Abstract
The riverine barrier hypothesis (RBH) posits that rivers comprise geographical barriers to gene flow for terrestrial organisms, thus promoting genetic differentiation between populations. Here, we explored the RBH on larviparous and pueriparous populations of the live-bearing fire salamander (Salamandra salamandra). While larviparous fire salamanders exhibit a semi-aquatic life cycle (females deposit pre-metamorphic larvae on water), pueriparous salamanders present a fully terrestrial life cycle (females deliver terrestrial juveniles) and, therefore, a greater independence from water for survival and reproduction. We performed a fine-scale sampling of opposite transects in 11 rivers (six and five for larviparous and pueriparous populations, respectively) to test the hypothesis that rivers are more effective barriers for pueriparous salamanders due to their terrestrial life cycle. We carried out individual- and population-based genetic analyses using 14 microsatellites and a mitochondrial marker to examine the extent to which rivers hinder short- and long-term gene flow. We found that rivers are semi-permeable obstacles for both larviparous and pueriparous salamanders, although they appear to be more effective barriers for the latter when rivers with similar attributes are compared. We also found that river width and possibly the presence of crossing structures may influence the genetic barrier effects of rivers in fire salamanders. This is one of the very few studies in amphibians showing how different reproductive strategies influence the barrier effects imposed by rivers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The riverine barrier hypothesis (RBH) posits that rivers can be geographical barriers to dispersal for terrestrial organisms, leading to a reduction/restriction of gene flow between individuals at both sides of the river, and thus enhancing genetic differentiation and, ultimately, allopatric speciation (Wallace 1852). The barrier effect of this landscape feature is pervasive across the world and applies to many taxonomic groups (e.g. insects, Hall and Harvey 2002; birds, Kopuchian et al. 2020; mammals, Nicolas et al. 2011; plants, Nazareno et al. 2017; reptiles, Pirani et al. 2019; amphibians, Waraniak et al. 2019).
The extent to which rivers affect dispersal depends mainly on their physical attributes, and on the phenotypic traits of organisms. Large rivers with strong water currents are often effective dispersal barriers that promote genetic population subdivision and lineage diversification (e.g. Funk et al. 2007; Frantz et al. 2010; Nicolas et al. 2011; Moraes et al. 2016; Godinho and da Silva 2018), although the RBH is often assumed when populations or lineages´ range boundaries coincide with the location of rivers. The barrier effects of rivers are expected to be lower in large-sized organisms that can swim and counteract water current, and in animals that exhibit great swimming/flight abilities (e.g. mammals, birds, some amphibians; Cushman et al. 2006; Pérez-Espona et al. 2008; Luqman et al. 2018; Kopuchian et al. 2020). Conversely, rivers generally comprise relatively impermeable barriers for small-sized and poor dispersers, such as small mammals, amphibians or reptiles (Funk et al. 2007; Nicolas et al. 2011; Fouquet et al. 2012; Pirani et al. 2019; Waraniak et al. 2019). Moreover, life-history also plays a crucial role in determining the barrier effects of rivers. For semi-aquatic species, which spend more time in water and evolved adaptations to an aquatic life style, such as limbs adapted for a more efficient swimming (Dunstone 2007; Moen 2019) or walking on water (Bush and Hu 2006), it is likely rivers comprise more permeable obstacles for these taxa compared to close-related and strictly terrestrial species. Hence, one may hypothesize the extent to which rivers influence dispersal and gene flow may have changed in those species that underwent major life-history modifications regarding the way they interact with aquatic systems, though this subject is still poorly explored.
Despite being broadly characterized as semi-aquatic group, amphibians present a high diversity of life-histories ranging from fully aquatic and semi-aquatic life cycles to fully terrestrial ones (Crump 2015). This vertebrate group comprises a good system in which to examine how changes in life-history traits influence the barrier effects of rivers. Throughout the deep evolutionary history of amphibians, many species shifted from a biphasic life-history (aquatic eggs and larvae develop into terrestrial adults) to a fully terrestrial one (Blackburn 2015), in which the aquatic stage is skipped and, therefore, oviposition (direct development; Wake and Hanken 1996) or the parturition of metamorphosed juveniles (pueriparity; Greven 2003) occurs in land. Such pronounced changes in their life cycle have been associated to a lack of suitable water bodies in their environments for depositing offspring (e.g. García-París et al. 2003; Beukema et al. 2010; Velo-Antón et al. 2012; Liedtke et al. 2017; Lion et al. 2019), but the effects of these aquatic features as potential barriers to dispersal in both reproductive modes (i.e. aquatic and terrestrial) have never been thoroughly assessed.
Here, we use the fire salamander (Salamandra salamandra, Linnaeus 1758) as study model to examine the genetic barrier effects of rivers in salamander populations. This species presents both semi-aquatic and fully terrestrial populations, which makes it a good system to infer how life-history influences the barrier effects of rivers. Salamandra salamandra presents two live-bearing (viviparous) reproductive strategies: (1) an ancestral larviparous reproduction, which occurs throughout most of its distribution and, in which females deliver aquatic larvae (ca. 20–80); and (2) a phylogenetically derived pueriparous reproduction, in which the aquatic larval stage is skipped and females give birth to ca. 1–35 fully developed terrestrial juveniles (Buckley et al. 2007; Velo-Antón et al. 2015; Alarcón-Ríos et al. 2020a). The latter is restricted to north-western Iberian Peninsula and evolved independently in the subspecies S. s. bernardezi during the Pleistocene in the Cantabrian Mountains (García-París et al. 2003), and more recently, in insular populations of S. s. gallaica during the Holocene (Velo-Antón et al. 2007).
A previous comparative landscape genetics study assessed how the landscape composition (i.e. land cover, topography, and vegetation) affect genetic connectivity in larviparous and pueriparous populations of S. salamandra (Lourenço et al. 2019). This study showed that water courses (both low order streams and rivers) significantly hinder gene flow only in pueriparous salamanders, thus implying the evolution of a fully terrestrial life cycle entailed lower crossing rates across these landscape features. However, despite the clear effect of water courses on the population connectivity of pueriparous salamanders, the study design carried out by Lourenço et al. (2019) at a regional level did not have sufficient spatial resolution to determine the importance of rivers in shaping gene flow in both larviparous and pueriparous salamanders. A fine-scale sampling design based on parallel transects along riversides should not only allow inferring more accurately the extent to which rivers hamper dispersal, but also better controlling for the confounding effects of other landscape features on genetic structure. Here, we used 14 microsatellite markers and a mitochondrial marker to assess the short- and long-term genetic barrier effects of 11 rivers in larviparous and pueriparous populations, respectively. We performed parallel sampling transects (< 1 km, only separated by rivers) and used both individual- and population-based genetic methods to evaluate the role of main rivers as barriers to gene flow. We expect a higher genetic structure and lower relatedness between pueriparous salamanders sampled in opposite sides of the river compared to their larviparous counterparts, thus providing greater support to the hypothesis that rivers impose greater barriers to pueriparous salamanders due to their terrestrial life-history (Lourenço et al. 2019). Nevertheless, we also expect the characteristics of rivers (e.g. river width) can also play a role in determining the barrier effect of these landscape features.
Methods
Study area and sampling design
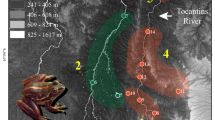
We studied populations of two S. salamandra subspecies distributed throughout north-western Iberian Peninsula: (1) S. s. gallaica, which is larviparous in mainland populations; and (2) the pueriparous S. s. bernardezi (Velo-Antón et al. 2015). We sampled lowland populations in six and five permanent rivers located within S. s. gallaica and S. s. bernardezi ranges, respectively (Fig. 1; Table 1). All these rivers fall within the Atlantic climate influence of the Iberian Peninsula and share a similar native vegetation (deciduous forests of Quercus spp.). While we tried to sample all major rivers across the north-western Iberia, this was not possible due to the absence or very low population densities of fire salamanders that have been recently affected by the severe habitat loss and fragmentation across this area (i.e. presence of agricultural fields and eucalyptus plantations along riversides; Deus et al. 2018; Lourenço et al. 2019). We performed sampling in sections of the rivers where patches of deciduous forests with high availability of shelter were present (Velo-Antón and Buckley 2015) to minimize the potential confounding effects of habitat fragmentation on individual dispersal (Cosgrove et al. 2018). We also tried to avoid sampling sections of the river containing crossing structures (e.g. bridges), which presumably may promote higher dispersal and gene flow rates across the river. Nevertheless, this was not possible in three of the studied rivers where nearby bridges were present (Table 1).
Map displaying the study area in north-western Iberia. The inset shows the distribution range of Salamandra salamandra. Within A, the distribution patterns of reproductive modes are shown in blue and red for larviparity (S. s. gallaica) and pueriparity (S. s. bernardezi), respectively. Sampled locations are named after the studied rivers
We sampled during rainy nights of early spring (from March to April of 2019) and autumn (October and November of 2018 and 2019), thus matching the peaks of activity of adult fire salamanders in northern Spain (Velo-Antón and Buckley 2015). We sampled adults to prevent spurious estimates of genetic structure arising from an excessive sampling of related individuals because larvae and, to a less extent juveniles, are often spatially clustered (Wang 2018; O’Connell et al. 2019). We collected toe clips from a total of 311 individuals (191 larviparous and 120 pueriparous salamanders) in 11 localities (mean ± SD: 28 ± 10.5 individuals per locality). All individuals were georeferenced and sexed through visual inspection of the cloaca (Velo-Antón and Buckley 2015). We performed parallel transects of an approximate average length of 563 m (range: 152–1180 m) to sample individuals along both sides of each river (Table 1). This sampling scheme avoids potential confounding effects of other landscape features affecting salamander dispersal, while the length was selected to assure the transects were longer than the width of the rivers, thus potentially preventing misleading results arising from a pattern of isolation by distance (Meirmans 2012).
Laboratory procedures and genotyping
We extracted genomic DNA using the Genomic DNA Tissue kit (Easy Spin), following manufacturer’s protocol. Quantity and quality of the extracted DNA was evaluated on an electrophoresis 0.8% agarose gel. A total of 14 microsatellites, distributed in four multiplexes (Steinfartz et al. 2004; Hendrix et al. 2010), were amplified through Polymerase Chain Reactions (PCRs) following optimized and previously tested conditions (see Lourenço et al. 2018a). PCR products were tested in 2% agarose gel and run on an ABI3130XL capillary sequencer (Applied Biosystems). Allele scoring was performed in GENEMAPPER 4.0 (Applied Biosystems). Since salamanders are capable of regenerating their toes, we used the option Multilocus Matches in GENALEX 6.5 (Peakall and Smouse 2012) to check for genotype matches, as some rivers were sampled during different seasons and multiple nights apart.
We randomly selected a mean number of six individuals from each riverside in each sampled locality to amplify a short fragment of the mitochondrial DNA (mtDNA) gene cytochrome b (cytb), using primers cytb-2 (Kocher et al. 1989) and MVZ 15 (Moritz et al. 1992). We followed the laboratory protocol described in Velo-Antón et al. (2007). In total, this mtDNA marker was amplified for 72 larviparous and 66 pueriparous individuals. DNA sequencing was outsourced to Genewiz Inc. (Leipzig, Germany). All the obtained chromatograms were verified, aligned and corrected by eye using GENEIOUS PRO version 11.1.4 (http://www.geneious.com/). The aligned cytb sequences were trimmed to avoid missing data, resulting in a consensus sequence of 315 bp.
Microsatellite quality control
We tested all microsatellite loci for potential deviations from Hardy Weinberg Equilibrium (HWE) and Linkage Equilibrium (LE) by performing exact tests in GENEPOP 4.2 (Rousset 2008; dememorization = 10,000, batch number = 5000; batch length = 10,000). We applied the false discovery rate (Benjamini and Hochberg 1995) to correct p-values from HWE and LE multiple exact tests. We calculated frequencies of null alleles in INEST 2.0 (Chybicki and Burczyk 2009) using the individual inbreeding model and setting a total of 200,000 iterations, thinned every 200 iterations, with an initial burn-in of 20,000 iterations. We calculated the Probability of Identity PIID (Waits et al. 2001) and the Probability of Identity accounting for sibs PIIDsibs in GENALEX 6.5 (Peakall and Smouse 2012) to assess the informative power of our loci to discriminate between individuals.
Genetic diversity
We estimated parameters of population genetic diversity in GENALEX 6.5 for all individuals sampled in each river and within each riverside separately. Specifically, we calculated the mean number of alleles per marker (NA), observed (HO) and expected (HE) heterozygosities, number of private alleles (PA), and the number of individuals containing private alleles (NPA). We estimated allelic richness (AR) corrected for the smallest locality's sample size using the package diveRsity (Keenan et al. 2013) in R (R Development Core Team 2017), and population mean inbreeding coefficient (F) in INEST 2.0.
Genetic differentiation between riversides
We used the cytb data to examine the potential long-term level of population differentiation between individuals sampled on each riverside. We generated a haplotype network using TCS v. 1.21 (Clement et al. 2000) with a 95% of probability of parsimony, and edited the haplotype network in TCSBu (Santos et al. 2015).
We further evaluated the contemporary genetic barrier effects of the sampled rivers using both individual- and population-based genetic approaches that relied on the microsatellite genotypes. We first analysed the spatial distribution of related individuals between riversides. We used COANCESTRY 1.0.1.9 (Wang 2011) to calculate two estimates of pairwise relatedness among individuals: (1) the triadic likelihood estimator (RTRI) (Wang 2007); and (2) the Queller and Goodnight’s point estimator (RGT) (Queller and Goodnight 1989). We did not account for genotyping errors or inbreeding, and 95% confidence intervals (CIs) were computed through 1000 bootstrap resamplings. We considered pairs of related salamanders those that exhibited a pairwise relatedness ≥ 0.125 for both metrics (the theoretical value of third-order relationships; e.g. cousins). We calculated the frequency of pairs of relatives located on opposite riversides (FINTER) for each sampled locality. We also performed a permutation test of the difference of means between two groups to assess the hypothesis that average relatedness among dyads separated by the river is lower than the average relatedness calculated for dyads sampled on the same riverside. Specifically, for each river, the pairwise relatedness values were split into two groups: (1) those involving pairs sampled on the same riverside (intra-riverside); and (2) those involving dyads located on different riversides (inter-riverside). The permutation tests were performed in COANCESTRY using a total of 10,000 permutations (α = 0.05).
We then evaluated contemporary genetic differentiation between individuals sampled on different riversides using population-based approaches. For each sampled locality, we grouped all individuals sampled on opposite transects as independent populations. We first calculated two indices of genetic differentiation: (1) the pairwise FST (Nei 1977); and (2) the pairwise Jost’s DEST (Jost 2008). Significance was tested (α = 0.05) in GENALEX by performing 9999 permutations of the data. Finally, we also examined how river width is associated with pairwise genetic differentiation between individuals sampled on opposite riversides. We used the non-parametric Kendall correlation (τR) test to perform a quantitative assessment of the relationship between both variables.
We also inferred and visualised patterns of contemporary population genetic structure for each sampled locality through two clustering methods: (1) the Bayesian approach implemented in STRUCTURE 2.3.4. (Pritchard et al. 2000); and (2) the multivariate method of discriminant analysis of principal components (DAPC) implemented in the R package adegenet 2.1.1 (Jombart et al. 2010). Both methods have previously shown decent performance in detecting the effects of linear and non-linear landscape barriers on genetic structure (Jombart et al. 2010; Blair et al. 2012; Sánchez-Montes et al. 2018). Because we are interested in assessing whether the study rivers comprise a genetic barrier, for these analyses, we grouped individuals from each riverside as a different “population” and ran the algorithms assuming only two genetic clusters (K = 2).
Preliminary analyses in STRUCTURE using default parameters (e.g. no sampling location information) showed genetic structure between sides of the river was very weak for several sampled localities. Under these circumstances, incorporating sampling information in STRUCTURE (i.e., including the LOCPRIOR parameter) enables a more accurate detection of subtle patterns of genetic structure (Hubisz et al. 2009). Hence, we carried out STRUCTURE analyses with a prior location information (LOCPRIOR parameter), admixture model, and correlated allele frequencies. We performed ten independent runs for K = 2 and each run was set to 500,000 iterations with a burn-in period of 10%. Moreover, the inclusion of related individuals in the analyses may overestimate population genetic structure (Wang 2018; O’Connell et al. 2019). Accordingly, we removed an individual from each pair of first-order relatives (RTRI ≥ 0.5, parent–offspring and full-sibs) in those localities that showed some evidence of genetic structure between riversides. In those cases, we then re-estimated population structure using the same input parameters.
DAPC is a multivariate method that summarizes the data to maximize the variation between groups while minimizing it within groups. Unlike STRUCTURE, it does not make any assumptions regarding HWE and LE. We did not follow the package manual’s guidelines regarding the choice of the optimum value of Principal Components (PCs). This is because the cross-validation method proposed in the manual would potentially lead to overfitting of the discriminant functions due to the relatively low sample sizes observed in some rivers (Jombart et al. 2010). To overcome this issue, we retained a number of PCs correspondent to one third of the sample size in given locality (e.g. Grummer and Leaché 2017).
Results
Marker validation and population genetic diversity
All sampled individuals in a river exhibited a minimum of six to nine allele mismatches between each other (Table 2). The probability of identity (PIID) and probability of identity when accounting for sibs (PIIDsibs) were low, ranging from 3.7 × 10−21 to − 6.7 × 10−13 and 2.2 × 10−7 to 1.2 × 10−5, respectively. Accordingly, we concluded that our 14 loci have high information content to discriminate individuals and, hence, we considered none of the sampled individuals were recaptures and all were retained for downstream analyses.
After accounting for multiple testing, we found evidence of deviations from LE in two loci (Sal3, SalE8) in three populations (Minho, Cávado and Anllóns). Three loci (Sal E2, Sal3, SST-C3) showed departures from HWE due to heterozygote deficiencies although deviations were not consistent across populations. Because we did not find a congruent pattern regarding deviations from HWE and LE, we retained all loci in subsequent analyses. No locus showed evidences for null alleles or large allele dropout.
Genetic diversity was overall high and relatively similar between larviparous and pueriparous populations (mean values for larviparous and pueriparous populations, respectively: HO: 0.73 vs. 0.73; HE: 0.78 vs. 0.79; NA: 10.06 vs. 9.82; PA: 0.63 vs. 0.52; NPA: 39.67 vs. 23.4; AR: 6.44 vs. 6.88; Table 2; “Appendix 1” for information per riverside). Mean inbreeding coefficients were low in both larviparous (mean F = 0.03) and pueriparous (mean F = 0.08) populations.
mtDNA analysis
The cytb dataset comprised 137 samples with a mean number of 12 samples (± 2.4) per river (“Appendix 1”). We found a total of 14 haplotypes, of which seven and five were exclusive of pueriparous and larviparous populations, respectively (“Appendix 5”). All samples from larviparous rivers were clustered in seven haplotypes, while nine haplotypes contained at least one sample from pueriparous populations. All rivers showed more than one haplotype, and at least one haplotype shared between riversides (“Appendix 6”). The larviparous population of Eume and the pueriparous populations of Nora and Narcea showed highly divergent shared haplotypes, and overall, pueriparous populations showed more haplotype diversity than larviparous populations with at least three haplotypes per river. Despite the low sample sizes, all rivers but Anllóns showed unique haplotypes at one riverside.
Individual-based analysis
COANCESTRY identified a total of 309 pairs of relatives (RTRI ≥ 0.125) out of 4793 dyads, of which 88 in pueriparous populations (out of 1546 dyads; 5.69%) and 221 in larviparous populations (out of 3247 dyads; 6.8%) (Table 3). The average intra-riverside relatedness was similar between reproductive modes (larviparous RTRI: 0.034 ± 0.005; pueriparous RTRI: 0.035 ± 0.006), while relatedness among inter-riverside dyads was on average lower than within riversides, particularly, in pueriparous populations (larviparous RTRI 0.023 ± 0.009; pueriparous RTRI 0.016 ± 0.009) (“Appendices 2, 3”). The frequency of pairs of relatives found in opposite sides of the river was on average higher in larviparous populations (mean FINTER = 0.021, range = 0.013–0.033) compared to their pueriparous counterparts (mean FINTER = 0.012, range = 0–0.029; Fig. 2a). Permutation tests showed significant differences in mean pairwise relatedness between intra- and inter-riverside groups in four larviparous (Minho, Cávado, Ulla, and Lérez) and two pueriparous populations (Navia and Narcea) (Table 3). The conclusions drawn when using RQG as a relatedness metric are similar to the ones obtained for RTRI (“Appendices 4, 7a”).
a Strip plot comparing the frequency of pairs of relatives (RTRI ≥ 0.125) found across riversides (each point represents a population). The blue and red dashed lines represent the mean frequency of relatives across riversides for larviparous and pueriparous populations, respectively. b Strip plot representing pairwise population FST values for individuals sampled across riversides. Dashed lines represent mean FST values per reproductive mode
Population-based analysis
Pairwise population values between individuals on opposite sides of the river were low or moderate, and were on average higher for pueriparous populations (mean FST = 0.042, range = 0.017–0.059; mean DEST = 0.124, range = − 0.001 to 0.261) than for larviparous ones (mean FST = 0.027, range = 0.015–0.041; mean DEST = 0.062, range = − 0.008 to 0.155) (Table 4 and Fig. 2b; “Appendix 7b”). The wider larviparous rivers (Ulla, Minho and Cávado) and two pueriparous rivers (Navia and Narcea) showed significant values on pairwise genetic differentiation for both distance measures. Indeed, the correlation between river width and population genetic differentiation was high for both reproductive modes (τR ≥ 0.60) but significant only in larviparous populations for DEST (τR = 0.97, p = 0.007) (Table 5; Fig. 3).
Plots of genetic distance (FST) between riversides relative to river width (measured in meters) for a larviparous, and b pueriparous populations. Each circle represents a population. Curves fitted using a second-order polynomial are displayed only to facilitate the visualisation of the relationship between the FST and river width
STRUCTURE was only able to recover a barrier effect (K = 2) in the larviparous populations of Cávado and Minho (Fig. 4), and in the pueriparous population of Navia (Fig. 5). The remaining populations show strong admixture levels between riversides. Among those rivers showing a strong signal of structure, we only found close relatives in Navia river (RTRI ≥ 0.5). We found an identical pattern of genetic structure after discarding an individual from each pair of relatives.
Images displaying STRUCTURE barplots as well as the individual pie charts representing cluster membership proportions to each riverside when K = 2 for larviparous populations. In each barplot, individuals are represented by vertical bars that are partitioned into two colours representing their estimated membership coefficient to each riverside. Vertical white lines in the barplots separate riversides
Images displaying STRUCTURE barplots as well as the individual pie charts representing cluster membership proportions to each riverside when K = 2 for pueriparous populations. In each barplot, individuals are represented by vertical bars that are partitioned into two colours representing their estimated membership coefficient to each riverside. Vertical white lines in the barplots separate riversides
DAPC revealed the larviparous populations of Cávado and Minho, and the pueriparous populations of Navia and Narcea showed clear patterns of genetic differentiation (“Appendices 8, 9”). The larviparous populations of Eume and Ulla, and the pueriparous population of Esva showed moderate genetic differentiation and admixture levels, while the remaining populations exhibited high genetic admixture between riversides (“Appendices 8, 9”).
Discussion
Dispersal and gene flow in amphibians are intimately linked with aquatic systems (Semlitsch 2008; Pittman et al. 2014). It is thus reasonable to hypothesize the evolution of terrestrial reproductive modes, which entail greater independence from water, may affect the way amphibians interact with aquatic systems, and ultimately, change patterns of dispersal across the landscape (see Lourenço et al. 2018a, 2019). We evaluated the RBH across NW Iberian rivers through a comparative framework, in which, a fine-scale sampling scheme and individual- and population-based genetic analyses were employed to examine the barrier effects of rivers in populations of S. salamandra that exhibit terrestrial (pueriparity) or semi-aquatic (larviparity) life-histories. Our results suggest that rivers are semi-permeable barriers to gene flow for both reproductive modes, yet they appear to be more effective in the terrestrial pueriparous salamanders. To the best of our knowledge, this work represents one of the very few studies using a fine-scale sampling design to explicitly assess the RBH in organisms with different reproductive modes, further emphasizing the importance of accounting for life-history to better understand the effects of rivers on dispersal and genetic structure.
Riverine barrier effect for aquatic-breeding and terrestrial-breeding fire salamander populations
Previous landscape genetic studies have demonstrated that rivers can negatively affect dispersal in amphibians exhibiting aquatic reproduction (e.g. Richardson 2012; Waraniak et al. 2019) and fully-terrestrial cycles (e.g. Fouquet et al. 2012, 2015). In agreement with those results, our genetic analyses suggest rivers comprise semi-permeable obstacles to gene flow for both larviparous and pueriparous salamanders. All sampled populations presented signs of genetic differentiation and genetic structure, and/or a lower frequency of relatives and average relatedness among individuals sampled in opposites sides of the rivers (Tables 3, 4; Figs. 2, 4, 5 and “Appendices 2, 7–9”). However, only the largest rivers (mainly Minho and Cávado for larviparous populations, and Navia and Narcea for pueriparous ones) consistently showed to impose great barrier effects to gene flow in fire salamanders. Indeed, the pairwise genetic differentiation values of the populations bisected by these rivers are comparable to those observed in some fire salamander populations that are ca. 10–20 km apart (and even up to 40–50 km in very few cases; see Lourenço et al. 2019).
Despite the negative effects of rivers on fire salamander dispersal, we found slight differences in those effects between reproductive modes. When comparing rivers with similar attributes (e.g. river width), the higher average pairwise genetic differentiation and lower frequency of relatives found across riversides in pueriparous salamanders indicate the barrier effects of these landscape features are to some extent greater when compared to their larviparous congeners (Figs. 2, 4, 5). These results corroborate the ones obtained by Lourenço et al. (2019) who found that water courses reduced genetic connectivity in pueriparous populations of S. salamandra. Because their terrestrial life cycle implies that aquatic systems are no longer necessary for reproduction, pueriparous salamanders may be less compelled to use and attempt to cross rivers. This behaviour can help explain the results obtained in other studies. For example, Fouquet et al. (2015) reported higher levels of mitochondrial divergence among individuals of terrestrial direct-developing Amazonian frog species separated by rivers compared to aquatic-breeding frog species, while Marsh et al. (2007) observed moderate levels of genetic differentiation between individuals located in opposite sides of small streams (width ≤ 7 m) in a direct-developer salamander (Plethodon cinereus). All this cumulative evidence reinforces the notion that rivers hinder dispersal to a larger extent in terrestrial-breeding amphibians compared to aquatic ones. Unfortunately, because some sampled rivers are not entirely comparable between reproductive modes (e.g. rivers with similar attributes in which nearby bridges are inexistent), we are unable to accurately infer in relative terms how much greater the barrier effects imposed by rivers are in pueriparous salamanders compared to their larviparous counterparts.
Despite the marked terrestrial life cycle of adults, larviparous fire salamanders inhabit aquatic systems (including rivers) during the larval stage, which lasts approximately three months in the focal region (Velo‐Antón and Buckley 2015). This larval stage, and the need of larviparous females to move into rivers (and other aquatic systems) to deposit offspring, likely create more opportunities for larviparous individuals to cross rivers. Both streams and rivers are associated with downstream larval drift in S. salamandra (Reinhardt et al. 2018; Veith et al. 2019) and the strong water current and drifting objects in both streams and rivers, together with the presence of predators, may cause high mortality rates (Segev and Blaustein 2014; Reinhardt et al. 2018; Wagner et al. 2020). However, considering that only a relatively small number of immigrants are required to prevent population genetic differentiation due to genetic drift (Lowe and Allendorf 2010), it is plausible that even a small proportion of successful dispersal events across rivers may minimize their barrier effects, even when fire salamander populations often present high densities (and potentially large effective population sizes; Velo‐Antón and Buckley 2015).
Besides variation in reproductive modes and other phenotypic traits (e.g. colouration), S. s. bernardezi is on average smaller (mean ± SD body size: 79 ± 8.4 mm) than S. s. gallaica larviparous salamanders (115 ± 9.1 mm) (Velo-Antón and Buckley 2015; Alarcón-Ríos et al. 2020b). Because body size is positively correlated with locomotor performance and dispersal distances, we cannot discard this phenotypic trait may enable larviparous salamanders overcoming more easily these aquatic barriers compared to pueriparous salamanders and, therefore, contributing for a lower genetic structure in the former. Indeed, a determinant role of body size on populations´ genetic connectivity was unveiled for some amphibian species (Paz et al. 2015; García et al. 2017).
Landscape barrier effects for fire salamander populations
The strong riverine barrier effects observed in this study for both larviparous and pueriparous salamanders are partially in agreement with those reported by Lourenço et al. (2019), who found evidence of strong effects of rivers on gene flow only in pueriparous populations. These partially contrasting results emphasize the importance of thorough sampling designs to evaluate specific landscape features as putative barriers to dispersal and gene flow. Furthermore, our study allows us inferring the role of other related factors that may also govern the effects of rivers on genetic variation in S. salamandra. Specifically, we argue that both (1) river width, and (2) crossing structures can also be determinant factors.
Wider rivers, such as Cávado (≈ 90 m) and Minho (≈ 190 m) in larviparous populations, and Navia (≈ 50 m) and Narcea (≈ 30 m) in pueriparous ones, appear to impose stronger barrier effects to fire salamanders given the observed low frequency of relatives and greater levels of genetic structure between riversides (Tables 3, 4; Figs. 4, 5 and “Appendices 2–4, 7–9”). In general, wider rivers exhibit stronger water currents and high water-flow velocities. The greater distance that salamanders must travel to successfully cross these rivers under these unsuitable aquatic conditions most likely hinder the swimming capacity of individuals, which eventually end up being dragged downstream and dying (Reinhardt et al. 2018). River width was also suggested as a determinant factor in shaping patterns of gene flow for an Amazonia frog (Adenomera andreae) by Fouquet et al. (2012) because the authors observed that populations of this species were more genetically differentiated in sections where the studied river was wider. Contrarily, in narrower rivers with low water-flow velocities, larvae (or adults) of S. salamandra may be able to actively resist the entrance on the current (Segev and Blaustein 2014) or to actively move against it (Veith et al. 2019).
There was one bridge bisecting the river where the pueriparous populations of Navia, Esva, and Nora were sampled. We cannot discard these structures may facilitate dispersal of fire salamanders across rivers, although our data does not enable us to infer accurately the role of bridges on genetic connectivity between riversides. Nevertheless, circumstantial evidence suggest the small stone Roman bridge located in the river Nora may be partially responsible for the high levels of genetic admixture and high relatedness among individuals on both riversides in this locality. Indeed, salamanders were observed crossing this bridge (located about 60 m away from the transect) during sampling nights (A. Lourenço and G. Velo-Antón personal observations), thus supporting our premise. We did not observe salamanders crossing road traffic bridges in other sites (e.g. Esva, whose bridge was located roughly 15 m away from the transect) although we cannot discard that occasional dispersal events may still occur. Fire salamanders, like many other amphibian species, move to roads and are subjected to high road mortality rates (Carvalho and Mira 2011; Beebee 2013). Hence, it is not unreasonable that at least a small proportion of salamanders could cross small traffic bridges. The river Navia is also bisected by a car bridge (located about 40 m away from the transect), though unlike the rivers Nora and Esva, there was a strong signal of genetic structure, thus implying that even if that bridge is used by fire salamanders at all, the number of immigrants is not sufficient to counteract the effects of genetic drift. It is possible the role of traffic bridges in promoting dispersal across rivers is complex because many interacting factors may be at play, such as bridge age and length, nocturnal traffic volume (which is positively correlated with road mortality), and the genetic effects imposed by the river itself. Collecting movement data can provide a clearer picture about this subject (e.g. capture-mark-recapture techniques such as PIT-tag based analyses; Hendrix et al. 2017).
We were not able to detect a clear signal of historical genetic structure based on the amplified fragment of a mtDNA marker. While this is congruent with the relatively shallow phylogeographic structure at a broader spatial scale shown by northwestern Iberian populations of S. s. gallaica (Velo-Antón et al. 2007; Lourenço et al. 2018b), it is not in agreement with the historical genetic structure and diversity of haplotypes found in S. s. bernardezi (Lourenço et al. 2019). These results are partially explained by possible artifacts resulting from the short amplified mtDNA fragment (315 bp), which prevents the detection of a higher number of unique haplotypes that might occur in each riverside. The use of high-resolution genomic-scale data (e.g. from reduced representation approaches such as RADseq; Burgon et al. 2021) can increase the power to detect both historical and subtle patterns of genetic population structure (e.g. McCartney-Melstad et al. 2018) and, hence, provide more accurate insights regarding the short- and long-term effects of rivers at fine spatial scales.
Conclusions
Unlike previous studies performed at a regional scale in S. salamandra (Lourenço et al. 2019), our individual- and population-based genetic analyses carried out at a fine spatial scale provided detailed insights on how rivers influence patterns of gene flow in larviparous and pueriparous populations of S. salamandra. Our study partially supports the RBH in both reproductive modes, highlighting the key effect of life-history (i.e. reproductive mode) and physical properties of rivers in explaining current patterns of genetic variation across riversides. Specifically, the comparative framework employed in this study somewhat reinforces the hypothesis that rivers comprise more effective obstacles to dispersal and gene flow in terrestrial-breeding amphibians compared to aquatic ones, although other factors (river width and potentially crossing structures) may also play a significant role. This study will stimulate future research to better understand how life history and landscape features govern population connectivity in a threatened group, such as amphibians.
Availability of data and materials
Sequences generated for the mtDNA cytb fragment have been submitted to GenBank and the dataset containing microsatellite data will be provided by the corresponding author upon request.
References
Alarcón-Ríos L, Nicieza AG, Lourenço A, Velo-Antón G (2020a) The evolution of pueriparity maintains multiple paternity in a polymorphic viviparous salamander. Sci Rep 10:14744
Alarcón-Ríos L, Nicieza AG, Kaliontzopoulou A, Buckley D, Velo-Antón G (2020b) Evolutionary history and not heterochronic modifications associated with viviparity drive head shape differentiation in a reproductive polymorphic species, Salamandra salamandra. Evol Biol 47:43–55
Beebee TJ (2013) Effects of road mortality and mitigation measures on amphibian populations. Conserv Biol 27:657–668
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Beukema W, De Pous P, Donaire D, Escoriza D, Bogaerts S, Toxopeus AG, de Bie CAJM, Roca J, Carranza S (2010) Biogeography and contemporary climatic differentiation among Moroccan Salamandra algira. Biol J Linn Soc 101:626–641
Blackburn DG (2015) Evolution of vertebrate viviparity and specializations for fetal nutrition: a quantitative and qualitative analysis. J Morphol 276:961–990
Blair C, Weigel DE, Balazik M, Keeley ATH, Walker FM, Landguth E, Cushman SAM, Murphy M, Waits L, Balkenhol N (2012) A simulation-based evaluation of methods for inferring linear barriers to gene flow. Mol Ecol Resour 12:822–833
Buckley D, Alcobendas M, García-París M, Wake MH (2007) Heterochrony, cannibalism, and the evolution of viviparity in Salamandra salamandra. Evol Dev 9:105–115
Burgon JD, Vences M, Steinfartz S, Bogaerts S, Bonato L, Donaire-Barroso D, Martínez-Solano I, Velo-Antón G, Vieites D, Mable BK, Elmer KR (2021). Phylogenomic inference of species and subspecies diversity in the Palearctic salamander genus Salamandra. Mol Phylogenet Evol 157:107063. https://doi.org/10.1016/j.ympev.2020.107063
Bush JW, Hu DL (2006) Walking on water: biolocomotion at the interface. Annu Rev Fluid Mech 38:339–369
Carvalho F, Mira A (2011) Comparing annual vertebrate road kills over two time periods, 9 years apart: a case study in Mediterranean farmland. Eur J Wildl Res 57:157–174
Chybicki IJ, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100:106–113
Clement M, Posada DCKA, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Cosgrove AJ, McWhorter TJ, Maron M (2018) Consequences of impediments to animal movements at different scales: a conceptual framework and review. Divers Distrib 24:448–459
Crump ML (2015) Anuran reproductive modes: evolving perspectives. J Herpetol 49:1–16
Cushman SA, McKelvey KS, Hayden J, Schwartz MK (2006) Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168:486–499
Deus E, Silva JS, Castro-Díez P, Lomba A, Ortiz ML, Vicente J (2018) Current and future conflicts between eucalypt plantations and high biodiversity areas in the Iberian Peninsula. J Nat Conserv 45:107–117
Dunstone N (2007) Adaptations to the semi-aquatic habit and habitat. In: Dunstone N, Gorman ML (eds) Behaviour and ecology of riparian mammals. Cambridge University Press, New York, pp 1–16
Fouquet A, Ledoux JB, Dubut V, Noonan BP, Scotti I (2012) The interplay of dispersal limitation, rivers, and historical events shapes the genetic structure of an Amazonian frog. Biol J Linn Soc 106:356–373
Fouquet A, Courtois EA, Baudain D, Lima JD, Souza SM, Noonan BP, Rodrigues M (2015) The trans-riverine genetic structure of 28 Amazonian frog species is dependent on life history. J Trop Ecol 31:361–373
Frantz AC, Pope LC, Etherington TR, Wilson GJ, Burke T (2010) Using isolation-by-distance-based approaches to assess the barrier effect of linear landscape elements on badger (Meles meles) dispersal. Mol Ecol 19:1663–1674
Funk WC, Caldwell JP, Peden CE, Padial JM, De la Riva I, Cannatella DC (2007) Tests of biogeographic hypotheses for diversification in the Amazonian forest frog, Physalaemus petersi. Mol Phylogenet Evol 44:825–837
García VOS, Ivy C, Fu J (2017) Syntopic frogs reveal different patterns of interaction with the landscape: a comparative landscape genetic study of Pelophylax nigromaculatus and Fejervarya limnocharis from central China. Ecol Evol 7:9294–9306
García-París M, Alcobendas M, Buckley D, Wake D (2003) Dispersal of viviparity across contact zones in Iberian populations of Fire salamanders (Salamandra) inferred from discordance of genetic and morphological traits. Evolution 57:129–143
Godinho MBC, Da Silva FR (2018) The influence of riverine barriers, climate, and topography on the biogeographic regionalization of Amazonian anurans. Sci Rep 8:3427
Greven H (2003) Larviparity and pueriparity. In: Jamieson BGM, Sever DM (eds) Reproductive biology and phylogeny of urodela. Science Publishers, Enfield, pp 447–475
Grummer JA, Leaché AD (2017) Do dams also stop frogs? Assessing population connectivity of coastal tailed frogs (Ascaphus truei) in the North Cascades National Park Service Complex. Conserv Genet 18:439–451
Hall JP, Harvey DJ (2002) The phylogeography of Amazonia revisited: new evidence from riodinid butterflies. Evolution 56:1489–1497
Hendrix R, Hauswaldt JS, Veith M, Steinfartz S (2010) Strong correlation between cross-amplification success and genetic distance across all members of ‘True salamanders’ (Amphibia: Salamandridae) revealed by Salamandra salamandra-specific microsatellite loci. Mol Ecol Resour 10:1038–1047
Hendrix R, Schmidt BR, Schaub M, Krause ET, Steinfartz S (2017) Differentiation of movement behaviour in an adaptively diverging salamander population. Mol Ecol 26:6400–6413
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Res 9:1322–1332
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodӧhl PA (2013) diveRsity: an R package for the estimation of population genetics parameters and their associated errors. Methods Ecol Evol 4:782–788
Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial-DNA evolution in animals - amplification and sequencing with conserved primers. PNAS 86:6196–6200
Kopuchian C, Campagna L, Lijtmaer DA, Cabanne GS, García NC, Lavinia PD, Tubaro PL, Lovette I, Di Giacomo AS (2020) A test of the riverine barrier hypothesis in the largest subtropical river basin in the Neotropics. Mol Ecol 29:2137–2149
Liedtke HC, Müller H, Hafner J, Penner J, Gower DJ, Mazuch T, Rӧdel M-O, Loader SP (2017) Terrestrial reproduction as an adaptation to steep terrain in African toads. Proc R Soc B 284:20162598
Lion MB, Mazzochini GG, Garda AA, Lee MT, Bickford D, Costa GC, Fonseca CR (2019) Global patterns of terrestriality in amphibian reproduction. Global Ecol Biogeogr 28:744–756
Lourenço A, Antunes B, Wang IJ, Velo-Antón G (2018a) Fine-scale genetic structure in a salamander with two reproductive modes: does reproductive mode affect dispersal? Evol Ecol 1(32):699–732
Lourenço A, Sequeira F, Buckley D, Velo-Antón G (2018b) Role of colonization history and species-specific traits on contemporary genetic variation of two salamander species in a Holocene island-mainland system. J Biogeogr 45:1054–1066
Lourenço A, Gonçalves J, Carvalho F, Wang IJ, Velo-Antón G (2019) Comparative landscape genetics reveals the evolution of viviparity reduces genetic connectivity in fire salamanders. Mol Ecol 28:4573–4591
Lowe WH, Allendorf FW (2010) What can genetics tell us about population connectivity? Mol Ecol 19:3038–3051
Luqman H, Muller R, Vaupel A, Brodbeck S, Bolliger J, Gugerli F (2018) No distinct barrier effects of highways and a wide river on the genetic structure of the Alpine newt (Ichthyosaura alpestris) in densely settled landscapes. Conserv Genet 19:673–685
Marsh DM, Page RB, Hanlon TJ, Bareke H, Corritone R, Jetter N, Beckman NG, Gardner K, Seifert DE, Cabe PR (2007) Ecological and genetic evidence that low-order streams inhibit dispersal by red-backed salamanders (Plethodon cinereus). Can J Zool 85:319–327
McCartney-Melstad E, Vu JK, Shaffer HB (2018) Genomic data recover previously undetectable fragmentation effects in an endangered amphibian. Mol Ecol 27:4430–4443
Meirmans PG (2012) The trouble with isolation by distance. Mol Ecol 21:2839–2846
Moen DS (2019) What determines the distinct morphology of species with a particular ecology? The roles of many-to-one mapping and trade-offs in the evolution of frog ecomorphology and performance. Am Nat 194:E81–E95
Moraes LJCL, Pavan D, Barros MC, Ribas CC (2016) The combined influence of riverine barriers and flooding gradients on biogeographical patterns for amphibians and squamates in south-eastern Amazonia. J Biogeogr 43:2113–2124
Moritz C, Schneider CJ, Wake DB (1992) Evolutionary relationships within the Ensatina eschscholtzii complex confirm the ring species interpretation. Syst Biol 41:273–291
Nazareno AG, Dick CW, Lohmann LG (2017) Wide but not impermeable: testing the riverine barrier hypothesis for an Amazonian plant species. Mol Ecol 26:3636–3648
Nei M (1977) F-statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet 41:225–233
Nicolas V, Missoup AD, Denys C, Kerbis Peterhans J, Katuala P, Couloux A, Colyn M (2011) The roles of rivers and Pleistocene refugia in shaping genetic diversity in Praomys misonnei in tropical Africa. J Biogeogr 38:191–207
O’Connell KA, Mulder KP, Maldonado J, Currie KL, Ferraro DM (2019) Sampling related individuals within ponds biases estimates of population structure in a pond-breeding amphibian. Ecol Evol 9:3620–3636
Paz A, Ibáñez R, Lips KR, Crawford AJ (2015) Testing the role of ecology and life history in structuring genetic variation across a landscape: a trait-based phylogeographic approach. Mol Ecol 24:3723–3737
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pérez-Espona S, Pérez-Barbería FJ, McLeod JE, Jiggins CD, Gordon IJ, Pemberton JM (2008) Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Mol Ecol 17:981–996
Pirani RM, Werneck FP, Thomaz AT, Kenney ML, Sturaro MJ, Ávila-Pires TC, Peloso PLV, Rodrigues MT, Knowles LL (2019) Testing main Amazonian rivers as barriers across time and space within widespread taxa. J Biogeogr 46:2444–2456
Pittman SE, Osbourn MS, Semlitsch RD (2014) Movement ecology of amphibians: a missing component for understanding population declines. Biol Conserv 169:44–53
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 43:258–275
R Development Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Reinhardt T, Baldauf L, Ilić M, Fink P (2018) Cast away: drift as the main determinant for larval survival in western fire salamanders (Salamandra salamandra) in headwater streams. J Zool 306:171–179
Richardson JL (2012) Divergent landscape effects on population connectivity in two co-occurring amphibian species. Mol Ecol 21:4437–4451
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Sánchez-Montes G, Wang J, Ariño AH, Martínez-Solano I (2018) Mountains as barriers to gene flow in amphibians: quantifying the differential effect of a major mountain ridge on the genetic structure of four sympatric species with different life history traits. J Biogeogr 45:318–331
Santos AM, Cabezas MP, Tavares AI, Xavier R, Branco M (2015) tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics 32:627–628
Segev O, Blaustein L (2014) Influence of water velocity and predation risk on fire salamander (Salamandra infraimmaculata) larval drift among temporary pools in ephemeral streams. Freshw Sci 33:950–957
Semlitsch RD (2008) Differentiating migration and dispersal processes for pond-breeding amphibians. J Wildl Manag 72:260–267
Steinfartz S, Kuesters D, Tautz D (2004) Isolation and characterization of polymorphic tetranucleotide microsatellite loci in the Fire salamander Salamandra salamandra (Amphibia: Caudata). Mol Ecol Resour 4:626–628
Veith M, Baubkus M, Kugel S, Kulpa C, Reifenrath T, Schafft M, Wagner N (2019) Drift compensation in larval European fire salamanders, Salamandra salamandra (Amphibia: Urodela)? Hydrobiologia 828:315–325
Velo-Antón G, Buckley D (2015) Salamandra común – Salamandra salamandra. In: Carrascal LM, Salvador A (eds) Enciclopedia Virtual de los Vertebrados Españoles. Museo Nacional de Ciencias Naturales, Madrid. http://www.vertebradosibericos.org/anfibios/salsal.html
Velo-Antón G, García-París M, Galán P, Cordero Rivera A (2007) The evolution of viviparity in holocene islands: ecological adaptation versus phylogenetic descent along the transition from aquatic to terrestrial environments. J Zool Syst Evol Res 45:345–352
Velo-Antón G, Zamudio KR, Cordero-Rivera A (2012) Genetic drift and rapid evolution of viviparity in insular fire salamanders (Salamandra salamandra). Heredity 108:410–418
Velo-Antón G, Santos X, Sanmartín-Villar I, Cordero-Rivera A, Buckley D (2015) Intraspecific variation in clutch size and maternal investment in pueriparous and larviparous Salamandra salamandra females. Evol Ecol 29:185–204
Wagner N, Harms W, Hildebrandt F, Martens A, Ong SL, Wallrich K, Lötters S, Veith M (2020) Do habitat preferences of European fire salamander (Salamandra salamandra) larvae differ among landscapes? A case study from Western Germany. Salamandra 56:254–264
Waits LP, Luikart G, Taberlet P (2001) Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol 10:249–256
Wake DB, Hanken J (1996) Direct development in the lungless salamanders: what are the consequences for developmental biology, evolution and phylogenesis? Int J Dev Biol 40:859–869
Wallace AR (1852) On the monkeys of the Amazon. Proc Zool Soc Lond 20:107–110
Wang J (2007) Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet Res 89:135–153
Wang J (2011) Coancestry: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour 11:141–145
Wang J (2018) Effects of sampling close relatives on some elementary population genetics analyses. Mol Ecol Resour 18:41–54
Waraniak JM, Fisher JDL, Purcell K, Mushet DM, Stockwell CA (2019) Landscape genetics reveal broad and fine-scale population structure due to landscape features and climate history in the northern leopard frog (Rana pipiens) in North Dakota. Ecol Evol 9:1041–1060
Acknowledgements
We thank Iria Pazos, Bernardo Antunes, and Barbara Correia for assistance during field work, and S. Lopes for laboratory assistance.
Funding
This work was supported by FEDER funds through the Operational Programme for Competitiveness Factors – COMPETE (FCOMP-01-0124-FEDER-028325 and POCI-01-0145-FEDER-006821); and by National Funds through FCT – Foundation for Science and Technology (EVOVIV: PTDC/BIA-EVF/3036/2012; SALOMICS: PTDC/BIA-EVL/28475/2017). GV-A and AL were supported by FCT (IF/01425/2014-CEECIND/00937/2018, PD/BD/106060/2015, respectively). The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Contributions
GV-A conceived the study. GV-A, CF-V, AL conducted field work. CF-V and AL performed laboratory work. CF-V and AL carried out data analysis. CF-V wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
Fieldwork for obtaining tissue samples was done with the corresponding permits from the regional and national administrations (Xunta de Galicia, Ref. 410/2015; Gobierno del Principado de Asturias, Ref. 2016/001092; Portugal Ref. 36490/2016/DRNCN/DGEFF). Sampling procedures were carried out following the Guidelines for Use of Live Amphibians and Reptiles in Field and Laboratory Research, 2nd Edition, revised by the Herpetological Animal Care and Use Committee (HACC) of the American Society of Ichthyologists and Herpetologists, 2004.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix 1
See Table 6.
Appendix 2
See Table 7.
Appendix 3
See Table 8.
Appendix 4
See Table 9.
Appendix 5
See Fig. 6.
Appendix 6
See Fig. 7.
Appendix 7
See Fig. 8.
a Strip plot comparing the frequency of pairs of relatives (RQG I ≥ 0.125) found across riversides (each point represents a population). The blue and red dashed lines represent the mean frequency of relatives across riversides for larviparous and pueriparous populations, respectively. b Strip plot representing pairwise population DEST values for individuals sampled across riversides. Dashed lines represent mean DEST values per reproductive mode
Appendix 8
See Fig. 9.
Images illustrating both DAPC barplots and individual pie charts representing cluster membership proportions to each riverside when K = 2 for larviparous populations. In each barplot, individuals are represented by vertical bars that are partitioned into 2 colours representing its estimated membership coefficient to each riverside. Vertical white lines in the barplots separate riversides. Top-left images represent the density of individuals from each riverside (depicted with different colors) throughout the discriminant function
Appendix 9
See Fig. 10.
Images illustrating both DAPC barplots and individual pie charts representing cluster membership proportions to each riverside when K = 2 for pueriparous populations. In each barplot, individuals are represented by vertical bars that are partitioned into two colours representing its estimated membership coefficient to each riverside. Vertical white lines in the barplots separate riversides. Top-left images represent the density of individuals from each riverside (depicted with different colors) throughout the discriminant function
Rights and permissions
About this article
Cite this article
Figueiredo-Vázquez, C., Lourenço, A. & Velo-Antón, G. Riverine barriers to gene flow in a salamander with both aquatic and terrestrial reproduction. Evol Ecol 35, 483–511 (2021). https://doi.org/10.1007/s10682-021-10114-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-021-10114-z