended that additional collections of melon genetic resources should be made in southern and eastern India as this could lead to the discovery of genetic diversity not present in the existing world collections of melon. A similar genetic picture of Indian melon germplasm has been portrayed by Akashi et al. (2002).
We have collected melon landraces belonging to two groups: var. acidulus Naudin and var. momordica (Roxb.) Duthie et Fuller. C. melo var. acidulus is native to the humid tropics of southern India. Its vines are monoecious with oval or elliptic fruits, skin is smooth, orange or light yellow in colour with or without green spots, and fruit flesh is white or cream light yellow, very firm and without sugar or aroma. This type of melon is grown in two southern states of India viz. Kerala and Tamil Nadu and the approximate area under its cultivation in these two states is 1,000 and 590 ha, respectively (T.K. Sam, personal communication.). It is cultivated as a mixed crop in coconut fields as well as in homesteads. In the local language, this melon is known as ‘Vellari’. Fruits have very long shelf life and tender fruits are consumed as salad or used for ‘Sambhar’ preparation. ‘Sambhar’, a vegetable stew based on a broth with tamarind and ‘toor dal’ (small lentils), is a dish common in south India and Sri Lanka Tamil cuisines. Ripe fruits are also eaten.
C. melo var. momordica is cultivated in India under the name of ‘Phut’. The group name is a reference to the genus Momordica of the Cucurbitaceae family whose fruits are cracking and opening at maturity. The plants are monoecious. The fruits are oval or elliptic, with a smooth skin, orange or light yellow in colour. Fruit flesh may be white, cream light yellow or light orange, mealy and without sugar or aroma. Immature fruits are cooked or pickled.
These landraces were collected from three agro-ecological regions of southern India representing six agro-ecological sub-regions dispersed over two states (Kerala and Tamil Nadu). These agro-ecological regions and sub-regions have been classified according to National Bureau of Soil Survey and Land Use Planning (NBSS & LUP) and National Agricultural Research Project (NARP) classification, respectively (Ghosh 1991; Sehgal et al. 1992). Each agro-ecological region has a uniform growing period, climate, land form, and soil type.
We used morphological and pest/pathogen resistance data, biochemical traits and mineral composition data to assess melon genetic diversity. The variation detected among melons originating in southern India, was compared to a reference group of melon accessions from diverse origins (Eastern Asia, Central Asia, Western Asia, Africa, Europe, other geographic parts of India) (Monforte et al. 2003, 2005) using a set of simple sequence repeat (SSR) markers which have proved valuable for melon germplasm characterisation (Katzir et al. 1996).
Materials and methods
Germplasm
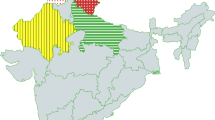
Fifty landraces of melon (Table 1) were collected from the two states in southern India, Kerala and Tamil Nadu, representing three agro-ecological regions (Sehgal et al. 1992) and six agro-ecological sub-regions (Ghosh 1991) (Fig. 1, Table 2). Accessions belonging to a broad range of melon horticultural types (Monforte et al. 2003, 2005) and Indian melon accessions belonging to north and eastern India (momordica, agrestis Naudin) were also included in the study as reference populations (Table 3). These 50 landraces of melon originating from southern India were evaluated for morphological and biochemical traits, mineral content and disease/pest resistance. Thirty-nine of these landraces representing all the three agro-ecological regions of Kerala and Tamil Nadu, together with accessions mentioned in Table 3 (reference genotypes) were used for SSR analysis. Original germplasm, maintained through sibing, was used for the molecular study and single plant selections from the S3 generation of each accession were used for rest of the evaluations.
Morphological evaluation
Fifty landraces of melon were evaluated for morphological traits and productivity in 2006 at the Punjab Agricultural University, Ludhiana, India. Accessions were sown in compost and seedlings at the three-leaf stage were transplanted to the field. Three replications containing ten plants of each accession were arranged in a randomised complete block design such that row spacing was 3.0 m and within row spacing was 0.45 m. Plants were furrow irrigated and fertilised using standard cultural practices. In each replication, five central plants of each accession were used for sampling. The following traits were recorded: (1) plant growth habit, (2) number of primary branches/vine, (3) stem shape, (4) leaf size of fully developed leaf as described by Srivastava et al. (2001), (5) fruit shape, (6) fruit skin primary colour, (7) fruit skin secondary colour, (8) pattern produced by secondary fruit skin colour, (9) mature fruit flesh colour, (10) flesh odour assessed through smelling the mature fruits, (11) marketable fruit number/vine, (12) marketable fruit weight, (13) days to marketable maturity i.e. the number of days taken from sowing to the harvest of first marketable fruit and (14) sex expression.
Biochemical assay
Fifty landraces of melon were evaluated for biochemical traits. Five fruits of each accession in each replication were harvested at marketable maturity (for ascorbic acid and carotene analysis) and at fruit maturity (for total soluble solids and acidity analysis). Total soluble solids (TSS, expressed as Brix, °B) were measured from fruit juice using a hand refractometer. Ascorbic acid was quantified as described by Bajaj and Kaur (1981). Titrable acidity (%) was determined by titration of a fruit juice sample with 0.05 N NaOH, using phenolphthalein as indicator. Total carotenoids were estimated by the method described by Thomas and Joshi (1977).
Mineral assay
P and K concentrations were measured in diacid digest of fruit samples by Spectrophotometer ‘Spectronic 21’ (Bausch & Lomb, USA) and Flame Photometer (BWB Technologies UK Ltd), respectively (Gupta 1999). Micronutrients (Fe, Zn) were determined with atomic absorption spectrometer (Varian AA 20). The analysis of variance of various morphological and biochemical characters was carried out using SAS (SAS Institute 2003) software. Means were compared using least significant differences (Steel and Torrie 1980).
Screening for Cucumber mosaic virus (CMV), root knot nematode (Meloidogyne incognita Chitwood) and leafminer (Liriomyza trifolii Burgess) resistance
The assessment for resistance to CMV and root knot nematode were carried out as described in Dhillon et al. (2007). Screening for CMV resistance was done under natural epiphytotic conditions in the field, during the rainy season (August–September) in 2007. The assessment for root knot nematode resistance was carried out in infested potted soil. The Liriomyza infestation in the field was on an epidemic scale. No plant protection measure was adopted to control this pest. For measuring resistance, a 1–5 scale was adopted for assessing individual leaves, 1 = no damage, resistant (R), 2 = 1–25% leaf area damaged, moderately resistant (MR), 3 = 26–50% leaf area damaged, moderately susceptible (MS), 4 = 51–75% leaf area damaged, susceptible (S), 5 = 76–100% leaf area damaged, highly susceptible (HS). When in the surrounding melon field, leaf damage from leafminer activity was at a maximum, 5 plants with 15 damaged leaves per plant were randomly selected from each accession for assessing pest damage.
Screening for Algerian watermelon mosaic virus (AWMV), Melon necrotic spot virus (MNSV), Moroccan watermelon mosaic virus (MWMV), Papaya ringspot virus watermelon strain (PRSV-W), Squash mosaic virus (SqMV), Zucchini yellow mosaic virus (ZYMV), Aphis gossypii Glover, Fusarium oxysporum f.sp. melonis Snyder et Hansen, and powdery mildew [Podosphaera xanthii (Castagne) U. Braun et Shishkoff] resistance.
Evaluation was made by artificial inoculation at the cotyledon stage for the different viruses and Fusarium wilt, at the first true leaf stage for A. gossypii and at the three leaves stage on leaf disks for powdery mildew (Pitrat et al. 1996).
DNA extraction
Leaf tissue from ten plants per accession was bulked for total genomic DNA extraction using the method described by Doyle and Doyle (1990), with modifications suggested by Garcia-Mas et al. (2000).
Microsatellite analysis
The 17 SSR markers used in this study (ECM50, ECM51, ECM52, ECM61, ECM65, ECM70, ECM80, ECM85, ECM109, ECM124, ECM125, ECM129, ECM130, ECM133, ECM134, ECM178, and ECM182) were developed by Fernandez-Silva et al. (2008). PCR reactions were performed in a final volume of 15 μl with 1× Taq buffer [10 mM Tris–HCl, 50 mM KCl, 0.001% gelatine, (pH 8.3)], 1.5–3.5 mM MgCl2, 166 μM dNTPs, 2 pmol of each forward and reverse primers, 0.66 pmol of IRD700- or IRD800-labelled oligonucleotide complementary to the 20-mer M13 sequence that was added to the forward primer (Fernandez-Silva et al. 2008), 2 U Taq DNA polymerase and 15 ng of genomic DNA. The cycling conditions were at 94°C 1 min, followed by 35 cycles at 94°C 30 s, 72°C 1 min, then 72°C 5 min. Electrophoresis were performed using LI-COR IR2 sequencer (Li-Cor Inc, Lincoln, NE, USA), using 25 cm plates with 6% acrylamide, 1× TBE (90 mM Tris–borate, 2 mM EDTA, pH 8.0 and 7.5 M urea) and electrophoresis was performed at 1,500 V, 35 mA and 31 W at 50°C until the PCR products were visible.
Thirty-nine accessions were analysed for SSR polymorphism. Due to the bulking of samples during DNA extraction, the observation of two or more SSR alleles in a single genotype could have resulted from the presence of several heterozygous plants, or homozygous plants for the alternative alleles or a combination of both. The PCR amplification that we used do not permit the quantification of the frequency of an SSR allele based on the band intensity or densitometry within pooled sample. Therefore, all the detected alleles were assumed to have a frequency of 1/n (n = number of alleles). Microsatellite allele sizes were estimated comparing their migration with the IRD-700 or -800, 50–350 bp size standards (Li-Cor, Inc.). Number of alleles, allele frequencies, polymorphism information content (PIC), Nei et al. (1983) genetic distances and Neighbour-Joining (NJ) tree were calculated with Powermarker (Liu and Muse 2005), NJ tree was plotted with MEGA 3.0 (Tamura et al. 2007). Factor Correspondence Analysis (FCA) was performed with NTSYSpc 2.11W (Exeter Software, Setauket, NY).
Correlations between qualitative traits were studied using the exact Fisher test, between quantitative traits by the correlation coefficient and between quantitative and qualitative traits by ANOVA.
Results
Morphological variations and field observations
A detailed description of the melon landraces used in the present study is provided in Tables 4 and 5. All the melon accessions were monoecious. The vines of these accessions were prostrate except two accessions (AM 5 and AM 6) which were of intermediate plant growth habit. Round and angular stems were observed in 26 and 74% of the accessions, respectively. Three sizes of leaf (large, medium and small) were found in 32, 66 and 2% of the accessions, respectively. The range of primary branches/vine was from 2.0 to 7.5. The highest number of primary branches was observed in two accessions (AM 5 and AM 7) from the Cudalore district of Tamil Nadu. Four types of fruit shapes were available in the germplasm viz. elongated, oblate, elliptical and pyriform (Table 4). The majority of the landraces belonged either to the elongated (42%) or oblate (40%) category; the types elliptical and pyriform were represented by 10 and 6% of the accessions, respectively. The majority of landraces (60%) had yellow primary skin colour, other accessions were orange (14%), light green (18%) or green (4%). The majority of the accessions had green (68%) or dark green (14%) secondary skin colour whereas only one accession (AM 39) was orange. No secondary skin colour was observed in four accessions and these accessions had either yellow or orange primary skin colour. Three kinds of pattern were produced by the secondary skin colour speckled (54%), spotted (22%), and striped (14%). Yellow orange (44%) to orange (56%) fruit flesh was observed amongst the accessions. Consumers had a preference for both these colours in both the states. Fruit cracking at maturity was observed in 62% of the accessions and these accessions had speckled (72%) and striped (27%) skin patterns. Farmers growing non-cracking type landraces sell the ripe fruits also and hence this trait was deliberately selected by them. Matured fruits emitted three kinds of odours viz. strong, mild and odourless. The majority (78%) of the accessions possessed a mild odour but a strong odour was recorded in 6% of the accessions. Sixteen percent of the accessions were odourless. The range of time to marketable maturity of fruit was from 50.1 to 77.2 days. The average number of fruits/vine ranged between 2.5 and 9.0. Average accession fruit weight ranged between 0.175 and 1.735 kg. Average yield per plant varied between 0.87 and 5.33 kg. The three accessions with highest fruit weight had yielded less number of fruits per vine (2.6–4.0). Furthermore, in the farmers’ fields, variability in melon landraces was also observed for fruit length, fruit breadth, flesh thickness, rind thickness, seed cavity size, fruit skin lustre (glossy, matt, intermediate) and fruit skin texture (smooth, wrinkled) (data not presented). According to the definition of acidulus and momordica given in the introduction, 21 accessions may be clearly classified as belonging to the acidulus group and 29 to the momordica group (Table 1).
Biochemical comparison
The carotenoid, ascorbic acid and titrable acidity values of the germplasm are shown in Table 6. The total carotenoids contents ranged between 30.8 and 146.3 μg/100 g of fruit flesh weight. Ascorbic acid and titrable acidity of mature fruits ranged between 1.4 and 9.0 mg/100 g of fresh fruit weight and 0.12–0.57% respectively. There was no significant difference for titrable acidity between accessions belonging to the acidulus and the momordica groups, although it was expected that the acidulus group had a lower acidity. Accessions AM 84 and AM 70 contained significantly (P < 0.05) more ascorbic acid than other accessions (9.0 and 8.4 mg/100 g of fruit weight, respectively). Accessions AM 8 (0.57%) and AM 52 (0.54%) were significantly (P < 0.05) more acidic than the other landraces. All the accessions were non sweet and their TSS ranged between 2.1 and 6.4 °B (data not presented).
Mineral content comparison
The P, K, Fe and Zn values of the 50 melon accessions are provided in Table 7. Their P and K ranged between 2.6 and 26.4 mg/100 g of fruit fresh weight and 19.7–294.3 mg/100 g of fruit fresh weight, respectively. The Fe and Zn content in the accessions ranged between 0.03 and 1.14 mg/100 g of fruit fresh weight and 0.12–1.29 mg/100 g of fruit fresh weight, respectively.
Evaluation for pathogen and pest resistance
Two melon accessions (AM 25, AM 82) were resistant to CMV. Five accessions (AM 25, AM 39, AM 41, AM 55, and AM 85) were moderately resistant to leafminer. All the accessions were susceptible to root knot nematode.
For PRSV resistance, two types of symptoms have been observed: mosaic as in the susceptible control Védrantais in most of the accessions or necrosis indicating an incompatible interaction in AM 4, AM 25, AM 27, AM 70, AM 78 and AM 100. After inoculation with MWMV, all the accessions exhibited necrotic symptoms. With SqMV, three accessions (AM 7, AM 29 and AM 48) exhibited light symptoms but were positive in ELISA indicating virus multiplication. All the accessions were susceptible to AWMV and MNSV. All the accessions were susceptible to ZYMV strain R5A but AM 87 that showed some level of resistance.
Two accessions were resistant to A. gossypii AM 5 and AM 52 and three were heterogeneous segregating with resistant and susceptible plants: AM 7, AM 78, and AM 86.
For powdery mildew, the following accessions were resistant to races 1 (strain Sm3) and 2 (strain S87-7): AM 5, AM 22, AM 24, AM 67, AM 86 and AM 90. More interestingly, AM 22 was also resistant to race 3 (strain 00Sm39) and AM 67, AM 86 and AM 90 to race 5 (strain 98Sm65).
AM 27 was the only accession with resistant plants to race 1 of Fusarium wilt but it was not homogeneous. Against race 2, some accessions were segregating with resistant and susceptible plants (AM 4, AM 5, AM 20, AM 21, AM 22, AM 24, AM 39, AM 41, AM 48, AM 52, AM 57, AM 67, AM 76, AM 80, AM 84, AM 89). Some accessions were resistant to race 2 with all the tested plants (usually 10 plants): AM 6, AM 27, AM 47, AM 87, AM 88 and AM 90. AM 27 was the most interesting as it was resistant to race 2 and segregating for resistance to race 1.
Characterization of microsatellite loci
A total of 114 alleles were found across the full set of melon accessions. The average number of alleles per microsatellite was 6.8. The average PIC value, a reflection of allele diversity and frequency among these was 0.544. The average observed heterozygosity for collected accessions was 0.23 whereas it was 0.13 for reference populations. Thirty-one alleles (24.2%) were present exclusively in the collected accessions and the reference population had the same proportion of unique alleles.
Genetic structure of the germplasm collection
Two approaches were applied to assess the genetic relationships of 39 collected melon landraces of Indian origin and a set of 20 reference accessions belonging to the different parts of the world. The NJ dendrogram based on 114 SSR alleles is shown in Fig. 2. Several major patterns of genetic association were observed. All the collected accessions of south-Indian origin, except AM 18 and AM 71, were well separated from the reference genotypes. The collected accessions were clustered in four major groups. There was a significant correlation (Prob = 0.02 for the exact Fisher test) between botanical groups and the clustering. Accessions belonging to the momordica group are significantly more frequent in clusters C and D and accessions belonging to the acidulus group are more frequent in clusters A and B. However, accessions AM 24 and AM 27 belonging to the momordica group were in cluster A and accessions AM 8, AM 22, AM 70, AM 83 and AM 87 belonging to the acidulus group were in clusters C and D.
Neighbour joining tree for the set of 39 melon accessions along with reference genotypes. A, B, C, D highlight the four clusters associated with the accessions collected in southern India and cluster with broken line contains mostly momordica accessions collected in northern India. Accessions belonging to the acidulus group are underlined and accessions belonging to the momordica group are in bold and italic characters
There was a higher correlation (Prob = 0.005) between the state where the accessions were collected and the SSRs clusters: accessions from Tamil Nadu are much more frequent in B and C clusters. The correlation was also significant between the zones and the SSR clusters (Prob = 0.01). However, accessions from zone 8 or from Tamil Nadu were present in the four clusters; accessions from Kerala (zone 19) were present in all clusters but B and the 3 accessions which were not in the four clusters (AM 18, AM 67 and AM 71) were also collected in Kerala.
Melon landraces from eastern India (SM 27, SM 28, SM 29, SM 31, and SM 32) grouped separately. Similarly melon landraces from north-India (AHK 200, KP-7, Chibbar wild, Ra Chibbar, and MOM) were grouped separately from the south-Indian landraces (exceptions were AM 18 and AM 71). Likewise, other reference populations of Spanish, Japanese, Korean, French, Israeli and Iraqi origin were positioned separately from the acidulus and momordica accessions of south-Indian origin. Interestingly, clusters B, C and D did not contain any reference genotype. One accession from the Maldives Islands, which is quite close South-East from Kerala, was in cluster A along with one accession from Zambia which is more surprising. The FCA (Fig. 3) is consistent in many aspects with the NJ dendrogram, most of the collected accessions are plotted in the left part of the graph, well separated by the first axis from the rest of the Indian and worldwide reference accessions.
Correlations between traits
A first group of correlations was observed between Botanical groups, Fruit flesh colour, Carotenoids content, and the SSR groups: On the one hand the acidulus group with white flesh, lower carotenoids content, and belonging to the A and B SSR clusters; On the other hand the momordica group with orange flesh, higher carotenoids content, and belonging to the C and D SSR clusters.
There were highly significant correlations between the primary fruit skin colour, the secondary colour and the repartition of these colours: For instance green primary colour was always associated with yellow secondary colour; dark green secondary colour was always associated with light green primary colour; all the accessions with no secondary colour were orange or yellow, never green or dark green.
Significant positive correlations were observed between the P, K, Fe and Zn concentration. Accessions AM 4 and AM 25, both belonging to the acidulus group, had higher concentrations and AM 7 and AM 31, both belonging to the momordica group, had lower concentrations for the four elements. However, the differences between the acidulus and the momordica groups were slighly significative only for K content (Prob = 0.046). Significative correlations were observed between P, K and Fe content and the States where the accessions were collected with a higher concentration of K and Fe (Prob = 0.008 and 0.04 respectively) and a lower concentration of P (Prob = 0.004) in Kerala than in Tamil Nadu.
As expected a negative highly significative correlation was observed between Fruit weight and Fruit number.
Among the surprising correlations: Zn concentration in the fruit and the angular or round stem shape (0.28 and 0.52 mg/100 g respectively, Prob = 0.002); The plants with orange primary fruit skin color had a higher number of primary branches (5.4) than the plants with light green, green or yellow primary skin color (3.9, 3.6 or 3.9 respectively, Prob = 0.02); The fruit weight was significantly correlated with the Fe concentration in the fruit but the correlation was not significative for K, P and Zn concentrations.
Discussion
Cultivation of melon dates back to 2000 BC in India, where various types of melons viz. vars. momordica, acidulus, and C. melo var. flexuosus (L.) Naudin are cultivated (Pitrat et al. 2000). These are non-sweet and consumed as vegetables and are clearly separated from other botanical varieties for instance the sweet types var. cantalupensis Naudin and var. inodorus Jacquin in Europe and the Middle East (Stepansky et al. 1999), which suggests that these melons developed independently in India. Melon var. acidulus is endemic to tropical humid southern India (states of Kerala and Tamil Nadu). This taxon has not been subjected to genetic diversity analysis previously. We collected a set of 50 melon landraces from three agro-ecological regions of southern India (comprising six sub-regions) in the two states of southern India which are the genetic origin of acidulus melon.
The accessions belonging to the acidulus group are mainly characterized by a very firm white flesh and the absence of sugar and aroma. Various types of growth habits (2), stem shape (2), and leaf sizes (3) were found in the germplasm. Similarly, different kinds of fruit shapes (4), primary fruit skin colours (4), secondary fruit skin colours (5), fruit skin patterns (4), exist in acidulus melons. The accessions belonging to the momordica group are characterized by the splitting of the fruit at maturity, a mealy flesh and the absence of sugar and aroma. Farmers grow locally bred cultivars of these melons. These landraces have different periods of fruit maturity. We were informed that the farmers in different parts of these two states had practiced selection for this trait deliberately so as to extend the availability of produce in the market and avoid glut. Landraces with white-yellow fruit flesh colour were dominant in the acidulus group whereas ones with orange fruit flesh colour were more prevalent in the momordica group. This is a consumer driven trait and farmers have developed varieties accordingly. Carotenoids produce pigmentation which results in the range of yellows and oranges in the flesh colour. The expression of colour in the fruit flesh is conditioned by the particular carotenoid type and concentrations which are influenced by genetic and environmental factors. Similarly fruit shape is also governed by consumer preference and the majority of the south Indian consumers like elongated or oblate fruits. Eighty-four percent of the germplasm collected by us had this trait in the accessions. Also fruit size preference varied between different localities in these two states and farmers have developed melon cultivars with small, medium and large fruits. The range of ascorbic acid and carotenoids is wide in the collected melons. More germplasm should be surveyed for higher content of these two antioxidants.
Wide variation occurs in this germplasm for concentration of P, K, Fe, and Zn. These concentrations are significantly correlated and accessions AM 39, AM 25, AM 77 and AM 4 had the higher amounts of the total mineral concentrations (P + K + Fe + Zn) with respectively 313.3, 255.2, 204.3, 183.2 mg/100 g of fruit flesh. Cucumber is a main salad crop in India. It contains negligible amount of carotenoids and 17, 149 and 0.3 mg/100 g edible portion of P, K and Fe respectively (Wehner and Maynard 2003). Using vitamin and mineral content variability present in this germplasm, it is possible to develop mineral and vitamin dense melon cultivars containing higher P, K, Fe, Zn, ascorbic acid and carotenoids than available in cucumber. Both these cucurbits are used in salad. Similarly, the genetic variation for Mn (0.007–0.108 mg/100 g of fruit flesh) and Cu (0.004–0.074 mg/100 g of fruit flesh) was also observed in the collected germplasm (data not presented). Fe, Zn and vitamin A have been reported by the World Health Organization as limiting for human health. Adequate intake of carotenoids found in vegetable species has been reported to reduce vitamin A deficiency in humans (Goldman 2003). We are testing the melon lines with higher carotenoid and mineral trait expression across different environments. Microenvironment variation for minerals, particularly Zn has been reported (Pfeiffer and McClafferty 2007). Interestingly, accessions containing high amount of P (AM 71, AM 25), K (AM 39, AM 25), Fe (AM 67) and Zn (AM 4) and ascorbic acid (AM 70) originated in a single agro-ecological region of Kerala (No 19). Future explorations aimed at collecting landraces with even higher amounts of minerals and ascorbic acid should be carried out in this agro-ecological region. Landraces with the highest amount of ascorbic acid (AM 84) and carotenoid (AM 79, AM 72) were located in a single agro-ecological region of Tamil Nadu (8.1).
The sugar/acid ratio determines the fruit flavour. Sweet melons possess low titrable acidity (0.12–0.2%). Independent genetic control of sugar and acid accumulation in sweet melon has been demonstrated (Burger et al. 2003) and it is possible to combine high sugar and acidity in one single genotype. Our survey of south Indian melon indicates high genetic variability for acidity in this species. Accessions AM 8 (0.57%) and AM 52 (0.54%) appear to be good parents for the genetic improvement of this trait in sweet melon. Previously, we have also isolated high acidity in var. momordica germplasm (0.57–0.61%) from India (Dhillon et al. 2007). Burger found high acidity (0.51%) in var. flexuosus (cv. Faqquous), which is a landrace developed in Israel (Yosef Burger, personal communication). It would be interesting to compare the genetic control of acidity in vars. momordica, acidulus and flexuosus. Two accessions (AM 25 and AM 82) segregated for high resistance to CMV. This has been confirmed through artificial inoculation as well as by growing the germplasm at the second location under epidemic conditions (data not presented). At least seven QTLs with recessive gene action have been reported to control CMV resistance in melons (Dogimont et al. 2000). Recently, Essafi et al. (2008) demonstrated that a single recessive gene is responsible for the resistance to P9 and P104.82 CMV strains but not to strains M6 and TL. The genetic control of CMV resistance found in AM 25 and AM 82 should be investigated to assess if other genes are involved and could be cumulated with those already described. Resistance to A. gossypii was already known in Indian accessions, for instance in PI 414723 of the momordica group. Accessions AM 5 and AM 52 were resistant and AM 7, AM 78 and AM 86 were segregating for resistance. As the gene controlling aphid resistance has been cloned (Dogimont et al. 2008), it would be interesting to compare the sequences and phenotypes of the alleles in those accessions. Resistance to ZYMV in AM 87 should be compared with the resistance of PI 414723 (Pitrat and Lecoq 1984) and of IC 274007 and IC 274014 (Dhillon et al. 2007). Accessions AM 22 which exhibited resistance to race 3 and AM 67, AM 86 and AM 90 to race 5 of powdery mildew should also be compared with other sources of resistance to powdery mildew.
SSR analysis supports the separation of the south Indian germplasm compared to the worldwide reference accessions. The position of the collected accessions in both the NJ dendrogram and FCA plot suggests that this germplasm is lateral to the rest of the melon accessions. McCreight et al. (2004) also determined that melon germplasm from southern and eastern India might contain allelic diversity not presently available in the germplasm collections held in various global genebanks. Dhillon et al. (2007) observed that a collection of snapmelon landraces from north India had a central position in a similar FCA plot, suggesting that the collection could represent a central melon origin from where Oriental and Occidental melon germplasm was developed and this concept has been supported by Luan et al. (2008). However, the lateral position of the current collection suggest that this germplasm has had a minimum role in the generation of Oriental and Occidental germplasm, representing a large pool of genetic diversity that has not been yet exploited by Oriental/Occidental traditional farmers or modern breeders. A larger survey of both south Indian and worldwide reference germplasm would be necessary to confirm the last hypothesis. South Indian accessions of the acidulus or the momordica groups cannot be separated by the SSR analysis. But the accessions from northern India belonging to the momordica group (for instance MOM and SM 27 to SM 32) are separated in Figs. 2 and 3 from the accessions from southern India of the momordica group. Molecular markers are better indicators of the geographical origin than of the belonging to a botanical group.
Lastly this unique SSR analysis of a germplasm collection has provided the opportunity to further study the feasibility of developing genetically superior F1 hybrids using genetically distant accessions. Presently, there is no hybrid commercial cultivar of acidulus or momordica melon in India. As both are monoecious, the seed production of F1 hybrids should be quite easy (Robinson 1999). Fortunately, these landraces of melon have not been displaced by bred cultivars as commercial breeding activity in these types is almost absent. But before the active commercial breeding takes place, it is essential to conserve (ex situ) these landraces from southern India. Clearly, farmers and consumers desperately need improved varieties of acidulus or momordica melon, with better yields and nutritional value.
References
Akashi Y, Fukuda N, Wako T, Masuda M, Kato K (2002) Genetic variation and phylogenetic relationships in East and South Asian melons, Cucumis melo L., based on the analysis of five isozymes. Euphytica 125:385–396
Bajaj LK, Kaur G (1981) Spectrophotometric determination of l-ascorbic acid in vegetables and fruits. Analyst 106:117–120
Burger Y, Sáar U, Distelfeld A, Katzir N (2003) Development of sweet melon (Cucumis melo) genotypes combining high sucrose and organic acid content. J Am Soc Hortic Sci 128:537–540
Dhillon NPS, Ranjana R, Singh K, Eduardo I, Monforte AJ, Pitrat M, Dhillon NK, Singh PP (2007) Diversity among landraces of Indian snapmelon (Cucumis melo var. momordica). Genet Resour Crop Evol 54:1267–1283
Dogimont C, Lecomte L, Périn C, Thabuis A, Lecoq H, Pitrat M (2000) Identification of QTLs contributing to resistance to different strains of cucumber mosaic cucumovirus in melon. Acta Hortic 510:391–398
Dogimont C, Chovelon V, Tual S, Boissot N, Rittener V, Giovinazzo N, Bendahmane A (2008) Molecular diversity at the Vat/Pm-w resistance locus in melon. In: Cucurbitaceae 2008, IXth EUCARPIA meeting on Genetics and Breeding of Cucurbitaceae, (Pitrat M, ed), INRA, Avignon (FRA), pp 219–227
Doyle JJ, Doyle JL (1990) Isolation of DNA from fresh tissue. Focus 12:13–15
Essafi A, Diaz-Pendon JA, Moriones E, Monforte AJ, Garcia-Mas J, Martin-Hernandez AM (2008) Dissection of the oligogenic resistance to Cucumber Mosaic Virus in the melon accession PI 161375. Theor Appl Genet 118:275–284
Fernandez-Silva I, Eduardo I, Blanca J, Esteras C, Picó B, Nuez F, Arús P, Garcia-Mas J, Monforte AJ (2008) Bin mapping of genomic and EST-derived SSRs in melon (Cucumis melo L.). Theor Appl Genet 118:139–150
Garcia-Mas J, Oliver M, Gomez-Paniagua H, De Vicente MC (2000) Comparing AFLP, RAPD and RFLP markers for measuring genetic diversity in melon. Theor Appl Genet 101:860–864
Ghosh SP (1991) Agroclimatic zones specific research—Indian perspective under NARP. ICAR, Pusa
Goldman IL (2003) Recognition of fruit and vegetables as healthful: vitamins and phytonutrients. Horttechnology 13:252–258
Gupta PK (1999) Soil, plant, water and fertilizer analysis. Agrobios, India, pp 123–288
Katzir N, Danin-Poleg Y, Tzuri G, Karchi Z, Lavi U, Cregan PB (1996) Length polymorphism and homologies of microsatellites in several Cucurbitaceous species. Theor Appl Genet 93:1282–1290
Liu K, Muse SV (2005) Powermaker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Luan F, Delannay I, Staub JE (2008) Chinese melon (Cucumis melo L.) diversity analyses provide strategies for germplasm curation, genetic improvement, and evidentiary support of domestication patterns. Euphytica 164:445–461
McCreight JD, Staub JE, Koppar NM, Srivastava UC (1993) Indo-U.S. Cucumis germplasm expedition. Hortscience 28:492
McCreight JD, Staub JE, López-Sesé A, Sang-Min C (2004) Isozyme variation in Indian and Chinese melon (Cucumis melo L.) germplasm collections. J Am Soc Hortic Sci 129:811–818
Monforte AJ, Garcia-Mas J, Arús P (2003) Genetic variability in melon based on microsatellite variation. Plant Breed 122:153–157
Monforte AJ, Eduardo I, Abad S, Arús P (2005) Inheritance mode of fruit traits in melon-heterosis for fruit shape and its correlation with genetic distance. Euphytica 144:31–38
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol 19:153–170
Pangalo KJ (1929) Critical review of the main literature on the taxonomy, geography and origin of cultivated and partially wild melons. Trendy Prikl Bot 23:397–442 [In Russian, and translated into English for USDA by G. Saad in 1986]
Pfeiffer WH, McClafferty B (2007) Harvest plus: breeding crops for better nutrition. Crop Sci 47:S88–S105
Pitrat M, Lecoq H (1984) Inheritance of Zucchini yellow mosaic virus resistance in Cucumis melo L. Euphytica 33:57–61
Pitrat M, Risser G, Bertrand F, Blancard D, Lecoq H (1996) Evaluation of a melon collection for diseases resistances. In: Gómez-Guillamón ML, Soria C, Cuartero J, Torès JA, Fernandez-Munoz R (eds). Cucurbits toward 2000. VIth EUCARPIA meeting on Cucurbit Genetics and Breeding, Málaga (ES), pp 49–58
Pitrat M, Hanelt P, Hammer K (2000) Some comments on infraspecific classification of cultivars of melon. Acta Hortic 510:29–36
Renner SS, Schaefer H, Kocyan A (2007) Phylogenetics of Cucumis (Cucurbitaceae): Cucumber (C. sativus) belongs in an Asian/Australian clade far from melon (C. melo). BMC Evol Biol 7:58–69
Robinson RW (1999) Rationale and methods for producing hybrid cucurbit seed. J New Seeds 1:1–47
Robinson RW, Decker-Walters DS (1997) Cucurbits. CAB International, Oxon (GB), 226 pp
SAS Institute (2003) SAS 9.1 for Windows. SAS Institute Inc., Cary
Schaefer H, Heibl C, Renner SS (2009) Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc R Soc B 276:843–851
Sehgal JL, Mandal DK, Mandal C, Vadivelu S (1992) Agroecological regions of India. National Bureau of Soil Survey and Land use Planning (ICAR), Nagpur, India
Srivastava U, Mahajan RK, Gangopadhyay KK, Singh M, Dhillon BS (2001) Minimal descriptors of agri-horticultural crops. Part II: vegetable crops. National Bureau of Plant Genetic Resources, Pusa Campus, New Delhi, India, pp 260
Staub JE, McCreight JD (2004) Report of the Cucumis germplasm collection expedition in the state of Rajasthan, Madhya Pradesh and Uttar Pradesh, India, Oct–Nov. 1992. 5 May 2004. http://www.ars-grin.gov/npgs/egelist.html#Cucurbit
Steel R, Torrie JH (1980) Principles and procedures of statistics: a biometric approach, 2nd edn. New York, McGraw-Hill, p 176
Stepansky A, Kovalski I, Perl-Treves R (1999) Intraspecific classification of melons (Cucumis melo L.) in view of their phenotypic and molecular variation. Plant Syst Evol 217:313–332
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thomas P, Joshi MR (1977) Total carotenoids. Potato Res 20:78
Wehner TC, Maynard DN (2003) Cucumbers, melons and other cucurbits. In: Katz SH (ed) Encyclopedia of food and culture. Scribner & Sons, New York 2014 p
Acknowledgments
Balvir Kaur was supported by a Fellowship from Punjab Agricultural University. N.P.S. Dhillon was supported by a fellowship for sabbatical stays from the Spanish Ministerio de Ciencia e Innovacion (MCINN). The work was funded by grant AGL2006-12780-C02-01/AGR (MCINN). We are thankful to Fuensanta Garcia for technical support. The authors thank Professor G.J. Jellis for helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Fergany and Balvir Kaur contributed equally to this work and are considered the first authors.
Rights and permissions
About this article
Cite this article
Fergany, M., Kaur, B., Monforte, A.J. et al. Variation in melon (Cucumis melo) landraces adapted to the humid tropics of southern India. Genet Resour Crop Evol 58, 225–243 (2011). https://doi.org/10.1007/s10722-010-9564-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-010-9564-6