Abstract
The sable (Martes zibellina) is widely distributed in the northern zone of Eurasia. M. zibellina survival requires a better ability to regulate its body temperature than southern-distributed marten species. The genetic basis underlying the adaptive fat mechanism in the sable is unclear. We screened candidate genes related to lipid metabolism and absorption and compared these with orthologous sequences from other carnivores. Our goal was to determine the molecular mechanism underlying the thinner fur layer and determine if the cold adaptation of sables is associated with evolutionary changes in these genes. We found that the PNLIP and ACSS2 genes have received positive selection and the GOT2 gene has undergone rapid evolution. We performed comparative transcriptome sequencing of the sable adipose tissue and found multiple genes with different expression levels. GO enrichment analysis on the differentially expressed genes showed that they were significantly enriched in GO entries, such as lipid metabolism, lipase activity, and glucose metabolism. KEGG enrichment analysis revealed that these genes were significantly enriched in metabolic pathways and fatty acid metabolism pathways, indicating significant differences in the metabolic level of adipose tissue in winter and summer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The sable (Martes zibellina) is widespread in the taiga zones of Eurasia, mainly in Russia, northern China and Mongolia, the Korean Peninsula, and the Hokkaido Island of Japan (Harrison et al. 2004). The sable population has decreased in Northeast China since the 1950s due to habitat loss and hunting (Kashtanov et al. 2015), and the sable is now listed as a national first-class protected animal in China. The sable is a omnivorous mammal, with a slender body that has a high surface-to-volume ratio. The sable’s body fat rate remains at about 8% even during summer and winter (Mustonen and Nieminen2006), and this seems beneficial for their hunting activities. However, sable habitats are cold, with an annual average temperature below 0 °C and minimum temperatures often below − 30 °C. These temperatures pose a challenge to survival. Sables have evolved a balanced fat mechanism to reduce the fat layer and to provide energy for the maintenance of body temperature. However, in contrast to the polar bear (Miller et al. 2012) and cetaceans (Dunkin et al. 2005), the subcutaneous fat of sables may not always provide adequate thermal insulation for resisting cold temperatures.
Animals have undergone selection pressures to adapt to food shortages and cold environments. This has driven many physiological and genetic adaptations (Li et al. 2014). Mammal exposure to cold climates has resulted in physiological adaptations that can change energy balance. For example, heat production by non-shivering thermogenesis (NST) leads to an increase in energy expenditure (Rawls2015). Pikas have evolved a high NST and resting metabolic rate (Sheafor 2003). Moreover, the expression of genes encoding proteins participating in glucose uptake, lipogenesis, and fatty acid combustion can be elevated, which plays a role in the adaptive thermogenic processes of cold-exposed mice (Yu et al. 2002). The Inuit inhabitants of Greenland live in a frigid Arctic environment and consume a diet rich in protein and fatty acids. The most prominent allele-frequency distinction between Inuit and other populations was located in FADS1, FADS2, and FADS3, a cluster related to fatty acid desaturases, which might contribute to Inuit adaptation to the cold climate (Fumagalli et al. 2015). Indigenous Siberians also live in an extremely cold environment. They have a low serum lipid level and an enhanced basal metabolic rate, and they have undergone missense mutations in three genes (ANGPTL8, PLIN1, and PLA2G2A) that participate in lipid metabolism. These changes might aid in adaptation to the local environment (Hallmark et al. 2019). Similarly, a polygenic selection scan of Siberians found three gene sets related to fat metabolism, which is consistent with adaptation to a fat-rich diet (PingHsun et al. 2017). Kumar et al. (2015) found that fat metabolism might play a role in the transformation of Arctic foxes (Vulpes lagopus) to freezing environments.
Animal fat deposition is a complex biological process involving regulation by many genes and transcriptional factors (William et al. 2011). There are two types of adipose tissues, brown adipose tissue (BAT) and white adipose tissue (WAT), which allow mammals to meet energy demands. BAT is specialized for heat production by metabolizing lipids via the adaptive thermogenesis process. This is an essential part of energy consumption caused by diet or cold exposure (Himms-Hagen 1990). WAT is specially engaged in energy storage and satisfies body energy demands during periods of energy deficit. This is an adaptive mechanism for food shortage requirements (Langin 2010). The essential function of adipose tissue is no longer viewed merely as a passive energy-storing tissue but is an endocrine organ with a prominent role in the metabolism of the entire body and energy balance (Vernon et al. 2001; Wozniak et al. 2009).
We studied the relationship between lipid metabolism and the body shape and cold adaptation of the sable. Coding sequences of the candidate genes involved in lipid metabolism were examined to determine the evolutionary patterns and their association with the slim body and cold adaptations of sables. We also performed comparative transcriptome sequencing of sable adipose tissue between winter and summer to evaluate the different expression levels of lipid metabolism genes. Our results reveal the molecular mechanisms underlying sable fat layer thinning during the cold acclimatization process (Table 1).
Materials and methods
Gene retrieval and alignments
The list of genes related to the human lipid metabolism pathway was retrieved from the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (https://www.kegg.jp/). There were 81 orthologous genes associated with triacylglycerol (TAG) and fat digestion and absorption. The gene list was used to find the gene IDs and search annotated coding sequences (CDS) for carnivores in the NCBI database (Table S1, http://www.ncbi.nlm.nih.gov). We also used a BLAST search to determine one-to-one orthologs genes in the genomes of 21 carnivore species to obtain a carnivore gene set associated with lipid metabolism. We sequenced the genome of the sable, and the genomes of remaining carnivores were downloaded from the NCBI database. PRANK v.170427, which has low false-positive rates compared to other alignment programs like MUSCLE and MAFFT (Fletcher and Yang 2010), was performed to align the candidate genes’ nucleotide sequences at the codon level. Codons with any ambiguity sites in these carnivore species genes were removed from the following analyses.
Molecular evolution analyses
The impact of natural selection on lipid metabolism-related genes was evaluated by calculating the ratio of nonsynonymous (dN) to synonymous (dS) substitutions (ω = dN/dS) with the PAML software package (Yang 2007). Briefly, ω > 1, ω = 1, ω < 1 correspond to positive selection, neutral selection, and negative (purifying) selection, respectively. First, we evaluated the positively selected sites (PSSs) acting on lipid metabolism-related genes from carnivores using the site model (M8 vs. M7). M8 presumes a beta-distribution for the ω range from 0 to 1 and permits sites with ω > 1, while M7 tests for neutral evolution (Swanson et al. 2003). To evaluate whether the possibility of positive selection was confined to a specific lineage, we compared the free-ratio model (diverse ω ratio for each lineage) with the one-ratio model (same ω ratio for all lineages) among the carnivores. We next implemented the branch model (M2) compared with the one-ratio model (M1) to study selection along the specific lineage of the sable. The branch-site model (Ma) assumes sites are under positive selection along specific foreground lineages with ω > 1, and the null model (Ma0) fixed ω = 1 was set as the compared model. Likelihood ratio tests (LRTs) with chi-squared test were executed on nested likelihood models. Sites determined by the site model and branch-site model were evaluated by Bayes empirical Bayes (BEB) analysis, with posterior probabilities of > 0.80. The species tree of carnivores downloaded from TimeTree (http://www.timetree.org/) was used as the guide tree in all molecular evolution analyses.
RNA-sequencing of adipose tissues in sables
Since the sable is critically endangered in China, adipose tissue samples were harvested from only two individuals. The winter adipose tissue sample was obtained from a natural death individual, while the summer sample was taken from a dead individual while wearing a collar. Sample collection was carried out according to the ethical standards of the Qufu Normal University Animal Care and Use Committee (Permit Number: QFNU2018-036). The fresh adipose tissues were placed in RNAprotect Animal Tissue Tubes (QIAGEN, USA), frozen in liquid nitrogen, and then stored at − 80 °C, before further extraction procedures. Total RNA was extracted using the RNAeasy Mini Kit (QIAGEN, USA) following the manufacturer instructions. An RNA 6000 Nano Assay Kit with the Bioanalyzer 2100 (Agilent Technologies, USA) and the Nanodrop 2000c (Thermo Scientific, USA) was applied to determine the integrity and concentration of RNA, respectively. The Illumina Tru-Seq RAN Low Sample (LS) protocol was used to generate the transcriptome libraries. Sequencing of the libraries was done using the Illumina HiSeq 2000 platform, generating 125-bp paired-end reads. Adapter sequences and low-quality reads were removed to retrieve clean transcriptome sequences and then the quality of the sequences was examined by the FASTQC program.
Analysis of differentially expressed genes and functional enrichment
The HTseq package was used to count the reads numbers mapped to each transcript. The mapped reads were normalized for RNA length to quantify transcripts expression by using the parameter Fragment Per Kilobase of exon per Million fragments mapped (FPKM) based on the mapped transcript fragment, length, and sequence depth. Differentially expressed genes (DEGs) analysis of samples from the two seasons was implemented using DEseq package (Anders and Huber 2010), using a negative binomial distribution-based model to confirm differential expression level in the digital gene expression data. Genes with a q-value < 0.05 and |log2 (FoldChange) > 1 found by DESeq were considered as differentially expressed. Functional Go ontology (GO) enrichment analysis of the DEGs was accomplished by applying GOseq based on Wallenius non-central hypergeometric distribution. KEGG pathway enrichment analysis was applied to detect metabolism pathways significantly enriched in DEGs using the KOBAS 2.0 software (Xie et al. 2011). After adjustment, only the results with corrected p-value < 0.05 were determined as significantly enriched in the DEGs.
Results
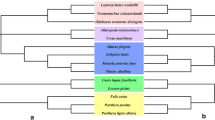
We retrieved a total of 81 lipid metabolism-related genes from the genome of 21 carnivore species. For the purpose of determining the possible roles of positive selection during the evolution of lipid metabolism-related genes in carnivore lineages, evolutionary analyses were first performed using the site model in the PAML program. Site model analysis of all carnivores showed that the M8 model fit the sequences significantly better than the M7 model for ABHD5, DGKE, DGKH, GPAM, GPAT2, GPAT4, PLCB4, PLCD4, PLIN5, PPARD, SLC27A1, SLC27A2, SREBF1, ABCG5, ABCG8, ACSS1, AGPAT2, APOB, DGAT1, PNLIP, and PNLIPRP2 lipid metabolism-related genes. Several PSSs were found with significant levels (Table 1). In contrast, no significant evidence of positive selection was found in the remaining genes (Table S2). The free-ratio model was used to gain insight into the positive selection for specific lineages in carnivores. LRT indicated that the free-ratio model fitted the data better than the one-ratio model at genes (i.e., ACSS2, ACSS3, AGPAT5, CD36, DGKA, DGKG, DGKI, FABP5, GNPAT, GOT2,GPAT4, MOGAT1, PDE3B, PLCD3, PLIN2, PNPLA2, SLC27A2, SLC27A3, SLC27A4, SREBF1, SREBF2, ABCG5,APOB, DGAT1, GAT 2, LIPF, NPC1L1, PLA2G12A, PLPP1, and PNLIPRP2). Specifically, evidence of positive selection was defined along the terminal branch of Vulpes lagopus and the lineage to the last common ancestor (LCA) of Leptonychotes weddellii and Neomonachus schauinslandi at LIPF. We detected positive selection evidence along the terminal branch of Ursus maritimus at APOB and GPAT4. We mapped the signals under positive selection in the carnivore phylogeny and found that positively selected signals were spread across a diverse phylogenetic tree, from the ancestral to the terminal branches of carnivores (Fig. 1). To further detect whether sable was subjected to sable during evolution, we applied the branch model and branch-site model in the analyses. The branch model analysis was set as a two-ratio model (M2), which allows a different ω value between the sable (foreground) and the remaining background lineages. We found that the M2 model fitted the data better than the M1 model at seven genes (DGKQ, GOT2, GPAT4, PLCB3, PLCD4, AGPAT2, and PNLIPRP2). Specifically, the evolutionary rates of GOT2 (ω1 = 0.05023; ω2 = 0.17427) and ACSS2 (ω1 = 0.09805; ω2 = 0.42843) in the sable were almost three times as high as those of the remaining background lineages (Table 2). The elevated ω estimated for GOT2 and ACSS2 genes relative to other branches indicates the accelerated evolution of these two genes in the sable. However, the evolutionary rates of DGKQ (ω1 = 0.05324; ω2 = 0.01867), GPAT4 (ω1 = 0.04913; ω2 = 0.00010), PLCB3 (ω1 = 0.05415; ω2 = 0.02571), PLCD4 (ω1 = 0.18010; ω2 = 0.02529), AGPAT2 (ω1 = 0.10713; ω2 = 0.00010), and PNLIPRP2 (ω1 = 0.34899; ω2 = 0.10429) were lower than those of the background lineages. We further implemented a more stringent branch-site model to investigate specific sites that underwent positive selection in sable lineages. Evidence for positive selection was detected along the sable lineage at multiple genes (i.e., ABHD5, AGPAT5, GPAT2, LIPH, LPL, MOGAT1, PDE3B, and PLCD3), while the LRT tests of these genes did not reach the level of significance. LRT tests showed strong positive selection at PNLIP and ACSS2 with several PSSs detected with a posterior probability > 0.8 (Table S3).
To determine the adaptive transcriptomic variation in sables for life in a cold climate, we sequenced the transcriptomes of the adipose tissue obtained from two seasons. A total of 55,866,908 and 53,011,440 clean reads were generated, after quality control, for summer and winter samples, respectively. The error rates of the two samples were less than 0.04, and the quality of the base value was Q20 > 94% and Q30 > 86% (Table 3), indicating that the data is reliable and meets the following analysis requirements. The error base distribution along the reads in two samples is shown in Figure S1. Eventually, 78.72% and 86.81% of clean reads in summer and winter samples, and greater than 77% of the reads in the two samples were uniquely mapped to the sable genome (Table 4). The bases content distribution along the reads in the two adipose tissue samples is shown in FigureS2.
It is essential to determine the DEGs of adipose tissue between two different seasons to investigate the molecular mechanism of lipid metabolism. A total of 3552 DEGs identified between two groups when |log2 (FoldChange) > 1 and q-value < 0.05 were applied as cut-off values. Of these, 2149 genes were upregulated in summer adipose tissue, and 1403 genes were upregulated in winter adipose tissue (Fig. 2). We conducted GO classification on DEGs to explore their functional distribution and identified 2601 DEGs enriched in 2754 GO terms categorized into three major groups: biological process, molecular function, and cellular component. Several significant GO terms were enriched (p-value < 0.05). GO terms such as lipid metabolic process (GO: 0006629), glucose metabolic process (GO: 0006006), glucose catabolic process (GO: 0006007), and glycolysis (GO: 0006096) belonged to BP; ATP binding (GO: 0005524), lipid binding (GO: 0008289), and lipase activity (GO: 0016298) belonged to molecular function (Fig. 3, Table S4). Based on KEGG analyses searching for metabolic or signal transduction pathways, we identified the DEGs enriched in 273 pathways, of which 15 pathways were significantly enriched. We mapped the scatterplot of the enriched KEGG pathways for upregulated genes in winter (Fig. 4). The metabolic pathway (ko:01100) was most enriched, while carbon metabolism (ko:01200), fatty acid metabolism (ko:01212), glycerolipid metabolism (ko:00561), glycerophospholipid metabolism (ko:00564), glycolysis/gluconeogenesis (ko:00010), and citrate cycle (ko:00020) were also significantly enriched. All genes involved in the enriched pathways were upregulated in winter (Table S5). These results demonstrate that the GO terms and KEGG pathway of genes with high expression levels between summer and winter adipose tissues are related to the metabolism process.
Volcano plot showing the number of differential expressed genes between the summer and winter samples. The x-axis corresponds to the log2 fold change value, and the y-axis displays the mean expression value of log10(q-value). The red dots represent the up-regulated genes; the green dots represent the down-regulated genes
Bar plot of the enriched Gene ontology (GO) terms for differential expressed genes in the adipose of sable. The x-axis shows the number of differential expressed genes, and the y-axis shows the enriched GO terms. Gene ontology categories include biological process (green), cellular component (orange) and molecular function (blue)
Scatterplot of enriched KEGG pathways for DEGs between the summer and winter samples in sables. The x-axis corresponds to the rich factor (the ratio of the DEG number to the total gene number in the pathway), and the y-axis shows the enciched KEGG pathways. The dot size and color represent the gene number and the range of the q-value, respectively
Discussion
Researchers have applied comparative genomic analyses to study the genetic bases underlying distinct adaptations in many species (Miller et al. 2012; Welch et al. 2014; Zhou et al. 2014). Although exploration of the molecular mechanisms underlying cold adaptation has been initiated in humans (Sazzini et al. 2014), polar bear (Liu et al. 2014), arctic fox (Kumar et al. 2015), Yakutian horses (Librado et al. 2015), and woolly mammoth (Schuster 2015), the adaptation mechanisms of sables have received little attention. The optimum temperature of the sable ranges from 5 to 10 °C in summer, and − 25 to 10 °C in winter (Bakeyev et al. 2003). A lustrous and silky pelage is important for sables’ survival in cold environments. However, the subcutaneous fat of sables may not provide adequate thermal insulation with a low amount of body fat (Mustonen et al. 2006), so it may face more energetic challenges than most other carnivores. The evolution of mammals has been associated with the evolution and metabolism of adipose tissue, which enables survival in cold climates (MacDougald and Lane 1995). Therefore, we propose that lipid metabolism has been an important component of the sable’s adaptation to cold environments. To determine if lipid metabolism-related genes have undergone adaptive evolution, we initially compared patterns of selection evolution across 81 genes involved in lipid metabolism among sables and closely related carnivores. Selection analysis provided significant evidence for positive selection at 20 of 81 (24.7%) lipid metabolism-related genes in the carnivore lineage, including ABHD5, DGKE, GPAM, GPAT2, GPAT4, PLCB4, PLCD4, PLIN5, PPARD, SLC27A1, SLC27A2, SREBF1, ABCG5, ABCG8, ACSS1, AGPAT2, APOB, DGAT1, PNLIP, and PNLIPRP2. The most powerful signal of positive selection came from APOB, with 15 codons that were determined as robust candidate sites that underwent positive selection. APOB, coding a large amphipathic glycoprotein, has been verified to play an essential role in human lipoprotein metabolism (Whitfield et al. 2004). The APOB protein promotes the transport of fat in lymph and blood plasma and combines with low-density lipoproteins (LDL), accelerating the movement of cholesterol into cells (Benn 2009). We also found evidence of positive selection acting on lipid metabolism-related genes in diverse carnivore species. This implied broad adaptation of lipid metabolism in carnivores. Our analyses provide evidence that several lipid metabolism-related genes have undergone adaptive evolution in carnivores. Free-model tests identified different positive selection signals in divergent carnivore lineages. These may be the principal driving force for the evolution of lipid metabolism-related genes to evolve, enhancing fat storage and digestion in carnivores. This result is not unexpected but indicates positive selection may be a major force in the evolution of lipid metabolism-related genes with respect to different functional dietary habits of carnivore species during evolution.
Branch and branch-site model analyses comparing the evolutionary rates of lipid metabolism-related genes in sables and other carnivores, suggested that PNLIP and ACSS2 have undergone significant positive selection in sables. PNLIP, a pancreatic triglyceride lipase (PTL) encoding gene, plays an essential role in dietary lipid absorption by hydrolyzing triglycerides into diglycerides and then into monoglycerides and free fatty acids (Mun et al. 2007). The absence of PTL can reduce absorbance of dietary lipids by 50–60% (Ghishan et al. 1984). PNLIP has also suffered specific amino acid changes in mammoths. These might have played essential roles in adapting woolly mammoths to the cold arctic climate (Schuster 2015). The positive selection at PNLIP might have a positive impact on the sable’s lipid absorption and digestion capacity, which satisfy its high energy demands in cold environments. ACSS2 encodes a cytosolic enzyme, which plays an essential role in synthesizing acetyl-CoA from acetate for lipid synthesis and energy generation (Xu et al. 2018). Acetyl-CoA is an essential intermediate of carbon sou that fuels ATP production through the TCA cycle and acts as an crucial component for the synthesis of sterols and fatty acids (Comerford et al. 2014). ACSS2 plays a potential role in the hypoxic adaptation of high-altitude cattle breeds (Zhao et al. 2021). Amino acid changes in the ACSS2 of woolly mammoths have also been documented (Schuster 2015). We suspected that the positive selected signals at ACSS2 in sables might contribute to their cold adaptation and energy storage lipogenesis during periods of food scarcity. The evolutionary rate of GOT2 in sables was about three times greater than other carnivore relatives, suggesting an accelerating evolution and a more obvious selective relaxation. GOT2 encodes glutamate oxaloacetate transaminase 2, a mitochondrial enzyme oxidizing glutamate to generate α-ketoglutarate in tandem and giving rise to the generation of ATP through the Krebs cycle (Honorat et al. 2017). The absence of functional GOT2 is probably fatal, as the GOT2 knock-out mouse is an embryonic lethal (Borst 2020). GOT2 enzyme, a member of the malate–aspartate shuttle, performs an essential function in the intracellular NAD(H) redox balance during glycolysis (Karnebeek et al. 2019). The relaxation of selective pressure in sables at GOT2 implies that sables can obtain energy in a shorter time by the metabolism of lipids or amino acids and increase their cold adaptation. We also detected that the evolutionary rates of DGKQ, GPAT4, PLCB3, PLCD4, AGPAT2, and PNLIPRP2 in sables were low compared with the background, suggesting that sables had suffered substantial selective pressure constraints at these genes associated with lipid metabolism. These results indicate that the candidate genes above had likely experienced particularly strong selective pressure related to the higher energetic demands of living in a cold environment.
Adipose is an endocrine and metabolic tissue that plays roles in fatty acid synthesis, fat storage, and maintenance of metabolic homeostasis. It participates in circulating free fatty acid and organizing lipid metabolism (O"Hea and Leveille, 1969). RNA-seq, a high-throughput sequencing innovation, is an effective way of obtaining transcriptome information to study gene expression (Marguerat and Bähler 2010). However, previous adipose tissue transcriptome studies have focused on human food animals, such as chickens (Resnyk et al. 2015), sheep (Yuan et al. 2019), cattle (Song et al. 2019), and pigs (Tao et al. 2017). In the present study, we applied high-throughput sequencing to characterize transcriptomic profiles differences of the adipose tissue in sables between two seasons and further analyzed how the transcriptome has coped with the cold environment. It is possible that the adipose tissue of the sable is better able to produce heat by enhancing its metabolic level in winter to resist cold. To obtain a better understanding of the molecular mechanism behind the seasonal differences of the adipose tissue transcriptomes in sables, we acquired 8.38G and 7.96G clean reads of summer and winter samples, respectively. Transcriptome analyses of sable adipose tissue from different seasons showed that many DEGs are involved in metabolism. A total of 3,552 DEGs were detected, of which 2149 were upregulated and 1,403 downregulated in the summer sample. We annotated the functions of these DEGs and found that the highly expressed genes were mainly involved in the Go term “lipid metabolic process,” “glucose metabolic process,” “lipid binding,” which are metabolic requirements of adaptation to cold weather. These DEGs may contribute to the adaptation of sables to cold winter conditions by adjusting the gene expression levels. Transcriptome analysis of summer and winter sables allowed the recognition of DEGs and contributed to a better understanding of what metabolic changes emerged in the adipose tissue of the sables adapting to a different climate. Enhancement of the metabolic level resulting from differentially expressed genes of adipose tissue in the sable might contribute to the increase in thermogenesis in the sable to prevent hypothermia during cold conditions. These results indicate that expression-level changes in genes associated with metabolic heat production contributed to cold adaption in sables.
In summary, we studied the molecular basis of the cold adaptation of sables by analysis of the adaptive evolution of genes involved in lipid metabolism pathways. We also compared the DEGs of sable adipose tissue between summer and winter. We found that PNLIP and ACSS2 underwent strong positive selection and GOT2 underwent rapid evolution in the sable lineage. The characterization of the adaptive evolution of lipid metabolism genes and the transcriptome profiles of adipose tissue from two seasons in sables reveal the low body fat mechanism and cold environment adaptations of sables. Based on genomic sequence analyses and transcriptomic analyses, we found adaptive signatures that reflect molecular adaptations associated with the cold-climate environment at the gene and transcriptional levels. In summary, positive selection and transcriptomic analyses in sables at the molecular level provided useful insights into their genetic adaptation to cold climate. This information enhances our understanding of the complex biological features of cold adaptation in sables.
Data availability
The lipid metabolism-related genes ID is provided in Table S1. The transcriptome sequencing data have been deposited into the Short Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra/) with Accession Code: PRJNA675656.
Code availability
Not applicable.
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Bakeyev N, Monakhov G, Sinitsyn A (2003) Sable (Martes zibellina)
Benn M (2009) Apolipoprotein B levels, APOB alleles, and risk of ischemic cardiovascular disease in the general population, a review. Atherosclerosis 206:17–30
Borst P (2020) The malate-aspartate shuttle (borst cycle): how it started and developed into a major metabolic pathway. IUBMB Life 72:2241–2259
Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, Horton JD, Hammer RE, McKnight SL, Tu BP (2014) Acetate dependence of tumors. Cell 159:1591–1602
Dunkin RC, McLellan WA, Blum JE, Pabst DA (2005) The ontogenetic changes in the thermal properties of blubber from Atlantic bottlenose dolphin tursiops truncatus. J Exp Biol 208:1469–1480
Fletcher W, Yang Z (2010) The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol 27:2257–2267
Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jørgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, Christensen C, Brandslund I, Jørgensen T, Huerta-Sánchez E, Schmidt EB, Pedersen O, Hansen T, Albrechtsen A, Nielsen R (2015) Greenlandic inuit show genetic signatures of diet and climate adaptation. Science 349:1343–1347
Ghishan FK, Moran JR, Durie PR, Greene HL (1984) Isolated congenital lipase-colipase deficiency. Gastroenterology 86:1580–1582
Hallmark B, Karafet TM, Hsieh P, Osipova LP, Watkins JC, Hammer MF (2019) Genomic evidence of local adaptation to climate and Diet in indigenous siberians. Mol Biol Evol 36:315–327
Harrison D, Fuller A, Proulx G (2004) In: Martens and Fishers (Martes) in Human-Altered Environments. Springer, New York
Himms-Hagen J (1990) Brown adipose tissue thermogenesis: interdisciplinary studies. FASEB J 4:2890–2898
Honorat JA, Nakatsuji Y, Shimizu M, Kinoshita M, Sumi-Akamaru H, Sasaki T, Takata K, Koda T, Namba A, Yamashita K, Sanda E, Sakaguchi M, Kumanogoh A, Shirakura T, Tamura M, Sakoda S, Mochizuki H, Okuno T (2017) Febuxostat ameliorates secondary progressive experimental autoimmune encephalomyelitis by restoring mitochondrial energy production in a GOT2-dependent manner. PLoS ONE 12:e0187215
Karnebeek C, Ramos R, Wen XY, Tarailo-Graovac M, Gleeson JG, Skrypnyk C, Brand-Arzamendi K, Karbassi F, Issa MY, Lee R (2019) Bi-allelic GOT2 mutations cause a treatable malate–aspartate shuttle-related encephalopathy. Am J Hum Genet 105:534–548
Kashtanov SN, Svishcheva GR, Lazebny OE, Kolobkov DS, Pishchulina SL, Meshchersky IG, Rozhnov VV (2015) Influence of anthropogenous factors on the genetic variety of the sable (Martes zibellina L.). Mol Biol 49:397–402
Kumar V, Kutschera VE, Nilsson MA, Janke A (2015) Genetic signatures of adaptation revealed from transcriptome sequencing of arctic and red foxes. BMC Genom 16:1–3
Langin D (2010) Recruitment of brown fat and conversion of white into brown adipocytes: strategies to fight the metabolic complications of obesity? Biochem Biophys Acta 1801:372–376
Librado P, Der Sarkissian C, Ermini L, Schubert M (2015) Tracking the origins of yakutian horses and the genetic basis for their fast adaptation to subarctic environments. Proc Nat Acad Sci 112:E6889–E6897
Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, Wray G, Li J, He W, Wang Z, Fu W, Xiang X, Morgan CC, Doherty A, O’Connell MJ, McInerney JO, Born EW, Dalén L, Dietz R, Orlando L, Sonne C, Zhang G, Nielsen R, Willerslev E, Wang J (2014) Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157:785–794
Li Y, Lasar D, Fromme T, Klingenspor M (2014) White, brite, and brown adipocytes: the evolution and function of a heater organ in mammals <sup>1</sup>. Can J Zool 92:615–626
MacDougald OA, Lane MD (1995) Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem 64:345–373
Marguerat S, Bähler J (2010) RNA-seq: from technology to biology. Cell Mol Life Sci CMLS 67:569–579
Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE, Tomsho LP, Ibarra-Laclette E, Herrera-Estrella L, Peacock E, Farley S, Sage GK, Rode K, Obbard M, Montiel R, Bachmann L, Ingólfsson O, Aars J, Mailund T, Wiig O, Talbot SL, Lindqvist C (2012) Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci USA 109:E2382-2390
Mun S, Decker EA, McClements DJ (2007) Influence of emulsifier type on in vitro digestibility of lipid droplets by pancreatic lipase. Food Res Int 40:770–781
Mustonen AM, Nieminen P (2006) Fatty acid composition in the central and peripheral adipose tissues of the sable (Martes zibellina). J Therm Biol 31:617–625
Mustonen AM, Puukka M, Saarela S, Paakkonen T, Aho J, Nieminen P (2006) Adaptations to fasting in a terrestrial mustelid, the sable (Martes zibellina). Comp Biochem Physiol Part a Mol Integr Physiol 144:444–450
O"Hea EK, Leveille GA (1969) Significance of adipose tissue and liver as sites of fatty acid synthesis in the pig and the efficiency of utilization of various substrates for lipogenesis. J Nutr 99:338–344
PingHsun H, Brian H, Joseph W, Tatiana K, Ludmila, (2017) Exome sequencing provides evidence of polygenic adaptation to a fat-rich animal diet in indigenous siberian populations. Mol Bio Evol 34:2913–2926
Rawls J, F., Canny, Torre, S.G.D.L. (2015) Baby, it’s cold outside: host-microbiota relationships drive Temperature adaptations. Cell Host Microb 18:635–636
Resnyk CW, Chen C, Huang H, Wu CH, Simon J, Le Bihan-Duval E, Duclos MJ, Cogburn LA (2015) RNA-seq analysis of abdominal fat in genetically fat and lean chickens highlights a divergence in expression of genes controlling adiposity, hemostasis, and lipid metabolism. PLoS ONE 10:e0139549
Sazzini M, Schiavo G, De Fanti S, Martelli PL, Casadio R, Luiselli D (2014) Searching for signatures of cold adaptations in modern and archaic humans: hints from the brown adipose tissue genes. Heredity 113:259–267
Schuster VLOB-RARMSDD-MGPWMS (2015) Elephantid genomes reveal the molecular bases of woolly mammoth adaptations to the arctic. Cell Rep 12:217–228
Sheafor BA (2003) Metabolic enzyme activities across an altitudinal gradient: an examination of pikas (genus Ochotona). J Exp Biol 206:1241–1249
Song C, Huang Y, Yang Z, Ma Y, Chaogetu B, Zhuoma Z, Chen H (2019) RNA-Seq analysis identifies differentially expressed genes insubcutaneous adipose tissuein qaidamford cattle, cattle-yak, and angus cattle. Animals 9:1077
Swanson WJ, Nielsen R, Yang Q (2003) Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol 20:18–20
Tao X, Liang Y, Yang X, Pang J, Zhong Z, Chen X, Yang Y, Zeng K, Kang R, Lei Y, Ying S, Gong J, Gu Y, Lv X (2017) Transcriptomic profiling in muscle and adipose tissue identifies genes related to growth and lipid deposition. PLoS ONE 12:e0184120
Vernon RG, Denis RG, Sørensen A (2001) Signals of adiposity. Domest Anim Endocrinol 21:197–214
Welch AJ, Bedoya-Reina OC, Carretero-Paulet L, Miller W, Rode KD, Lindqvist C (2014) Polar bears exhibit genome-wide signatures of bioenergetic adaptation to life in the arctic environment. Genome Biol Evol 6:433–450
Whitfield AJ, Barrett PH, van Bockxmeer FM, Burnett JR (2004) Lipid disorders and mutations in the APOB gene. Clin Chem 50:1725–1732
William C, Stewart L, A., Pearcy, Z., Elizabeth, Floyd, Jacqueline, (2011) STAT5A expression in swiss 3T3 cells promotes adipogenesis in vivo in an athymic mice model system. Obesity 19:1731–1734
Wozniak SE, Gee LL, Wachtel MS, Frezza EE (2009) Adipose tissue: the new endocrine organ? a review article. Dig Dis Sci 54:1847–1856
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316-322
Xu H, Luo J, Ma G, Zhang X, Yao D, Li M, Loor JJ (2018) Acyl-CoA synthetase short-chain family member 2 (ACSS2) is regulated by SREBP-1 and plays a role in fatty acid synthesis in caprine mammary epithelial cells. J Cell Physiol 233:1005–1016
Yang Z (2007) Paml 4: phylogenetic analysis by maximum likelihood. Mol Bio Evolut 24:1586–1591
Yuan ZH, Xiang RD, Li WH, Li FD, Yue XP (2019) Transcriptomic analyses revealed common tailed and perirenal adipose differentially expressed genes in four Chinese indigenous sheep breeds. Livest Sci 230:103832
Yu XX, Lewin DA, Forrest W, Adams SH (2002) Cold elicits the simultaneous induction of fatty acid synthesis and beta-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. FASEB J 16:155–168
Zhao H, Gao YP, Jiang Q, Wang JP, Liu WH, Ju ZH, Wang XG, Wei XC, Gao YD, Huang JM (2021) Molecular characterization of the ACSS2 gene involved in adaptation to hypoxia in high-altitude cattle breeds. Anim Biol 71:49–66
Zhou X, Wang B, Pan Q, Zhang J, Kumar S, Sun X, Liu Z, Pan H, Lin Y, Liu G, Zhan W, Li M, Ren B (2014) Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat Genet 46:1303–1310
Funding
This work was supported by the Special Fund for Forest Scientific Research in the Public Welfare (201404420) and the National Natural Science Foundation of China (31872242, 31672313).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Handling editor: Allan McDevitt.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xia, T., Zhang, L., Sun, G. et al. Insights into cold tolerance in sable (Martes zibellina) from the adaptive evolution of lipid metabolism. Mamm Biol 101, 861–870 (2021). https://doi.org/10.1007/s42991-021-00135-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42991-021-00135-0