Abstract
Carnivores exhibit various fat contents and energy reserves to adapt to their environments. However, the molecular mechanisms underlying lipid metabolic differences among carnivores have not been well explored. Long-chain acyl-CoA synthetases (ACSLs) catalyze the initial step in lipid metabolism by activating fatty acids (FAs), and they drive acyl-CoAs toward anabolic lipid synthesis or catabolic β-oxidation. We identified the sequences of the genes of the ACSL family (ACSL1, ACSL3, ACSL4, ACSL5 and ACSL6) in the sable (Martes zibellina) via transcriptome sequencing. The ACSL gene sequences of 13 other carnivores were obtained from NCBI. Phylogenetic results showed that unlike the widely accepted carnivore phylogeny, Canidae and Felidae tend to group together based on ACSL4 and ACSL6. The evolutionary analyses identified a series of positively selected amino acid residues in ACSL1, ACSL4 and ACSL5. Two radical amino acid substitutions detected in sable suggested potential insights into the molecular mechanism underlying the relatively low fat content in this animal. This is the first study to investigate the molecular mechanisms underlying the adaptive evolution of fat metabolism in carnivores. Overall, the ACSL genes were under different evolutionary forces in carnivores, and some genes have undergone adaptive evolution in lipid metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carnivora is a diverse order of placental mammals that includes 280 species widely distributed in the world (Nowark 1999). During evolution, carnivores have developed diverse adaptabilities to distinct habitats, such as selva, steppe, Arctic and aquatic environments (Eisenberg 1989). The carnivores in these habitats exhibit different fat contents and energy reserves that are adapted to their habitats and their behavior. For example, pinnipeds, which are marine mammals, exhibit relatively thick subcutaneous fat that contributes to maintaining body temperature, energy storage and increased swimming efficiency (Pond 1978; Knutsen and Born 1994; Berta 2002; Budge et al. 2004). However, another marine mammal in Carnivora, the sea otter (Enhydra lutris), has developed other strategies, such as exceptionally thick fur and a high metabolic rate, to keep warm instead of thickened blubber (Thometz et al. 2014). Further, the American marten (Martes americana) and sable (Martes zibellina) maintain a lean body with a low body fat content, which may be helpful for hunting and other activities (Harlow 1994; Mustonen et al. 2006; Nieminen et al. 2007). Thus, carnivores can be used for the research on the adaptive evolution of fat metabolism and can contribute to illustrating the evolutionary process and the associated molecular mechanisms.
In mammals, acyl-CoA synthetases (ACSs) catalyze the initial step in lipid metabolism (Lopes-Marques et al. 2013). The fatty acids (FAs) are activated and converted to acyl-CoAs by these enzymes in an ATP-dependent pathway and then integrated into metabolic pathways such as β-oxidation and complex lipid biosynthesis (phospholipids, diacylglycerol and triacylglycerol) (Suzuki et al. 1990, 1995; Teodoro et al. 2017). Long-chain acyl-CoAs are formed by synthetases of the long-chain ACS (ACSL) family, which preferentially activate FAs with 12–20 carbons (C12–C20, the most abundant in the diet) and are then involved in a variety of metabolic pathways (Schneiter and Kohlwein 1997; Van Horn et al. 2005; Li et al. 2010). The ACSL family consists of five distinct isoforms, ACSL1, ACSL3, ACSL4, ACSL5, and ACSL6, and these proteins are characterized by varying subcellular localization, fatty acid substrate selectivity and tissue specificity (Soupene and Kuypers 2008; Rajkumar et al. 2016). It is suggested that the five ACSLs can be divided into two subfamilies depending on their substrates and their sequence similarity, with ACSL1, ACSL5, and ACSL6 constituting one family, and ACSL3 and ACSL4 constituting the other (Mashek et al. 2004; Watkins et al. 2007; Soupene and Kuypers 2008). ACSL1 is highly expressed in the liver, muscle, and adipose tissue, in which various fatty acids are used for energy production and storage (Suzuki et al. 1990). ACSL3 and ACSL6 were reported to be expressed primarily in the brain, testis and muscle (Fujino et al. 1996; Teodoro et al. 2017). ACSL4 is expressed abundantly in the adrenal gland and liver, and ACSL5 is primarily expressed in brown adipose tissue, small intestine, and liver (Glick and Rothman 1987; Kang et al. 1997; Oikawa et al. 1998; Mashek et al. 2006). Previous studies have reported that the ACSL isoforms are important regulators of whole-body energy metabolism and are implicated in triacylglycerol (TAG) synthesis and fat deposition, especially in the cases of ACSL1 and ACSL5 (Li et al. 2015; Bowman et al. 2016; Senkal et al. 2017).
Although the ACSL family has attracted much attention in various studies, its evolutionary history has seldom been investigated previously (Lopes-Marques et al. 2013). Positive selection has been detected in the ACSL1, ACSL5 and ACSL6 genes of cetaceans during their adaptation to the aquatic environment, which implied an association with enhanced TAG synthesis and thickened blubber (Wang et al. 2015). In polar bears, a fixed variant was found in ACSL6, which may be related to their fatty acid profile (Miller et al. 2012). Considering the variety of fat contents in carnivores and the pivotal role of ACSLs in lipid metabolism, in this study, we focused on the evolutionary patterns of the ACSL genes and explored their potential adaptive evolution in carnivores. Our study represents the first attempt to contextualize the adaption of lipid accumulation in carnivores.
Methods
Identification of the ACSL genes and phylogenetic analyses
The transcriptome sequencing of the subcutaneous fat and intestine of the sable (Martes zibellina) (unpublished data) was previously performed in our laboratory. A total of 103 million paired reads with a length of 150 bp were generated from transcriptome libraries. Clean reads (96.95%) were obtained after removal of adapters and low-quality reads and were then de novo assembled using Trinity software with default parameters (Grabherr et al. 2011). We identified the ACSL genes in generated high-quality sequence datasets and utilized the sequences to perform the phylogenetic and evolutionary analyses. The genome assemblies of sea otter (Enhydra lutris, ASM228890v2), lesser panda (Ailurus fulgens, ASM200746v1) and African wild dog (Lycaon pictus, LycPicSAfr1.0) were downloaded from the GenBank database in September 2017. ACSL gene sequences in these species were identified and annotated by comparing known dog sequences to genomic contigs using BLASTN and TBLASTN in BLAST v2.6.0 with an E-value cut-off of 1E-5 (Additional file S1) (Altschul et al. 1997). Meanwhile, from GenBank, we obtained the ACSL sequences of 10 carnivores, namely, the ferret (Mustela putorius furo), giant panda (Ailuropoda melanoleuca), polar bear (Ursus maritimus), walrus (Odobenus rosmarus divergens), Hawaiian monk seal (Neomonachus schauinslandi), Weddell seal (Leptonychotes weddellii), cat (Felis catus), leopard (Panthera pardus), Siberian tiger (Panthera tigris altaica) and dog (Canis lupus familiaris). The accession numbers and sequences of the ACSL family used in this study are presented in Additional file S1.
Multiple sequence alignment at the amino acid level was performed for ACSL family members using Multiple Sequence Comparison by Log-Expectation (MUSCLE) with the default settings (Edgar 2004) (Additional file S2). The phylogenetic relationships were reconstructed using the neighbor-joining (NJ) and maximum likelihood inference (ML) methods with 1000 bootstrap replicates in MEGA6.0, with all positions in each alignment considered for phylogenetic calculation. In these analyses, which were conducted with ModelGenerator (Keane et al. 2006), the Jones-Taylor-Thornton (JTT) + G (ACSL1, ACSL5) and JTT + I (ACSL3, ACSL4 and ACSL6) models were determined to be the best.
Tests of selection
Calculations of nonsynonymous/synonymous substitution ratios (ω = dN/dS) have been widely performed to evaluate the positive selection of protein-coding genes in molecular evolution studies (Ohta 1992). In this study, we first used site models implemented in the CODEML program in PAML version v4.8 (Yang 2007) to perform analyses of selective pressure at each codon position for ACSL genes. Two alternative models (M7 and M8) implemented in CODEML were used to conduct positive selection analysis on each ACSL gene, and the likelihood ratio test (LRT) with 2 degrees of freedom was then used to compare the nested models. Using the Bayes empirical Bayes (BEB) approach, the sites with a posterior probability > 0.9 were considered candidates for selection. Meanwhile, we analyzed the ACSL genes through three ML methods (SLAC, REL, and FEL) applied in the HyPhy package available from the Datamonkey server (Pond and Frost 2005; Pond et al. 2005), and the best fitting nucleotide substitution model was determined by the automatic model selection tool. The site with significance levels less than 0.1 for SLAC and FEL, and Bayes Factors larger than 50 for REL were considered to be under positive selection.
To detect the independent evidence for each branch among the carnivores, we used the branch-specific model to calculate the ω ratio for each branch and test whether the ω was significantly different among carnivores that inhabit different habitats. In the branch-specific models, a ‘free-ratio’ model that assumed separate ω ratios for all branches was run with the CODEML program in PAML version v4.8. We also used the branch-site model implemented in CODEML to detect the potential positive selection of ACSL genes. The species with less subcutaneous fat (Canidae, Felidae, and Mustelidae) and those with thick blubber (Pinnipedia and Ursidae) were considered as foreground branches to identify the signal of positive selection (Pond 1978; Knutsen and Born 1994; Budge et al. 2004; Mustonen et al. 2006; Thometz et al. 2014; Shero et al. 2015). To determine the amino acid changes at positively selected sites, the ancestral amino acid sequences of each ACSL were reconstructed via a Bayesian method with ANCESTOR (Zhang and Nei 1997).
Results
Phylogenetic analyses
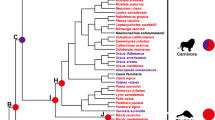
To investigate the phylogenetic relationship of ACSL genes within carnivores, phylogenetic trees were constructed with the NJ and ML methods. These analyses yielded similar topologies (Fig. 1, Additional files S3–S7), which were basically consistent with a widely accepted relationship among carnivores (Flynn et al. 2005; Yu et al. 2011). The phylogenetic analyses support Ailuridae as the only sister taxon to Mustelidae, although the clades within Mustelidae were different. The monophyly of Pinnipedia, Ursidae and Musteloidea was strongly supported; however, the trichotomy of these lineages remains unresolved. The animals in Caniformia tended to cluster together, but Canidae and Felidae were grouped in the analyses based on the ACSL4 and ACSL6 genes, with robust support (Fig. 1b), which was quite different from the widely accepted hypothesis of the carnivore phylogeny (Flynn et al. 2005). The phylogenetic trees of each ACSL are provided in Additional files S3–S7. We used one of the most widely accepted phylogenies as the working topology in the subsequent analyses.
Positive selection at carnivore ACSL genes
First, we used the site model incorporated in PAML to detect the signals of positive selection among the carnivores (Table 1, Additional file S8). The positively selected sites were detected in the carnivores’ ACSL4 and ACSL5, with significant LRT values (13.28 and 13.83, p < 0.01). There were 1 and 18 specific codons identified by the BEB approach with a posterior probability of 90% in ACSL4 and ACSL5, respectively. In the analyses with Datamonkey, 2 and 6 codons were detected by FEL and REL in ACSL4, and 9 and 20 codons were detected by FEL and REL in ACSL5, with 2 codons also confirmed by the SLAC method (Table 1). In addition, the FEL method reported positive selection in 5 codons of ACSL1, while 5 codons were found by REL in ACSL6. There was no positively selected site identified in ACSL3 by these four ML methods. When the branch-site model was used to detect the potential positively selected sites of ACSL genes on each branch, only two codons (219, 266) were identified in ACSL1 of the sable (Table 2, Additional file S9). The LRT p value of this branch was < 0.05, indicating that model A fit the data better than did model M1a. The BEB values of the positively selected sites were 0.598 and 0.902 (Table 2). These two sites showed radical amino acid changes in terms of size, polarity, or electric charge, which indicated that the amino acid substitutions might affect the function of this protein (Yampolsky and Stoltzfus 2005). In the branch-specific model analyses, we ran a “free-ratio” model to detect the independent ω ratio for each branch. The ω was significantly lower than 1 in all cases except for the branches including the seal (ω = 1.59), cat (ω = 1.36) and Panthera spp. (ω = 1.13) in ACSL5, with significant statistical support (p < 0.01) (Table 2), which indicated that with the exception of ACSL5, the ACSL family among these species had faced strong purifying selection (Additional file S10).
Discussion
Fat serves as the major form of energy storage in mammals and plays important roles in normal metabolic regulation (Wang et al. 2015). Lipid metabolism disorders and excessive accumulation of fat underlie the development of obesity and other related diseases (Ellis et al. 2010). The carnivores have significantly different fat contents. For example, the polar bear and seal develop relatively thick subcutaneous fat that comprises more than 30% of their body mass, which is far greater than the percentages of dissectible adipose tissue found in other wild carnivores (Pond 1978; Mustonen et al. 2006; Shero et al. 2015). ACSLs catalyze the initial step in lipid metabolism, and they are important regulators of whole-body energy metabolism (Lopes-Marques et al. 2013). Strong signals of positive selection detected in ACSL genes could provide novel insights into the molecular mechanism of lipid metabolic differences among the carnivores.
Phylogenetic analyses of the carnivores based on ACSL genes yielded topologies similar to those of previous studies, with the trichotomy of Pinnipedia, Ursidae and Musteloidea remaining unresolved (Flynn et al. 2005; Yu et al. 2011). Thus, the ACSL genes were highly conserved in evolution, both in carnivores with thick subcutaneous fat and in species with lean body types. Although belonging to the Caniformia, the canine (dog and African wild dog) clustered with felines instead of with other caniform carnivores in the phylogenetic tree based on ACSL4 and ACSL6, which was significantly different from the widely accepted phylogeny (Wyss and Flynn 1993). This distinction implied that these species have developed adaptations related to lipid metabolism in response to the environment during evolution.
In the present study, we used three models implemented in PAML and three ML methods from the Datamonkey server to test the potential positive selection of the ACSL gene family among the carnivores. Comparison of the ω values revealed distinct overall purifying selection for ACSL genes across the carnivores, and ACSL5 possessed the highest ω values. The most positively selected sites detected in ACSL5 showed stronger evidence of positive selection than was observed for the other genes, which indicated that this gene might be closely related to differences in lipid metabolism among the carnivores. Positive selection was also detected in the carnivores’ ACSL1 and ACSL4 genes. Evolutionary analyses of the carnivores’ ACSL genes revealed the absence of a consistent pattern of positive selection across the carnivore phylogeny.
In the ACSL1 gene, the branch-site model tests showed that two positively selected codons (219, 266) were determined in the lineage of sable. Although the ω value of this branch was less than 1 (ω = 0.20), the radical amino acid changes from a nonpolar Pro to a polar Ser and a small, nonpolar, and neutral Ala to a polar and positively charged Arg may affect the function of the protein. The three-dimensional (3D) structure of ACSL1 were predicted by the I-TASSER server (Additional file S11) (Zhang 2008), and it was reported that these sites were localized in the topological domain of ACSL1 (http://www.uniprot.org/). The function of ACSL1 in highly oxidative tissues, such as adipose, heart and skeletal muscle, is to activate FAs destined for β-oxidation (Ellis et al. 2010; Li et al. 2015). ACSL1 in brown adipose tissue metabolizes FAs for heat production to maintain a normal body temperature and degrades FAs for contractile energy in the heart (Ellis et al. 2010, 2011). Ellis et al. found that an adipose-specific knockout of ACSL1 displayed reduced adipose FA oxidation and marked cold intolerance (Ellis et al. 2010), and an ACSL1 deficiency in skeletal muscle showed a failure to switch to the use of FAs for oxidation, with enhanced glucose oxidation and muscle protein degradation, which ultimately impaired the capacity for endurance exercise (Li et al. 2015). In the liver, however, ACSL1 specifically directs FAs toward TAG synthesis and lipid storage (Wu et al. 2011). It has been reported that SNPs in the ACSL1 gene are associated with dysregulated fasting glucose and diabetes, most likely by disturbing fatty acid metabolism (Phillips et al. 2010; Manichaikul et al. 2016). The sable is a inhabitant of the Palearctic taiga with a low level of body fat (8.0%) (Mustonen et al. 2006). Sables may confront harshest weather conditions in winter, while subcutaneous fat may not provide thermal insulation, such as cetaceans, pinnipeds and polar bears (Pond 1978; Miller et al. 2012; Wang et al. 2015). The positive selection of the ACSL1 gene in sable may contribute to FA oxidation and increase the capacity for cold tolerance in this environment. Meanwhile, it is plausible that FAs destined for β-oxidation may decrease fat storage and result in a low level of body fat. The sable is an adroit, tireless, and strong predator that inhabits in dense coniferous taiga forests, flatlands, and mountainous areas, and the lean body shape may be helpful for the well-developed hunting skills and avoiding predators (Monakhov 2011). It was reported that a low body fat content in a congener of sable, American marten (Martes americana), is an adaptation to maintain the lean body shape required for hunting in the burrows of rodents (Harlow 1994). It seems that this is the common mechanism underlying adaptation in Martes. Another explanation for the low body fat content is that the limited capacity of the small gastric ventricle makes it difficult to consume large amounts of food, and the high-protein diet and energetic costs of foraging may also decrease fat storage (Nieminen et al. 2006). In addition, ACSL1 is also expressed in macrophages and plays a functionally distinct role in the innate immune response (Rubinow et al. 2013).
Another strong signature of positive selection was detected along the branches, including those of the seal, cat and Panthera (leopard and tiger) in the ACSL5 gene. Although there was no positively selected codon identified, the ω values of these branches were greater than 1, suggesting the accelerated evolution of the ACSL5 gene in these species. ACSL5 is an important regulator in fatty acid channeling between anabolic lipid synthesis and the catabolic β-oxidation pathway. Overexpression of ACSL5 in hepatoma cells primarily increases the conversion of fatty acids to triacylglycerol, while knockdown of ACSL5 in isolated rat hepatocytes reduces triglyceride accumulation and increases fat oxidation (Mashek et al. 2006; Bu and Mashek 2010). Ablation of ACSL5 in mice also displayed increased energy expenditure, reduced fasting glucose, serum triglyceride and adiposity (Bowman et al. 2016). In addition, previous studies have reported that a SNP in the ACSL5 gene resulting in elevated transcription of ACSL5 in skeletal muscle was associated with more rapid diet-induced weight loss, most likely by increasing fat oxidation (Teng et al. 2009; Rajkumar et al. 2016). In pinnipeds, the subcutaneous fat known as blubber is particularly thick and serves as both an energy reserve and thermal insulation (Berta 2002), and seals (lipid content > 30%) reportedly have more fat than walruses of similar size (Pond 1978; Knutsen and Born 1994; Shero et al. 2015). The positive selection on the branch of the seal may contribute to the thickness of the blubber by activating FAs destined for lipid synthesis. The ACSL5 was also determined to have undergone positive selection in cetaceans, which suggests that cetaceans possess the ability to enhance triacylglycerol synthesis during their adaptation to a fully aquatic life (Wang et al. 2015). It has been reported that members of Felidae do not deposit significant quantities of subcutaneous fat (Pond 1978), and the ACSL5 of cat and Panthera may have the completely opposite function of driving the ACSL products toward lipid β-oxidation.
Some positively selected sites were detected in ACSL genes, with codon 133 in ACSL4 and codons 41, 60, 68, 74, 80, 292, 370 and 498 in ACSL5 showing the relatively strong evidence of selection because they were detected by multiple ML methods. ACSL4 preferentially utilized arachidonic acid and eicosapentaenoic acid as substrates and was abundant in steroidogenic tissues (Kang et al. 1997). Previous studies have reported that polymorphisms of the ACSL4 gene were significantly associated with liver and intramuscular fat content (Rusc et al. 2011; Corominas et al. 2012). These positively selected sites were spread across the lineages among the carnivores, which suggested that these species have developed different adaptations of lipid metabolism. Furthermore, there was no evidence of positive selection in the ACSL3 gene among the carnivores, and for ACSL6, the positively selected sites were identified only by the REL method, which indicated that ACSL3 and ACSL6 faced strong purifying selection and that the carnivores’ ACSL genes were under different evolutionary forces. ACSL3 has been reported to mediate hepatic lipogenesis through transcriptional regulation of lipogenic gene expression (Bu et al. 2009). ACSL6 has been reported to drive ACSL products toward lipid synthesis and storage in skeletal muscle and its mRNA level is modulated by nutritional status (Teodoro et al. 2017). Downregulation of ACSL6 could improve lipid degradation and fatty acid oxidation through the AMPK/PGC1-α pathway (Teodoro et al. 2017). These two genes likely contribute little to the functional differences in lipid metabolism among carnivores.
Conclusion
In this study, our data suggested that the carnivore ACSL1, ACSL4 and ACSL5 genes have undergone adaptive evolution may related to lipid metabolism and fat deposition in carnivores. Two radical amino acid changes in ACSL1 provide new insights into the molecular mechanism underlying the relatively low fat content in the sable. Positive selection along two lineages suggested that ACSL5 may contribute to FAs anabolism in seals and to FAs catabolism in cat and Panthera. In addition, the positively selected sites implied that carnivores have developed diverse adaptations related to lipid metabolism. Nonetheless, additional genes involved in lipid metabolism and more species of carnivores should be investigated to discover the molecular mechanisms underlying the adaptive evolution of fat metabolism.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Berta A (2002) Pinnipedia, overview. Academic Press, San Diego, pp 903–911
Bowman TA, O’Keeffe KR, D’Aquila T, Yan QW, Griffin JD, Killion EA, Salter DM, Mashek DG, Buhman KK, Greenberg AS (2016) Acyl CoA synthetase 5 (ACSL5) ablation in mice increases energy expenditure and insulin sensitivity and delays fat absorption. Mol Metab 5:210–220. https://doi.org/10.1016/j.molmet.2016.01.001
Bu SY, Mashek DG (2010) Hepatic long-chain acyl-CoA synthetase 5 mediates fatty acid channeling between anabolic and catabolic pathways. J Lipid Res 51:3270–3280. https://doi.org/10.1194/jlr.M009407
Bu SY, Mashek MT, Mashek DG (2009) Suppression of long chain acyl-CoA synthetase 3 decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. J Biol Chem 284:30474–30483. https://doi.org/10.1074/jbc.M109.036665
Budge SM, Cooper MH, Iverson SJ (2004) Demonstration of the deposition and modification of dietary fatty acids in pinniped blubber using radiolabelled precursors. PBZ 77:682–687. https://doi.org/10.1086/420945
Corominas J, Ramayo-Caldas Y, Castello A, Munoz M, Ibanez-Escriche N, Folch JM, Ballester M (2012) Evaluation of the porcine ACSL4 gene as a candidate gene for meat quality traits in pigs. Anim Genet 43:714–720. https://doi.org/10.1111/j.1365-2052.2012.02335.x
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Eisenberg JF (1989) An introduction to the Carnivora. In: Gittleman JL (ed) Carnivore behavior, ecology, and evolution. Cornel Univ. Press, Ithaca
Ellis JM, Li LO, Wu PC, Koves TR, Ilkayeva O, Stevens RD, Watkins SM, Muoio DM, Coleman RA (2010) Adipose Acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab 12:53–64. https://doi.org/10.1016/j.cmet.2010.05.012
Ellis JM, Mentock SM, Depetrillo MA, Koves TR, Sen S, Watkins SM, Muoio DM, Cline GW, Taegtmeyer H, Shulman GI, Willis MS, Coleman RA (2011) Mouse cardiac acyl coenzyme a synthetase 1 deficiency impairs Fatty Acid oxidation and induces cardiac hypertrophy. Mol Cell Biol 31:1252–1262. https://doi.org/10.1128/MCB.01085-10
Flynn JJ, Finarelli JA, Zehr S, Hsu J, Nedbal MA (2005) Molecular phylogeny of the carnivora (mammalia): assessing the impact of increased sampling on resolving enigmatic relationships. Syst Biol 54:317–337. https://doi.org/10.1080/10635150590923326
Fujino T, Kang MJ, Suzuki H, Iijima H, Yamamoto T (1996) Molecular characterization and expression of rat acyl-CoA synthetase 3. J Biol Chem 271:16748–16752
Glick BS, Rothman JE (1987) Possible role for fatty acyl-coenzyme A in intracellular protein transport. Nature 326:309–312. https://doi.org/10.1038/326309a0
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Harlow HJ (1994) Trade-offs associated with the size and shape of American martens. Cornell University Press, Ithaca
Kang MJ, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto TT (1997) A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc Natl Acad Sci 94:2880–2884
Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6:29
Knutsen L, Born EW (1994) Body growth in atlantic walrus (Odobenus rosmarus rosmams) from greenland. J Zool 234:371–385
Li LO, Klett EL, Coleman RA (2010) Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochim Biophys Acta 1801:246–251. https://doi.org/10.1016/j.bbalip.2009.09.024
Li LO, Grevengoed TJ, Paul DS, Ilkayeva O, Koves TR, Pascual F, Newgard CB, Muoio DM, Coleman RA (2015) Compartmentalized acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis. Diabetes 64:23–35. https://doi.org/10.2337/db13-1070
Lopes-Marques M, Cunha I, Reis-Henriques MA, Santos MM, Castro LF (2013) Diversity and history of the long-chain acyl-CoA synthetase (Acsl) gene family in vertebrates. BMC Evolut Biol 13:271. https://doi.org/10.1186/1471-2148-13-271
Manichaikul A, Wang XQ, Zhao W, Wojczynski MK, Siebenthall K, Stamatoyannopoulos JA, Saleheen D, Borecki IB, Reilly MP, Rich SS, Bornfeldt KE (2016) Genetic association of long-chain acyl-CoA synthetase 1 variants with fasting glucose, diabetes, and subclinical atherosclerosis. J Lipid Res 57:433–442. https://doi.org/10.1194/jlr.M064592
Mashek DG, Bornfeldt KE, Coleman RA, Berger J, Bernlohr DA, Black P, DiRusso CC, Farber SA, Guo W, Hashimoto N, Khodiyar V, Kuypers FA, Maltais LJ, Nebert DW, Renieri A, Schaffer JE, Stahl A, Watkins PA, Vasiliou V, Yamamoto TT (2004) Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J Lipid Res 45:1958–1961. https://doi.org/10.1194/jlr.E400002-JLR200
Mashek DG, Li LO, Coleman RA (2006) Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J Lipid Res 47:2004–2010. https://doi.org/10.1194/jlr.M600150-JLR200
Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE, Tomsho LP, Ibarra-Laclette E, Herrera-Estrella L, Peacock E, Farley S, Sage GK, Rode K, Obbard M, Montiel R, Bachmann L, Ingolfsson O, Aars J, Mailund T, Wiig O, Talbot SL, Lindqvist C (2012) Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci USA 109:E2382–E2390. https://doi.org/10.1073/pnas.1210506109
Monakhov VG (2011) Martes zibellina (Carnivora: Mustelidae). Mammal Species 43:75–86
Mustonen AM, Puukka M, Saarela S, Paakkonen T, Aho J, Nieminen P (2006) Adaptations to fasting in a terrestrial mustelid, the sable (Martes zibellina). Comparative biochemistry and physiology. Part A. Mol Integr Physiol 144:444–450. https://doi.org/10.1016/j.cbpa.2006.03.008
Nieminen P, Rouvinen-Watt K, Collinsb D, Grant J, Mustonen AM (2006) Fatty acid profiles and relative mobilization during fasting in adipose tissue depots of the American marten (Martes americana). Lipids 41:231–240
Nieminen P, Rouvinen-Watt K, Saarela S, Mustonen AM (2007) Fasting in the American marten (Martes americana): a physiological model of the adaptations of a lean-bodied animal. J Compar Physiol B Biochem Syst Environ Physiol 177:787–795. https://doi.org/10.1007/s00360-007-0175-2
Nowark R (1999) Walker’s mammals of the world. Johns Hopkins Univ. Press, Baltimore, pp 632–793
Ohta T (1992) The nearly neutral theory of molecular evolution. Ann Rev Ecol Syst 23:263–286
Oikawa E, Iijima H, Suzuki T, Sasano H, Sato H, Kamataki A, Nagura H, Kang MJ, Fujino T, Suzuki H, Yamamoto TT (1998) A novel acyl-CoA synthetase, ACS5, expressed in intestinal epithelial cells and proliferating preadipocytes. J Biochem 124:679–685
Phillips CM, Goumidi L, Bertrais S, Field MR, Cupples LA, Ordovas JM, Defoort C, Lovegrove JA, Drevon CA, Gibney MJ, Blaak EE, Kiec-Wilk B, Karlstrom B, Lopez-Miranda J, McManus R, Hercberg S, Lairon D, Planells R, Roche HM (2010) Gene-nutrient interactions with dietary fat modulate the association between genetic variation of the ACSL1 gene and metabolic syndrome. J Lipid Res 51:1793–1800. https://doi.org/10.1194/jlr.M003046
Pond CM (1978) Morphological aspects and the ecological and mechanical consequences of fat deposition in wild vertebrates. Annu Rev Ecol Evol Syst 9:519–570
Pond SL, Frost SD (2005) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. https://doi.org/10.1093/bioinformatics/bti320
Pond SL, Frost SD, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. https://doi.org/10.1093/bioinformatics/bti079
Rajkumar A, Lamothe G, Bolongo P, Harper ME, Adamo K, Doucet E, Rabasa-Lhoret R, Prud’homme D, Tesson F (2016) Acyl-CoA synthetase long-chain 5 genotype is associated with body composition changes in response to lifestyle interventions in postmenopausal women with overweight and obesity: a genetic association study on cohorts Montreal-Ottawa New Emerging Team, and Complications Associated with Obesity. BMC Med Genet 17:56. https://doi.org/10.1186/s12881-016-0320-4
Rubinow KB, Wall VZ, Nelson J, Mar D, Bomsztyk K, Askari B, Lai MA, Smith KD, Han MS, Vivekanandan-Giri A, Pennathur S, Albert CJ, Ford DA, Davis RJ, Bornfeldt KE (2013) Acyl-CoA synthetase 1 is induced by gram-negative bacteria and lipopolysaccharide and is required for phospholipid turnover in stimulated macrophages. J Biol Chem 288:9957–9970. https://doi.org/10.1074/jbc.M113.458372
Rusc A, Sieczkowska H, Krzecio E, Antosik K, Zybert A, Kocwin-Podsiadla M, Kaminski S (2011) The association between acyl-CoA synthetase (ACSL4) polymorphism and intramuscular fat content in (Landrace x Yorkshire) x Duroc pigs. Meat Sci 89:440–443. https://doi.org/10.1016/j.meatsci.2011.05.008
Schneiter R, Kohlwein SD (1997) Organelle structure, function, and inheritance in yeast: a role for fatty acid synthesis? Cell 88:431–434
Senkal CE, Salama MF, Snider AJ, Allopenna JJ, Rana NA, Koller A, Hannun YA, Obeid LM (2017) Ceramide is metabolized to acylceramide and stored in lipid droplets. Cell Metab 25:686–697. https://doi.org/10.1016/j.cmet.2017.02.010
Shero MR, Costa DP, Burns JM (2015) Scaling matters: incorporating body composition into Weddell seal seasonal oxygen store comparisons reveals maintenance of aerobic capacities. J Compar Physiol B Biochem Syst Environ Physiol 185:811–824. https://doi.org/10.1007/s00360-015-0922-8
Soupene E, Kuypers FA (2008) Mammalian long-chain acyl-CoA synthetases. Exp Biol Med 233:507–521. https://doi.org/10.3181/0710-MR-287
Suzuki H, Kawarabayasi Y, Kondo J, Abe T, Nishikawa K, Kimura S, Hashimoto T, Yamamoto T (1990) Structure and regulation of rat long-chain acyl-CoA synthetase. J Biol Chem 265:8681–8685
Suzuki H, Watanabe M, Fujino T, Yamamoto T (1995) Multiple promoters in rat acyl-CoA synthetase gene mediate differential expression of multiple transcripts with 5′-end heterogeneity. J Biol Chem 270:9676–9682
Teng AC, Adamo K, Tesson F, Stewart AF (2009) Functional characterization of a promoter polymorphism that drives ACSL5 gene expression in skeletal muscle and associates with diet-induced weight loss. FASEB J 23:1705–1709. https://doi.org/10.1096/fj.08-120998
Teodoro BG, Sampaio IH, Bomfim LH, Queiroz AL, Silveira LR, Souza AO, Fernandes AM, Eberlin MN, Huang TY, Zheng D, Neufer PD, Cortright RN, Alberici LC (2017) Long-chain acyl-CoA synthetase 6 regulates lipid synthesis and mitochondrial oxidative capacity in human and rat skeletal muscle. J Physiol 595:677–693. https://doi.org/10.1113/JP272962
Thometz NM, Tinker MT, Staedler MM, Mayer KA, Williams TM (2014) Energetic demands of immature sea otters from birth to weaning: implications for maternal costs, reproductive behavior and population-level trends. J Exp Biol 217:2053–2061. https://doi.org/10.1242/jeb.099739
Van Horn CG, Caviglia JM, Li LO, Wang S, Granger DA, Coleman RA (2005) Characterization of recombinant long-chain rat acyl-CoA synthetase isoforms 3 and 6: identification of a novel variant of isoform 6. Biochemistry 44:1635–1642. https://doi.org/10.1021/bi047721l
Wang Z, Chen Z, Xu S, Ren W, Zhou K, Yang G (2015) ‘Obesity’ is healthy for cetaceans? Evidence from pervasive positive selection in genes related to triacylglycerol metabolism. Sci Rep 5:14187. https://doi.org/10.1038/srep14187
Watkins PA, Maiguel D, Jia Z, Pevsner J (2007) Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J Lipid Res 48:2736–2750. https://doi.org/10.1194/jlr.M700378-JLR200
Wu M, Cao A, Dong B, Liu J (2011) Reduction of serum free fatty acids and triglycerides by liver-targeted expression of long chain acyl-CoA synthetase 3. Int J Mol Med 27:655–662. https://doi.org/10.3892/ijmm.2011.624
Wyss AR, Flynn JJ (1993) A phylogenetic analysis and defnition of the Carnivora. In: Szalay FS, Novacek M, McKenna M (eds) Springer, New York, pp 32–52
Yampolsky LY, Stoltzfus A (2005) The exchangeability of amino acids in proteins. Genetics 170:1459–1472. https://doi.org/10.1534/genetics.104.039107
Yang ZH (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. https://doi.org/10.1093/molbev/msm088
Yu L, Luan PT, Jin W, Ryder OA, Chemnick LG, Davis HA, Zhang YP (2011) Phylogenetic utility of nuclear introns in interfamilial relationships of Caniformia (order Carnivora). Syst Biol 60:175–187. https://doi.org/10.1093/sysbio/syq090
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bionf 9:40
Zhang J, Nei M (1997) Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol 44(Suppl 1):S139–S146
Acknowledgements
This work was supported by Special Fund for Forest Scientific Research in the Public Welfare (201404420), National Natural Science Fund of China (31372220, 31672313).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2019_57_MOESM1_ESM.doc
Additional file S1. Table S1 Accession numbers and sources of the retrieved ACSL sequences for the species included in this study (DOC 143 KB)





Additional file S10. Table S4 Likelihood ratio tests of branch models for the ACSL genes (XLS 42 KB)
10709_2019_57_MOESM11_ESM.png
Additional file S11. Spatial distribution of positively selected sites in the three-dimensional (3D) structure of ACSL1 (PNG 224 KB)
Rights and permissions
About this article
Cite this article
Zhao, C., Liu, G., Shang, S. et al. Adaptive evolution of the ACSL gene family in Carnivora. Genetica 147, 141–148 (2019). https://doi.org/10.1007/s10709-019-00057-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-019-00057-3