Abstract
With climate change and ever-increasing population growth, the pace of varietal development needs to be accelerated in order to feed a population of 10 billion by 2050. Non-invasive high-throughput plant phenotyping (HTP) using advanced imaging technology has capabilities to boost the varietal development process. The tremendous data generated with sensor aided HTP have created the big data and problem in the downstream data analysis pipeline. The higher-level abstraction achieved on high dimensional data by multiple hidden layers for function approximation have made deep learning applications in HTP of significant interest. Application of deep learning models to enhance image-based throughput in phenotyping is an emerging and dynamic area of research in plant phenomics. In this comprehensive review we highlighted the recent developments in the field of deep learning application for HTP. The deep learning principles are described and contextualized relative to machine learning and conventional computer vision algorithms. Novel and emerging deep learning applications are identified. Recommendations are provided with the intent of choosing the most suitable models and training strategy for the capturing and predicting sensor-based phenotyping traits. It also includes steps and suggestions for the development and eventual deployment of such models for multi-task phenotyping. Public datasets have been identified and these datasets are reported which can be used for model training and benchmarking. Overall, this study provided a comprehensive overview of deep learning, it’s application in plant phenomics, potential barriers and scope of improvement.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
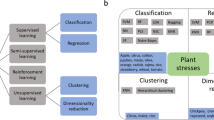
A principal objective of crop improvement is to enhance our understanding of phenotypic characteristics such as growth, yield, disease resistance and agronomic robustness of crops. Multitude of factors related to genotype and environment interact in complex ways which is manifested in form of phenotypic variation. This makes it essential to capture extensive phenotypic data if one wants access to the 'genotype–phenotype’ map (Nyine et al. 2018). However there exists a lag in throughput of plant phenotyping relative to genotypic characterization. Phenomics is the acquisition of high-dimensional phenotypic data on an organism-wide scale (Fahlgren et al. 2015). It is the enabling force to trace causal links between genotypes and environmental factors, studying the genotype–phenotype map, identifying the genetic basis of complex traits (Nyine et al. 2018); (Sandhu et al. 2020) (Sandhu et al. 2021b). As the high dimensional data is inherent to phenomics, it becomes very intensive for modelling and deriving phenotypic information. To relieve the phenotyping bottleneck, high throughput phenotyping is necessary. Here the term high throughput refers to capacity and speed that enables the current phenotyping system to acquire phenotypic data of hundreds of genotypes in a short period of time. Such capabilities inherent to the high-throughput plant phenotyping (HTP) systems allow for screening of mutant populations and detection of quantitative trait loci (QTLs) (Fahlgren et al. 2015). HTP is generally achieved by acquisition of images to quantify traits over a whole growth cycle of crop plants (Ampatzidis and Partel, 2019). The throughput combined with the multitude of imaging sensors causes accumulation of big data. Images are information rich and as such deriving information from vast arrays of images is becoming the new bottleneck in the task of plant phenotyping (Minervini et al. 2015). It has become the rate limiting step in throughput and clog the data analysis pipeline that translates data acquisition to information and finally to insight (Schmidhuber, 2015). Deep learning (DL) application in plant phenomics (Fig. 1) has been emerging as the much-needed catalyst to relieve this bottleneck in HTP. The multidimensional nature of data demands for such models which can extract and model phenotypic traits utilizing the hidden relationships and patterns in the data (Sandhu et al. 2020); (LeCun et al. 2015). This capability of high-level abstraction from data to generate valuable phenotypic information can only be achieved by application and use of DL models for data analysis pipeline in plant phenotyping (Sandhu et al. 2021c).
Illustration of deep learning applications in various areas of plant phenotyping with examples reviewed in this paper. It is obvious that deep learning has already penetrated the research and workflow in HTP across a wide range of applications which ranges from image reconstruction (Li et al. 2017; Yuan et al. 2017), genotype classification (Taghavi Namin et al. 2018), root tips (Yasrab et al. 2019), spikelets (Pound et al. 2018), spike (Hasan et al. 2018), fruits (Sa et al. 2016), hypocotyl length (Sa et al. 2016) and instance segmentation of plant organs (Douarre et al. 2018; Yamamoto et al. 2017; Sadeghi-Tehran et al. 2019; Ghosal et al. 2019)
Earlier the task of computer vision was confined to detection of corners, edges, circles, etc. and features were handcrafted. But with the advancement in powerful computers and ubiquitous nature of imaging technology the computer vision needed to be advanced. Since the last few decades this much needed boost is provided by DL. This includes convolution neural network (CNN), multilayer perceptron (MLP), recurrent neural network (RNN) and generative adversarial networks (GAN) (Wang et al. 2020a, b); (LeCun et al. 2015). There are multiple reviews and surveys about machine learning, DL, and application of machine learning in agriculture (Kamilaris and Prenafeta-Boldú, 2018; Liakos et al. 2018; Magomadov, 2019; Zhu et al. 2018) but it is quite lacking in DL applications for HTP. Also reviews related to phenotyping are present (Araus et al. 2018; Li et al. 2020; Minervini et al. 2015; Singh et al. 2018) but it lacks an exhaustive overview of DL based principles and application in plant phenotyping. The objective of this review is to show why and how DL has been integrated in the fabric of HTP as well as to give exposure to the existing well-organised, annotated, and public datasets and tools to have a head start in this field. Recommendation of models for task specific purpose with its advantage attributed to unique network architecture has been elucidated for deeper understanding into how model architectures are designed, modified, or selected for HTP analysis pipeline.
Deep learning concept and models
DL is a branch of machine learning which comprises a complex model that enables higher-level abstraction in data through multiple nonlinear transformations (Bengio et al. 2015). The word "deep" in "deep learning" emphasises the multitude of hidden layers (i.e., substantial credit assignment path or CAP depth) in DL algorithms through which the data is transformed. The CAP depth refers to a chain of transformations which occurs on input data to yield output. For a feedforward neural network, it is calculated by using the count of hidden layers in the network plus one (Schmidhuber 2015). Deep learning evolved from simple neural networks as a need to achieve a higher level of abstraction to capture intricate structure inherent in the big data made possible by multiple hidden layers as processing units (Bengio et al. 2015; Voulodimos et al. 2018). Percepton is the basic structural unit of such networks which is analogues to biological neurons. Mathematically individual percepton (`f) can be expressed in terms of weight w, bias term b and nonlinear activation function a, which together transforms input x.
\(\prime f = a(wx + b)\)
Stacks of such perception form a layer and the interconnected layer forms a network. When all the nodes are connected then it is called fully connected neural networks (FCNN). Another important type of network is the CNN characterized uniquely by convolution operation which considers the spatial data for learning (Fig. 2). Unlike FCC having a dense weight matrix the CNN is very sparse due to the concept of tied weights (Bengio et al. 2015; Voulodimos et al. 2018). The CNN architecture often features a convolution layer as an automatic feature extraction unit and a fully connected layer to perform classification, regression on the feature extracted (Bresilla et al. 2019; Schmidhuber 2015). The network topology can be understood as a combination of multiple layered operation which includes but not restricted to convolution, non-linear activation and subsampling layer (Jarrett et al. 2009). This layered structure makes network architecture flexible and customizable to use cases (Oquab et al. 2014). This analogous structure design makes DL networks capable of extraction of useful features from locally correlated data points. Three by three window size (3 × 3) convolution kernels has been most popularly (Lin and Guo 2020; Sa et al. 2016) (Sandhu et al. 2020) used but the size of the convolution kernel can be customized with target object size for which we want to preserve the features (Chollet, 2017). Convolution kernel slides over image and produces output image which is then passed to a non-linearly processing layer of the network. The introduction of non-linearity with the use of activation functions in the network leads to a nonlinear feature space that helps in learning abstraction of input data as well as activation patterns (Lin and Guo 2020; Sandhu et al. 2020); (Bresilla et al. 2019).
Network architecture of deep learning networks includes a deep neural networks having input, multiple hidden layers and a final output layer. Each layer consists of multiple neurons which are interconnected to neurons in the subsequent layers, resulting in the formation of a deep network; b pipeline for convolutional neural networks which consist of convolutional, max pool, flatten, dense and output layers. Information flows using multiple filters in the convolutional layer by using input images to provide the results at the output layer using various activation functions and dropouts
CNN has automatic feature extraction capabilities from images for which one of the most important operations is convolution operations performed by the convolution kernel on the image matrix. Kernels are flexible but generally used in odd pixel size dimensions (DeChant et al. 2017). These kernels can be simply understood as filters in the network. As the image passes through these convolution layers, features get extracted which acts as input to a fully connected layer for prediction which can be either discrete (classification) or continuous (regression). The backpropagation during training allows for selection of appropriate features which are relevant for the learning process and keep in check the overfitting. Complex regression tasks can also be accomplished using CNN based models (Sandhu et al. 2020).
Application of deep learning in plant phenotyping
DL includes a multitude of network architecture for example- deep neural networks (DNN), CNN and RNN. In recent years, CNNs have shown usefulness in a large variety of natural language processing and computer vision applications, including segmentation and image classification, and often surpassed state-of-the-art techniques (Mikolov et al. 2013; Szegedy et al. 2017). HTP relies heavily on non-destructive image-based phenotyping (Das Choudhury et al. 2019). A digital image is mathematically a matrix of digital numbers arranged in rows and columns resulting in extensive data. The major challenge is deriving information from such bigdata (Gnädinger and Schmidhalter 2017). In recent years DL based solutions for HTP is an emerging field finding its application in numerous tasks. It has become the go-to state-of-the-art technology for multi-task phenotyping (Michael P. Pound et al. 2017a, b). Deep learning integration in the phenomics image analysis pipeline to achieve high-throughput is depicted in Fig. 3.
An illustration of deep learning applications integrated in the image analysis pipeline to achieve higher-throughput plant phenotyping. One of the first steps includes a data collection, labelling and curation to make it suitable for input. Data obtained can be from multiple sources like RGB, hyperspectral, multispectral, NIR, fluorescence, and LIDAR sensors. Next step involves b model selection and suitable optimization algorithms for faster and efficient model training. Suitable model development for task specific application in iterative process. The back arrow represents tweaking the model selection, architecture and hyperparameters for better accuracy. Once assured of model performance, c model is used for inference on new phenotypic data obtained non-invasively using non-invasive sensors. The back-arrow represents the iterative nature of the model training process because models are not developed in one day, but it needs multiple training with finer adjustment in model hyperparameters for achieving better accuracy reflected performance measures like-MSE, RMSE, F1 score, precision, and recall. Finally, d the inferences are subjected to various data analysis techniques like GWAS, QTL mapping, broad sense heritability etc.as well characterization of plant stress tolerance response based on deep learning model’s inference derived traits. In this way instead of conventional manual labour-based plant phenotyping, deep learning-based application has been streamlined and very deeply integrated into workflow for enhancing the throughout high throughput phenotyping. This ultimately helps translate large phenotypic data into informative phenotypic data for crop improvement
Image classification for plant phenotyping
CNN is simply the best state of the art model than any other for classification tasks (LeCun et al. 2015). It can achieve higher accuracy even in smaller datasets. This is due to the fact that it is capable of automatic extraction of relevant features by convolution operation after training instead of hand-crafted features. The image classification task in plant phenotyping ranges from feature classification (awn, root tips, leaf tip, spike), growth stage detection, genotype classification to classification of various diseases.
Awn and awn-lessness is an important phenotypic trait. Screening genotypes for presence of awn using conventional methods had been challenging. For this purpose, Pound et al. 2017b developed an ACID dataset (Annotated Crop Image Dataset) with 520 wheat crop images of dimension containing a total of 4100 ears and 48,000 spikelets. The images were also tagged for presence or absence of awned phenotype. Awned plants made one third portion of the total ACID dataset. This dataset was then used for training multitask CNN architecture with hourglass structure which classified genotypes along with localizing spikelets and spikes. The stacked hourglass structure allowed for helping in combining hierarchical features from multiple scales which helps to preserve spatial resolution in the network output. At the resolution of 512 × 512 the network achieved a near perfect accuracy of 99% while lowering resolution of input data to 256 × 256 caused accuracy loss (98.39%) (Pound et al. 2017a, b). Wang et al. (2018) used resampled hyperspectral images as input to the CNN model (ResNet) which achieved higher accuracy than traditional machine learning approach for classification of damaged blueberries (Cyanococcus spps) from healthy (Wang et al. 2018). CNN can also perform classification using RGB images of sour lemons (Citrus limon) from the healthy ones at a higher accuracy on images running from low resolution to high resolution images (Jahanbakhshi et al. 2020). Availability of automated remote sensor-based phenotyping systems make non-invasive data collection throughout the life cycle. Such a multitemporal data set has a big advantage that it captures the phenological stages and change in crop over its whole life cycle (Sandhu et al. 2021a, b, c). The only problem is deriving information from such a huge amount of data. This bottleneck can be addressed by CNN-LSTM based networks which can take and derive meaning from multitemporal data (Ghosal et al. 2018; Taghavi Namin et al. 2018). Long short-term memory (LSTM) fully connected layers can solve the problem of long-term dependencies which is helpful to assimilate plant development multi-temporal data. For genotype/phenotype classification CNN_LSTM model proved very useful as it has CNN derived automatic deep feature extraction from images which is then used for classification using LSTM that showed improved classification over time with an accuracy of 0.93 (Taghavi Namin et al. 2018). Phenological stages are important for plant phenotyping. CNN classification model was shown to predict phenological stages CNN based approach for the classification of different phenological stages of multiple crop which included wheat, barley (Hordeum vulgare L.), lentil (Lens culinaris L.), cotton (Gossypium hirsutum L.), pepper (Piper nigrum L.) and maize (Zea mays L.) at different critical stages of crop life cycle (Yalcin 2017). Pretrained AlexNet was used in this study for classification which is fine tuned on a dataset of 400 images sampled in patches of 227 × 227 dimension. The trained model showed higher accuracy for wheat 83.4% as compared to manually extracted textural features trained machine learning model. Similar trend of higher accuracy for CNN was seen in all other crops.
Other than spike, the quantification of traits like number of leap tips, shoot tips and leaf base are important for detailed phenotypic studies. The automated robotic data acquisition demands also a computer vision based automated feature extraction. To attain this automation Pound et al. (2017a, b) used a dataset of root and lead tip labelled images. Image dimensions of 32 × 32 and 64 × 64 features were taken for training classification networks with annotations for its tip and base. Such small dimension images reduce training time as well provide enough feature details. A total number of 43 641 root and 62 118 shoot images were split in a ratio of 80:20. CNN layered architecture with multiple convolution and pooling layers finally culminating into a fully connected layer was used. More than 99% accuracy achieved for both shoot and root classification network. The network was then deployed on the whole image in stride mode for subpixel level classification inorder to localize the tips and based on root and leaves in the whole image. Also the quantitative data thus obtained was then used for QTL discovery for which many QTL regions were successfully identified. Wang et al. (2019) developed WheatNet DL model for detecting multiple phenotypic parameters of wheat under field condition using multi-array assembly of RGB cameras capable of collecting georeferenced proximal images of wheat canopy. WheatNet model consisted of ResNet18 as a base classifier for classifying single images divided in patches followed by a voting layer for prediction of heading percentage, awn and awnless phenotype. The model showed near perfect accuracy of 98.6% for discrimination of awned and awnless phenotype. Predicted traits from CNN showed high broad-sense heritability with a value of 0.99 for heading percentage and 0.51 for rate of heading (Wang et al. 2019).
Regression problems in high-throughput plant phenomics
Unlike classification where outcomes are categorical, regression problems involve continuous value as output. Regression is a very important technique in research for modeling and prediction of traits from training data in plant phenotyping. Earlier it was confined to only numerical data but with CNN image data has been regressed to important plant traits like length, biomass etc. (Zhou et al. 2020). Emergence counting under field condition particularly of wheat is not possible with conventional computer vision algorithms because wheat being a monocot plant has narrow overlapping leaves. Aich et al. (2018) used convolutional blocks which consisted of simple convolution operation followed by local response normalization and rectified nonlinearity with simplified residual blocks to design CNN architecture for biomass estimation and emergence counting on segmented wheat field image (Aich et al. 2018).
The phenotypic data collected on hundreds of genotypes are generally multidimensional in nature. In addition, with the development of inexpensive imaging sensors and Unmanned Aerial Vehicle (UAV) platforms, the task of data acquisition throughput crop life-cycle has become easier (Sankaran et al. 2019). It resulted in a shift of focus from data acquisition to data analysis – extracting significant features and recognizing underlying patterns paving way to significant insights. Conventional data analysis techniques are not able to exploit the benefits of huge and multidimensional data. Dataset from multiple targets (example crop genotype, soil, weather etc.) can be easily assimilated in a DL model for yield prediction for genotypes under varied environment conditions (Khaki and Wang 2019; Sandhu et al. 2020). Hyperspectral dataset are unique in the sense that the images capture over hundred of wavelengths giving its high dimensionality. The spectral data are converted into various indices which are derived from mathematical operations on significant spectral bands that reflect physiological status and health of the plant. Earlier studies used only index-based phenotyping (Zhu et al. 2020), but such index-based phenotyping was not able to get the benefits of DL applications on high-dimensional spectral data as well as some indices like Normalized Difference Vegetation Index (NDVI) gets saturated problem for higher leaf area index (Gitelson, 2004). Recent studies on hyperspectral data spectrally unmixing for spike and leaf pixels as input to DNN (multilayer perceptron) networks showed promising in prediction of crop yield parameters with high correlation (0.79) and low normalized root mean square 0.24 at sub-plot scale (Moghimi et al. 2020). Itakura et al. (2018) used CNN based regression model on fluorescence spectroscopy images for prediction of Brix ratio which is an indicator of fruit maturity. Seed content in individual pods is also an important plant parameter determining yield in podded crops (Itakura et al. 2018). CNN classification architecture was repurposed for regression by using softmax activation function in the output layer to ensure continuous output for regressing pea (Pisum sativum L.) pod image to seed count (Uzal et al. 2018). Similarly, ResNet50 as based model was repurposed for scoring of coffee (Coffea arabica) leaves with an accuracy of 97% for severity estimation (Esgario et al. 2020). Such a system when deployed on edge computing devices might prove to be a suitable tool for real time phenotyping under field conditions (Partel et al. 2019). Genome wide prediction is another area at which the function approximation capacity of CNN model can be exploited because of the high dimensional nature of marker data which is used to predict phenotypes. Sparse one dimensional CNN with l1-norm regularization was found to provide accurate and computationally efficient to estimate phenotypes with accuracy improvement of 25% on simulated data over east Absolute Shrinkage and Selection Operator method (LASOO) (Waldmann et al. 2020).
Feature localisation and counting
Yield determining crop features which are localized in nature include number of spikes, spikelets, number of ears and fruits are of central importance for the task of phenotyping. Feature localization of specific plant organs can be achieved by computer vision and image processing techniques that deals with detecting instances of target semantic objects by predicting coordinates or regions of objects of interest based on annotated training dataset (Gao et al. 2020; Majeed et al. 2020). Popular feature localization algorithms are you-only-look-once (YOLO) (Redmon et al. 2016), single-shot detector (SSD) (Liu et al. 2016) R-CNN (Girshick et al. 2014), and Faster R-CNN (Ren et al. 2017). The YOLO family and the SSD framework are one-stage models which makes it faster but with an added disadvantage that it suffers with extreme imbalances in number of objects detected as well as many times evades detection of small target objects. However, region based CNN (R-CNN), encoder-decoder type networks use region proposal layers which output multiple regions that are subjected to classification for instatance detection resulting in high accuracy(Girshick et al. 2014; Zhang et al. 2018). However, R-CNN has relatively higher training and inference time requirement due to the process of proposing and classifying thousands of regions per image which involved multiple convolutions. This drawback was overcome with the development of Faster-RCNN (F-RCNN) which does only one convolution for region proposal, making it faster and suitable for real-time application while maintaining higher localization accuracy (Gao et al. 2020; Ren et al. 2017). Itzhaky et al. (2018) showed tiny-YOLOv3 (ADAM optimizer with a learning rate of 0.00001) real time inference with higher accuracy (F1 score = 0.94) which was comparable to F-RCNN for leaf detection and counting application on Arabidopsis (Arabidopsis thaliana) (Itzhaky et al. 2018). Stein et al. (2016) used faster R-CNN for light detection and ranging (LIDAR) based detection of fruit for yield estimation of mango (Mangifera indica L.) orchards (Stein et al. 2016). Koirala et al. (2019) designed MangoYOLO with 33 layered architecture (as compared to YOLOv3 having 106 layers) to preserve details of smaller and darker mangoes on tree and achieved a F1 score of 0.968 and average precision of 0.983 on test dataset (refer to Table 1). MangoYOLO(s)-512 achieved the highest average precision (AP = 0.986), with similar results achieved for MangoYOLO(pt) (AP = 0.983), and SSD-300 (AP = 0.982), while the lowest AP was associated with Faster R-CNN-VGG-original (AP = 0.917). The accuracy is not only determined by choice of algorithm but also the input data (Koirala et al. 2019). A study done by Sa et al. (2016) using image fusion techniques for generating fused training data from different imaging sensors showed significant improvement in model performance. It was reflected in higher F1 scores values of models trained on late fused (RGB and near-infrared (NIR)) dataset (0.83) in comparison models trained separately on RGB (0.816) and NIR (0.797) images. The time of fusion in the network also showed to influence detection accuracy with the later stage fusion showering higher accuracy than the early fusion network architecture (F1 score = 0.799) (Sa et al. 2016).
In the case of wheat, the number of spikes is an important yield determining factor. Detection and counting of spikes by non-destructive methods proved to be a significant step forward in the area of HTP (Michael P. Pound et al. 2017a, b). The state-of-the art DL models have shown high detection accuracy avoiding tedious field phenotyping and achieving high-throughput. For control conditions, proximal sensing of a single plant allows for higher resolution images which ultimately can be used to derive more detailed phenotyping traits. Spike and spikelet count with a near perfect accuracy 95.91% and 99.66% can be obtained by just using a simple consumer grade 12 MP camera (Pound et al. 2017a, b). The model used was the encoder and decoder architecture network containing four stacked hourglasses including intermediate supervision. This unique network architecture design allowed for conservation of resolution and relevant features during down-sampling and up-sampling operation. Leaves are the plant's most important organ as it is the seat of photosynthetic processes and dubbed as a food factory for plants. So, the number of leaves is an important phenotyping parameter which determines vegetative growth of plants as well as productivity. However, leaf counting in rosette plants has been a challenging task for computers. Various pioneering studies have been done recently using CNN models for counting plant leaves (Ubbens and Stavness 2017); (Buzzy et al. 2020; Itzhaky et al. 2018). Ubbens and Stavness 2017 used multiple datasets having two arabidopsis (A1 and A2) and one tobacco dataset (A3) image with leaf annotations. The layered CNN architecture with stacked convolution and pooling layers followed by a fully connected layer and ADAM optimizer was trained on RGB images. It showed appreciable counting performance with mean absolute difference (MAD) value of 0.61 for arabidopsis dataset and tobacco dataset. However MAD score was shown to depreciate with A1 dataset due to compact and overlapping nature of leaf phyllotaxy for that accession of arabidopsis. Itzhaky et al. 2018 designed a network with two approaches for leaf counting tasks. The one network architecture was based on regression while the other on detection of keay-point. The regression network was designed with FPN backbone and a count regression submodel. The key-point detection model consisted of detection subnetwork and counting sub-network. Both models optimized with ADAM, showed higher precision of 95% which was improved over its previous counterparts. One of the recent studies has shown the potential of Real time phenotyping for leaf counting with inference time of 0.01 s using TinyYOLO v3 has been shown to achieve a F1 score of 0.94 (Buzzy et al. 2020). The inference time was 1000 times faster to Faster R-CNN (F1 score 0.89).
Segmentation in plant phenotyping
Image segmentation is one of the most fundamental and indispensable image pre-processing steps which allows for masking of images so that only the region of interest remains in images. It can be done for an instance or to segment a semantic in images. Conventionally segmentation was achieved using threshold-based masking which fails when background is complex, particularly under field conditions. CNN comes to the rescue under such conditions as it can reap the benefits of exploiting spatial relationships among pixels using higher level abstraction (Ramcharan et al. 2019). In the plant phenotyping process certain targets in the image, particularly thin and slender objects of interest like root and seedlings are hard to segment from the background. U-Net deep CNN has become a very popular algorithm for segmentation tasks in the plant phenotyping community (Mohanty et al. 2016). The strength of UNet lies in the fact that it is excellent for finding and segmenting thin objects like root and shoot (Dobos et al. 2019). The capability can be seen from the fact that U-Net CNN architecture with smooth Dice coefficient loss function was used for segmentation of seedling’s hypocotyl of different species (Dobos et al. 2019). The segmented skeletal output image was then utilized for hypocotyl length estimation. CNN models are also revolutionizing the throughput of root phenotyping tasks. Earlier root system analysis software was used to be semiautomatic like RootNAV 1.0, SmartRoot (Lobet et al. 2011). It has reached a new height with the development of RootNAV 2.0. RootNAV was originally semi-automatic but with the release of version 2.0 it became powered by a DL algorithm that automates the task of extracting root architecture and makes it ten times faster without sacrificing accuracy (Yasrab et al. 2019). It can generate automatic masks for first and second order roots as well as heatmaps for tip localization. This approach was effective as it seamlessly made it possible to combine the local pixel information to the global scene information (Yasrab et al. 2019).
DL based pipelines for determination of image-based traits in HTP systems have been gaining traction. High throughput capability of such a pipeline using F-RCNN for stalk counting and FCN coupled with ellipse fitting for width determination of the segmented stalk for sorghum (Sorghum bicolor L.) genotypes which achived a accuracy of 0.88 for stalk count and low mean absolute error for stem width determination of 2.77 mm (Baweja et al. 2018). This pipeline was advantageous as F-RCNN is an improvement over the RCNN (Ren et al. 2017), due to inclusion of specialized architecture called region proposal networks (RPN) which predicts object position based on features from the previous layer of VGG-16 network. Similarly, Mask R-CNN when coupled with DBSCAN clustering algorithm allowed automatic segmentation of overlapped poplar seedling leaves (Liu et al. 2020).
It can perform in real time when implemented on a graphics processing unit (GPU) based DL system. A major limitation is that many times it is not able to detect highly overlapping instances of objects due to non-maximal suppression rejection which is prevalent in images acquired under field conditions (Gao et al. 2020). Fully automated measurements from such a pipeline were proved to be accurate to within 10% of human validation measurements for stalk count and 2.76 mm on average measure for stalk width proves robustness of DL for HTP workflow. Such application of automated phenotypic data acquisition can be extended to other crops like sorghum, sugarcane (Saccharum officinarum), maize (Zea mays L). The application of these networks is not limited to RGB images but rather it can be extended to hyperspectral images and LIDAR data (Tsoulias et al. 2019). Feng et al. (2020) reported DL based neural network architecture for semantic segmentation of plants from hyperspectral images for plant phenotyping to have outperformed conventional approaches as revealed by higher value of segmentation performance measures like symmetric best dice (SBD) score for DL pipeline (Feng et al. 2020). However, use of DL for hyperspectral image segmentation might prove to be an overkill when simple machine learning techniques can do a better job due to higher content of information per pixel in hyperspectral images. Miao et al. (2020) used simple machine learning techniques like support vector machine, linear discriminant analysis, and partial least-squares discriminant analysis to achieve pixel based organ-level semantic segmentation of leaf, stalk and panicle of sorghum and maize genotypes to identify genes controlling variations of these phenotyping parameters (Miao et al. 2020). LIDAR images are unique in a way that it gives 3D point cloud information having a depth map of the target object. F-RCNN on compressed 2D versions derived from 3D LIDAR images can be used for individual maize segmentation (Jin et al. 2018). DL is proving to be essential for plant phenotyping for biotic stress tolerance. Root phenotyping is challenging considering the factors that there is very less contrast between soil and roots. Douarre et al. (2018) used transfer learning on X-ray computed tomography images to segment roots from the soil. Thus while semantic segmentation is useful for masking the whole object from the background, the instance based segmentation serves the purpose of specified plant parts like spike, fruits, roots (Douarre et al. 2018); (Yamamoto et al. 2017). Combining the best features of pre-existing models to make a new model for achieving multitasking phenotyping capabilities and achieving better accuracy have become a new trend. Misra et al. (2020) leveraged the Local Patch extraction Network (LPNet) and Global Mask refinement Network (GMRNet) to design SpikeSegNet with capabilities for segmentation of spike from wheat RGB image grown in controlled phenotyping facility in pots (Misra et al. 2020). The network trained on RGB data of wheat grown in pots for 200 epochs achieved a near perfect accuracy of 0.99 for spike segmentation and 0.95 for spike counting.
The model accuracy, precision and recall all depends on the data provided to the model. Image characteristics like contrast, resolution, quantization and number of bands play an important part in determining the accuracy of the trained CNN model (Yamamoto et al. 2017). To enhance contrast, sometimes only RGB imaging does not prove sufficient. This is often the case with field phenotyping using only RGB sensors. However, multiple image fusion techniques have been devised so that information from a multitude of sensors can be used in unison to achieve better predictions. The fusion of RGB image with multispectral image sensor provided much more contrast and thus performed better in segmentation (Lu et al. 2017).
Deep learning-based field phenotyping
Spike characteristic is a proxy measure for grain production. Under field conditions performing this task become complex with traditional methods due to overlapping of features as well as variability in visibility conditions. The R-CNN model has been found to perform with high accuracy (93.4%) and F1 score (0.95) for the detection and counting of spikes under field conditions (Hasan et al. 2018). Such DL techniques on RGB images are crucial for high-throughput quantitative assessment of a multitude of traits. As the data so is the model performance, got reflected in fact that model trained on higher colour contrast dataset between the object of interest and the background (yellow plant and green spike stage of wheat) showed better accuracy, over dataset having lower contrast between plant and spike (when both spike and plant were green and in later stages when both plant and spike were yellow). The case of field phenotyping is totally different because the background under field condition is very different (Ghosal et al. 2018). Simple linear iterative clustering (SLIC) which helps to develop masks using super-pixels is becoming an algorithm of choice for masking vegetation in field conditions with low computational cost. Output masked images can be fed to DL networks for counting. Sadeghi-Tehran et al. (2019) with SLIC and binary cross-entropy loss function leveraged the U-Net model with VGG model without fully connected layers as its encoder mechanism to develop DeepCount model for quantification of spikes under field conditions for large-scale breeding programs (Sadeghi-Tehran et al. 2019). These models outperformed the handcrafted image-based methods with a correlation coefficient value of 0.94 and root mean square of 25.1 which was attributed to DL model ability to handle the complexity of movements, shadow and overlap under field conditions. UAV is becoming a popular platform for field based plant phenotyping (Sankaran et al. 2019); (Ghosal et al. 2018). Lin and Guo (2020) used U-NET CNN model for segmentation of sorghum panicles based on UAV acquired data followed by its counting using open-source computer vision library (OpenCV) contour based functions. The U-Net architecture is so popular in biological application as its contracting part captures context and an asymmetric expanding path enables precise localization (Lin and Guo 2020). The model concatenates the encoder feature maps to up-sampled feature maps from the decoder at every stage. Other encoder and decoder type models are also very popular in such tasks. Field phenotyping calls for integration of DL with robotics and other computer vision techniques for development of DL based phenotyping solutions. Complex pipelines involving multiple algorithms are becoming a new trend to solve real world field phenotyping challenges for example Stalknet (Baweja et al. 2018). The Stalknet pipeline comprises Faster R-CNN, FCN and OpenCV based algorithms. Data flow through Faster R-CNN to give tiller count number, FCN acts as a masking module for the stalk while OpenCV ellipse fitting algorithms determined the width of the masked stalk using stereo images and the output of the masked stalk images from the FCN layer (Gao et al. 2020). This higher level of integration among algorithms allowed for synergistic effect allowing for multiple tasks like counting and width estimation with higher accuracy. In sorghum head detection and counting using UAV acquired images RetinaNet with ResNet-50 backbone was shown to give close estimates to the true count that too achieved with less training dataset (only 40 labelled images) for semi-trained CNN model (Ghosal et al. 2018). The model on transfer learning over only 40 epochs showed a high correlation coefficient of 0.88 and mean average precision of 0.94 for sorghum head labelling and counting respectively.
DL backed phenotypic data acquisition and hyperspectral image reconstruction
Other than the general trend of acquisition and prediction of plant traits there are numerous applications due to the inherent flexibility of DL models. It becomes obvious with the work of Wu et al. (2019) which showed that the decision making process for next optimal viewpoints of multiple robotic arms deployed simultaneously (for 3D images based plant phenotyping) can be driven by point completion network (PCN) based DL with high accuracy (Wu et al. 2019). So, DL driven robots can accelerate the data acquisition process making it autonomous and efficient (Ferentinos, 2018). Hyperspectral and multispectral imaging proved to be indispensable for plant phenotyping as it reveals the physiological characteristics of plants which RGB sensors cannot achieve at that scale (Yendrek et al. 2017) Zhao et al. (2020) used UAV acquired multispectral imagery for modelling vegetation index from RGB image (Zhao et al. 2020). The reconstruction of hyperspectral data from the down sampled version of the data can acts as opportunity to derive hyperspectral images form multispectral sensor (Feng et al. 2020) and also derive super-resolution hyperspectral images from low resolution data ((Li et al. 2017); (Yuan et al. 2017). Xiong et al. (2017) used HsCNN model for hyperspectral image recovery under sampled projections. The HsCNN model features an encoder-decoder layer with skips connections to allow for image reconstruction without loss of low-level details. The trained HsCNN model for 150 epochs achieved normalized root mean square error value of 0.38. RGB images are ubiquitous and affordable while hyperspectral sensors are expensive and require technical expertise (Xiong et al. 2017). So many hyperspectral image reconstruction from RGB cameras using sparse coding (Arad and Ben-Shahar, 2016) and DL (Shi et al. 2018) were attempted with appreciable accuracy. Shi et al. (2018) designed HsCNN + model pair (HsCNN-R and HsCNN-D) for hyperspectral reconstruction which achieved a mean relative absolute error of 0.014. The HsCNN-R having residual block allowed for propagation of smaller details as well as eliminated the requirement of designing hand-crafted down-sampling. In the HsCNN-D the network architecture featured densely connected structure allowing for going more deeper for high fidelity hyperspectral reconstruction (refer to Table 1). A major limitation of using RGB images as input to DL-CNN models for hyperspectral reconstruction is that RGB images are tuned for human vision making their spectral responses not optimal for hyperspectral reconstruction. To overcome this limitation, Nie et al. (2018) used DL derived spectral response functions instead of spectral response for hyperspectral image reconstruction from RGB data (Nie et al. 2018).
Strategy for development and deployment of deep learning models for high throughput plant phenotyping
DL models are complex in its architecture. Inorder to develop and deploy such models’ various strategies are available. It ranges from using pre-trained models directly to designing custom model architecture. The strategy to choose depends on the application and type of dataset in HTP. If datasets are almost similar, then trained models directly or transfer learning can be used. However, when datasets on which model is developed and on which inference is to be made are not similar then customized model architecture needs to be designed. Furthermore, DL model choice (listed in Table 2) is an important factor which is usually based on dataset, computing power available and task specific application. Some of the examples of recent studies as well as technical details with advantage and limitations are elucidated below.
Use of pre-trained models directly
From recent studies, we found a trend of sharing pre-trained DL models for specific tasks. One such example is for canola flower and rosette leaf counting in a proper software format named “Deep plant phenomics” (Ubbens and Stavness 2017). Sharing a CNN is easy with new repository and version management tools like github. Vegetation segmentation network is publicly available with the capability to achieve automatic segmentation of vegetation of interest from background pixels and thus speeding up the process of image pre-processing using DL. Other specialized task contribution from the research community involves Hypocotyl UNet model (Dobos et al. 2019) which can be downloaded and easily deployed for high-throughput estimation of hypocotyl length in seedling (https://github.com/biomag-lab/hypocotyl-UNet) (Dobos et al. 2019).
Transfer learning from pretrained models
DL networks as of now are very task specific based on the training dataset and intended use. Such trained models often cannot be used directly but instead, can be used as a starting point for DL application in plant phenomics on similar datasets (Liu et al. 2017). This is possible as the network architecture can be customized, and trainable parameters can be updated in the network such that it easily maps input to output for the new dataset. The changes and updates are usually at the end of the network in FCC layer with respect to number (based on number of class) and type of outputs (discrete or continuous) at the FCC layer and the rest of the CNN network are generally kept frozen for using it as just feature extractors. The term transfer refers to the fact that a major portion of model weight and trainable parameters are frozen that help to exploit previously learned features (Ward and Moghadam 2020). In addition to this, a plant phenotyping expert can choose from a wide variety of network architecture based on specific task, computational power available and nature of input data (Canziani et al. 2016). To give a few example-MLP for regression and classification, UNet for image segmentation task, ResNet (He et al. 2016), Inception-ResNet (Szegedy et al. 2017), AlexNet (Krizhevsky et al. 2012), GoogleNet (Szegedy et al. 2017), MobileNet (Howard et al. 2017) and VGG (Simonyan and Zisserman 2014) for image classification, RNN (Mandic and Chambers 2001) and LSTM for time-series data, Faster RCNN (Mao et al. 2018), YOLO (Redmon et al. 2016) and SSD (Liu et al. 2016) for object detection.
MLP models are the most basic deep neural networks which are composed of a series of fully connected layers (Collobert et al. 2011); (Sandhu et al. 2020) with hidden layers greater than four which help to learn from higher level features to solve regression and classification problems. U-net is revolutionizing image segmentation with its unique ability to segment even hard to segment objects using autoencoder-decoder like hourglass structure (Ronneberger et al. 2015). Autoencoders are unique DL architectures which use a contracting and an expanding branch to find representations of the input of a lower dimensionality (Vincent et al. 2008). In short it can be understood as analogous to principal component analysis but with non-linearity included. The ResNet became a very popular model for image classification tasks due to the fact that it overcomes the problem of accuracy less with increase in network depth by using residual blocks (Sladojevic et al. 2016). The residual blocks improve model performance to a large extent while allowing to make deeper networks by acting as a shortcut to data flow through the DL network architecture and also behave like ensembles of classifiers (Veit et al. 2016). GoogleNet is also an advanced model architecture as it features inception blocks that permits convolution and pooling operation in parallel to model locality and abstraction (Szegedy et al. 2015). Also, the network learns which path of convolution and pooling operation to take for solving computer vision problems at hand. For making predictions on sequential data (time-series data) RNNs can be used as it takes care of long term dependencies in the input data by use of state variables that permits cells to have memory and model any finite state machine (Mandic and Chambers 2001). However, learning those long-term dependencies is challenging for RNN. To solve this problem LSTM networks (Hochreiter and Schmidhuber 1997) and gated recurrent units (GRU) were designed which have long term memory learning inherent in the network architecture. So based on application, input data and network unique structural design one should choose appropriate models (Chung et al. 2014).
Deep-CNN features are more generic in early layers and more dataset-specific in later layers, there are four major scenarios. So, when the dataset is small and contrasts with the original dataset on which the model is trained, which is often the case for phenomics dataset, we should go for fine tuning only the last few layers for the model to be efficient. Tapas (2016) used Inception model v3 for classifying Arabidopsis and tobacco (Nicotiana tabacum L.) plants with an accuracy of 98% on a small dataset of only 284 images (Tapas 2016). RootNAV 2.0 software for root phenotyping was developed using a DL model trained on a high-resolution wheat root database. The trained model used in RootNav 2.0 when retained using transfer learning approach on images of Brassica napus from a hydroponic assay, demonstrated good accuracy despite the small dataset (Yasrab et al. 2019). The application can be further extended to high-throughput root cross section studies. F-RCNN model trained on rice (Oryza sativa) root cross-section images was able to accurately detect root, stele and late metaxylem objects (Wang et al. 2020a, b). Ghosal et al. (2019) deployed ResNet for counting sorghum heads in UAV acquired images with just 40 labelled images. This shows that transfer learning breaks the myth that deploying DL based solutions for phenotyping tasks will require a large amount of training dataset (Ghosal et al. 2019). The added advantage of transfer learning aloowed for less training time and lower computational requirements as the network converged over 40 epochs and achived appreciable performance mean average precision (mAP = 0.94) (Pan and Yang 2010). On the other hand, it is disadvantageous as transfer learning for a given DL model can only be used for similar types of problems, not for radically different tasks. Also, the availability of large-scale public dataset and pre-trained models are not exhaustive and extensive till now and need further research, development and archiving of data.
Deep learning model development from scratch
It might sound easy and efficient to use transfer learning in all possible cases but when the training dataset available is large and different in content from the original dataset (on which the model was trained), training network from scratch has been proved to be more beneficial (Zhuang et al. 2020). DL from scratch requires lots of data for function approximation as well as network design customized to data. This has been shown to add advantage for the model robustness to the new datasets. The major drawback is that acquiring and pre-processing such a large dataset is a tedious job. However, with image augmentation techniques applied on input dataset by applying distortions (brightness, contrast, cropping, and flipping) help in generation of more images with slight variations. This allows the network to see new images every time during training making the resultant trained model more general by preventing the overfitting. Most of the training strategy followed the 80–20 train-test split which helps in “early stopping” and thus prevent overfitting (Kingma and Welling 2019). Data augmentation also can be achieved by harnessing high fidelity simulation for target objects and can help in generating training data. Ubbens et al. (2018) proved the pragmatic characteristic of this approach by using synthetic plant data for training set data augmentation to ensure robustness of the DL model development for counting leaves on model plant (Arabidopsis) (Ubbens et al. 2018). The successful generation of supplementary 2D data training images from the 3D model of the plant improved the accuracy of the resultant model. Aich et al. (2018) designed a network which included convolution blocks for feature extraction and similar to inception network for faster training as well to allow for deeper layers without loss of accuracy for biomass and emergence estimation (Aich et al. 2018). Koirala et al. (2019) designed new architecture called MangoYOLO. For this YOLO architecture was restructured to constitute only 33 layers for achieving optimal tradeoff between higher inference speed as well as accuracy tradeoff compared to tiny-YOLOv2 (16 layered) and YOLOv3 (166 layered) for detection of mangoes (Koirala et al. 2019).
Tool for development and deployment of deep learning model for high throughput plant phenotyping
In recent decades, many tools have been developed for development and deployment of DL models. Some of the earlier libraries were Theano (Bastien et al. 2012) and Caffe (Jia et al. 2014) characterized by modular design. However the clear winner based on use in recent studies has been the tensorflow framework (Abadi et al. 2016) and higher level libraries like keras (Chollet, 2017). Higher level application programming interfaces are very popular among the plant phenotyping communtiy as they are easier to use and understand. On top of that Ubbens and Stavness (2017) developed higher level framework based on keras (Deep Plant Phenomics) having models customized for performing HTP tasks like rosette leaf counting, vegetation segmentation etc. with some pretrained models available (https://deep-plant-phenomics.readthedocs.io/en/latest/Tools/) (Ubbens and Stavness 2017). Other popular frameworks used are Microsoft Cognitive Toolkit (CNTK, (Seide and Agarwal 2016), Pytorch (Team, 2017) etc. Also ready to use software like RootNAV 2.0 (Yasrab et al. 2019) and pretrained models available in the public domain can be deployed quickly and easily with little knowledge of computers for relieving the data analysis bottleneck in phenomics. Nowadays AutoML and neural architecture search (NAS) algorithms are paving the way for computing power based neural architecture design which has the ability to generate optimized model architecture on the fly during training (Zhang et al. 2019). Hence it eliminates the need for manually designing network architecture as network architecture design optimization can now be done by computers itself (Abdelfattah et al. 2020). However, it has limitation that a very high computing power is required to train such model.
Publicly available datasets for high throughput plant phenotyping
Labelling, annotating, organising, and preprocessing images are labour and time intensive tasks which limit new researchers in participation and quick hands in DL based application for HTP. However, availability of organised and labelled datasets (listed in Table 3) from the research community overcomes this limitation. It enabled new researchers to have direct first hand experience of DL based solutions for plant phenotyping. The Annotated Crop Image Database (ACID) database is one such database. As the name database suggests, it consists of hundreds of annotated crop images. It has image datasets for root system architecture and annotations in root system markup language of wheat seedlings grown hydroponically for root tip detection, images of hydroponic wheat, oilseed rape and Arabidopsis plates roots (Michael P Pound et al. 2017a, b) as well as wheat spike dataset with 500 images and its json annotation for spike as well as spikelets (Pound et al. 2018). Other public datasets available are dominated by Arabidopsis plant images (Bell and Dee 2016; Minervini et al. 2015). The yield related plant organ like fruit and spike is also a popular category of dataset available. Many attempts have been made in recent studies to provide access to wheat spike dataset annotated for detection (Hasan et al. 2018) as well as for spike segmentation (Sadeghi-Tehran et al. 2019). The DeepFruit dataset is important for phenotyping of horticulture crops as it contains annotated dataset of various fruit types (Sa et al. 2016). Next group of dataset are seedling datasets consisting of various species (Giselsson et al. 2017) as well as specialized parts like hypocotyl (Dobos et al. 2019). From a variety of datasets one can choose any dataset for DL based applications in plant phenotyping, model development and finally benchmarking. Furthermore, freely available dataset are acting as fuel, accelerating the pace of integration of DL in HTP.
Challenges in deep learning for plant phenotyping
DL may seem lucrative in every scenario, but it is challenging in situations where the amount of training data is less. Data augmentation techniques alone are not enough to overcome such limitations. Furthermore, preparation of training dataset particularly for feature localization and counting as well as image segmentation is a very tedious and intensive process. It requires special tools and several hours to organize the dataset in proper format.
DL based application in plant phenomics has its unique challenges which are specific to this field. One of the major challenges for new researchers from plant breeding and agriculture is the requirement of a high level of technical and computing skills required for application of deep learning in plant phenotyping. Some studies attempted to ease the adoption of technology with the development of graphical user interface (GUI) tools for example RootNav2 (Yasrab et al. 2019). However there is a lack of such easy to use GUI tools for quick development and deployment of DL models. Hence translating research to mass adoption in phenomics facilities around the world for real time inference is still lagging. It is further complicated by the fact that DL model deployment for inference requires various dependencies which are related to both hardware and software. Some efforts have been made in development of dedicated hardware for quick DL deployment on Nvidia edge computing devices (Yamamoto et al. 2017); (Milioto et al. 2018; Partel et al. 2019). However, application of such ready to use devices and alternative options needs to be further explored for potential real time application to achieve higher throughput in plant phenotyping.
Another unique challenge specific to this discipline is the nature of sensor and datasets acquired. Majority of the studies in deep learning is confined to RGB image data. Hundreds of algorithms suited for such a dataset were specifically designed for better accuracy and computational efficiency as discussed in this review. In phenomics, multiple sensors like fluorescence, multispectral and hyperspectral are used which acquire datasets that have different features and information than simple RGB images (Signoroni et al. 2019). On one side hyperspectral data are high-dimensional having both spectral and spatial information while Fluorescence data has temporal information of fluorescence induction curve which captures plant physiological parameters. Exploiting such unique information for attaining better accuracy as well as with computational efficiency required designing of novel CNN architecture. This area needs further research so that data from all the sensors can be assimilated in the DL network for a comprehensive phenotyping. Phenomics image data are captured throughout the crop growth cycle which makes it essential for using the multitemporal dataset for model training. But only few studies were found that used temporal data for HTP (Taghavi Namin et al. 2018). Further there are only a few studies using image fusion techniques for DL based HTP. But integration of image from different sources is necessary to model complex traits in plant phenotyping like fresh biomass, yield etc. (Sa et al. 2016). Another major challenge is that most of research is done in controlled conditions and thus escape challenges associated with field environments, for example- occlusion, variable light conditions amongst several others (Fahlgren et al. 2015). Benchmarking of models after is an essential task for comparison and standardization to ensure wider adaptability among the plant phenomics community and performance. But benchmarking for better comparison and standardization is still a challenging task due to lack of availability of exhaustive publicly available datasets (Giselsson et al. 2017; Minervini et al. 2015). It is going to become a more pivotal issue for wider adoption of DL models among diverse plant phenomics communities which are growing faster than ever before.
Conclusion and future prospects
DL has expanded to almost all the areas of plant phenotyping. It has solved most of computer vision-based plant phenotyping problems which was thought to be impossible a few years back. Its accuracy and practicality on multi-dimensional data (images and multiple sensor data) has led to a recent surge in DL based HTP pipeline to derive useful information from big data culminating to novel insight in plant phenotyping. The level of automation and phenotypic information extracted using DL models is colossal in accelerating the throughput of current phenotyping systems.
In this paper we have identified not only DL applications in plant phenotyping but also elucidated the principle and unique architectural features associated with DL models as well as suggestions and best practices regarding selection criteria of DL models for task specific applications in HTP. We recommend using the latest model like F-RCNN over RCNN, ResNet over VGG, HsCNN + over HsCNN for serving benefits from the latest development in model architecture. Furthermore, datasets should also be captured in such a way that it accounts for variability in environmental conditions. This paradigm shift has made imaging and multiple sensor technology more inclusive to achieve the end goal of releaving data analysis bottleneck to achieve high-throughput phenotyping. Application of DL based prediction of yield, biophysical and biochemical parameters from multidimensional data acquired from multiple sensors (RGB, hyperspectral, thermal) and in real time on various phenotyping platforms (like UAV or greenhouse phenotyping facilities) need to be further explored. GPU intensive stream data processing pipelines need to be explored for near-real time applications.
However, pace of adoption has continued to accelerate with the popularization of transfer learning due to less requirement of dataset size, training time and custom model architecture, which have made it more and more inclusive for everyone. These pretrained models can be easily deployed and transferred in any corner of the world enhancing the dissemination of DL based solutions in HTP. However, standardization and benchmarking of the model will ensure wider adaptability and uses. DL based plant phenotyping is also a gateway to DL based smart agriculture. It will also pave the way for DL based smart agriculture solutions. The simultaneous development in DL driven computer vision, GPU computing, big data and plant phenotyping make it a very interesting time for further research in development in deployment of DL based solutions to achieve high throughput in plant phenotyping.
References
Abadi M, Barham P, Chen J, et al (2016) Tensorflow: A system for large-scale machine learning. In: 12th {USENIX} symposium on operating systems design and implementation ({OSDI} 16) 265
Abdelfattah MS, Dudziak L, Chau T, et al (2020) Best of both worlds: automl codesign of a CNN and its hardware accelerator. In: 2020 57th ACM/IEEE Design Automation Conference (DAC). IEEE, pp 1–6
Aich S, Josuttes A, Ovsyannikov I, et al (2018) DeepWheat: Estimating Phenotypic Traits from Crop Images with Deep Learning. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV). IEEE, pp 323–332
Ampatzidis Y, Partel V (2019) UAV-based high throughput phenotyping in citrus utilizing multispectral imaging and artificial intelligence. Remote Sens (basel) 11:410. https://doi.org/10.3390/rs11040410
Arad B, Ben-Shahar O (2016) Sparse recovery of hyperspectral signal from natural RGB images. In: Leibe B, Matas J, Sebe N, Welling M (eds) Computer vision – ECCV 2016. Springer International Publishing, Cham, pp 19–34
Araus JL, Kefauver SC, Zaman-Allah M et al (2018) Translating high-throughput phenotyping into genetic gain. Trends Plant Sci 23:451–466. https://doi.org/10.1016/j.tplants.2018.02.001
Bastien F, Lamblin P, Pascanu R, et al (2012) Theano: new features and speed improvements. arXiv preprint arXiv:12115590
Baweja HS, Parhar T, Mirbod O, Nuske S (2018) StalkNet: a deep learning pipeline for high-throughput measurement of plant stalk count and stalk width. In: Hutter M, Siegwart R (eds) Field, and service robotics. Springer International Publishing, Cham, pp 271–284
Bell J, Dee HM (2016) Aberystwyth leaf evaluation dataset. Zenodo. https://doi.org/10.5281/zenodo.168158
Bengio Y, Goodfellow IJ, Courville A (2015) Deep learning, book in preparation for mit press (2015). Disponıvel em http://www.iroumontrealca/bengioy/dlbook
Bresilla K, Perulli GD, Boini A et al (2019) Single-shot convolution neural networks for real-time fruit detection within the tree. Front Plant Sci 10:611. https://doi.org/10.3389/fpls.2019.00611
Buzzy M, Thesma V, Davoodi M, Mohammadpour Velni J (2020) Real-time plant leaf counting using deep object detection networks. Sensors. https://doi.org/10.3390/s20236896
Canziani A, Paszke A, Culurciello E (2016) An analysis of deep neural network models for practical applications. arXiv preprint arXiv:160507678
Chollet F (2017) Xception: Deep Learning with Depthwise Separable Convolutions. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, pp 1800–1807
Chung J, Gulcehre C, Cho K, Bengio Y (2014) Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:14123555
Collobert R, Weston J, Bottou L et al (2011) Natural language processing (almost) from scratch. J Mach Learn Res 12:2493–2537
Das Choudhury S, Samal A, Awada T (2019) Leveraging image analysis for high-throughput plant phenotyping. Front Plant Sci 10:508. https://doi.org/10.3389/fpls.2019.00508
DeChant C, Wiesner-Hanks T, Chen S et al (2017) Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning. Phytopathology 107:1426–1432. https://doi.org/10.1094/PHYTO-11-16-0417-R
Dobos O, Horvath P, Nagy F et al (2019) A deep learning-based approach for high-throughput hypocotyl phenotyping. Plant Physiol 181:1415–1424. https://doi.org/10.1104/pp.19.00728
Douarre C, Schielein R, Frindel C et al (2018) Transfer learning from synthetic data applied to soil-root segmentation in X-Ray tomography images. J Imaging 4:65. https://doi.org/10.3390/jimaging4050065
Esgario JGM, Krohling RA, Ventura JA (2020) Deep learning for classification and severity estimation of coffee leaf biotic stress. Comput Electron Agric 169:105162. https://doi.org/10.1016/j.compag.2019.105162
Fahlgren N, Gehan MA, Baxter I (2015) Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr Opin Plant Biol 24:93–99. https://doi.org/10.1016/j.pbi.2015.02.006
Feng X, Zhan Y, Wang Q et al (2020) Hyperspectral imaging combined with machine learning as a tool to obtain high-throughput plant salt-stress phenotyping. Plant J 101:1448–1461. https://doi.org/10.1111/tpj.14597
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Comput Electron Agric 145:311–318. https://doi.org/10.1016/j.compag.2018.01.009
Gao F, Fu L, Zhang X et al (2020) Multi-class fruit-on-plant detection for apple in SNAP system using Faster R-CNN. Comput Electron Agric 176:105634. https://doi.org/10.1016/j.compag.2020.105634
Ghosal S, Blystone D, Singh AK et al (2018) An explainable deep machine vision framework for plant stress phenotyping. Proc Natl Acad Sci USA 115:4613–4618. https://doi.org/10.1073/pnas.1716999115
Ghosal S, Zheng B, Chapman SC, et al (2019) A weakly supervised deep learning framework for sorghum head detection and counting. Plant Phenomics. https://doi.org/10.34133/2019/1525874
Girshick R, Donahue J, Darrell T, Malik J (2014) R-CNN Rich feature hierarchies for accurate object detection and semantic segmentation. In: IEEE Conference on Computer Vision and Pattern Recognition. IEEE, pp 580–587
Giselsson TM, Jørgensen RN, Jensen PK, et al (2017) A public image database for benchmark of plant seedling classification algorithms. arXiv preprint arXiv:171105458
Gitelson AA (2004) Wide dynamic range vegetation index for remote quantification of biophysical characteristics of vegetation. J Plant Physiol 161:165–173. https://doi.org/10.1078/0176-1617-01176
Gnädinger F, Schmidhalter U (2017) Digital counts of maize plants by unmanned aerial vehicles (uavs). Remote Sens (basel) 9:544. https://doi.org/10.3390/rs9060544
Hasan MM, Chopin JP, Laga H, Miklavcic SJ (2018) Detection and analysis of wheat spikes using convolutional neural networks. Plant Methods 14:100. https://doi.org/10.1186/s13007-018-0366-8
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, pp 770–778
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9:1735–1780. https://doi.org/10.1162/neco.1997.9.8.1735
Howard AG, Zhu M, Chen B, et al (2017) Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:170404861
Itakura K, Saito Y, Suzuki T et al (2018) Estimation of citrus maturity with florescence spectroscopy using deep learning. Horticulturae 5:2. https://doi.org/10.3390/horticulturae5010002
Itzhaky Y, Farjon G, Khoroshevsky F, et al (2018) Leaf counting: Multiple scale regression and detection using deep CNNs. BMVC 328
Jahanbakhshi A, Momeny M, Mahmoudi M, Zhang Y-D (2020) Classification of sour lemons based on apparent defects using stochastic pooling mechanism in deep convolutional neural networks. Sci Hortic 263:109133. https://doi.org/10.1016/j.scienta.2019.109133
Jarrett K, Kavukcuoglu K, Ranzato MA, LeCun Y (2009) What is the best multi-stage architecture for object recognition? In: 2009 IEEE 12th International Conference on Computer Vision. IEEE, pp 2146–2153
Jia Y, Shelhamer E, Donahue J, et al (2014) Caffe: convolutional architecture for fast feature embedding. In: Proceedings of the ACM International Conference on Multimedia - MM ’14. ACM Press, New York, New York, USA, pp 675–678
Jin S, Su Y, Gao S et al (2018) Deep learning: individual maize segmentation from terrestrial lidar data using faster R-CNN and regional growth algorithms. Front Plant Sci 9:866. https://doi.org/10.3389/fpls.2018.00866
Kamilaris A, Prenafeta-Boldú FX (2018) Deep learning in agriculture: a survey. Comput Electron Agric 147:70–90. https://doi.org/10.1016/j.compag.2018.02.016
Khaki S, Wang L (2019) Crop yield prediction using deep neural networks. Front Plant Sci 10:621. https://doi.org/10.3389/fpls.2019.00621
Kingma DP, Welling M (2019) An introduction to variational autoencoders. FNT in Machine Learning 12:307–392. https://doi.org/10.1561/2200000056
Koirala A, Walsh KB, Wang Z, McCarthy C (2019) Deep learning for real-time fruit detection and orchard fruit load estimation: benchmarking of ‘MangoYOLO.’ Precision Agric. https://doi.org/10.1007/s11119-019-09642-0
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Commun ACM 60:84–90. https://doi.org/10.1145/3065386
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/nature14539
Liakos KG, Busato P, Moshou D et al (2018) Machine learning in agriculture: a review. Sensors. https://doi.org/10.3390/s18082674
Lin Z, Guo W (2020) Sorghum panicle detection and counting using unmanned aerial system images and deep learning. Front Plant Sci 11:534853. https://doi.org/10.3389/fpls.2020.534853
Liu W, Anguelov D, Erhan D et al (2016) SSD: Single shot multibox detector. In: Leibe B, Matas J, Sebe N, Welling M (eds) European Conference on Computer VIsion (ECCV). Springer International Publishing, Cham, pp 21–37
Liu W, Wang Z, Liu X et al (2017) A survey of deep neural network architectures and their applications. Neurocomputing 234:11–26. https://doi.org/10.1016/j.neucom.2016.12.038
Liu X, Hu C, Li P (2020) Automatic segmentation of overlapped poplar seedling leaves combining Mask R-CNN and DBSCAN. Comput Electron Agric 178:105753. https://doi.org/10.1016/j.compag.2020.105753
Li Y, Hu J, Zhao X et al (2017) Hyperspectral image super-resolution using deep convolutional neural network. Neurocomputing 266:29–41. https://doi.org/10.1016/j.neucom.2017.05.024
Li Z, Guo R, Li M et al (2020) A review of computer vision technologies for plant phenotyping. Comput Electron Agric 176:105672. https://doi.org/10.1016/j.compag.2020.105672
Lobet G, Pagès L, Draye X (2011) A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol 157:29–39. https://doi.org/10.1104/pp.111.179895
Lu J, Hu J, Zhao G et al (2017) An in-field automatic wheat disease diagnosis system. Comput Electron Agric 142:369–379. https://doi.org/10.1016/j.compag.2017.09.012
Magomadov VS (2019) Deep learning and its role in smart agriculture. J Phys: Conf Ser 1399:044109. https://doi.org/10.1088/1742-6596/1399/4/044109
Majeed Y, Zhang J, Zhang X et al (2020) Deep learning based segmentation for automated training of apple trees on trellis wires. Comput Electron Agric 170:105277. https://doi.org/10.1016/j.compag.2020.105277
Mandic D, Chambers J (2001) Recurrent neural networks for prediction: learning algorithms, architectures and stability. Wiley
Mao H, Yao S, Tang T et al (2018) Towards real-time object detection on embedded systems. IEEE Trans Emerg Top Comput 6:417–431. https://doi.org/10.1109/TETC.2016.2593643
Miao C, Pages A, Xu Z, et al (2020) Semantic segmentation of sorghum using hyperspectral data identifies genetic associations. Plant Phenomics. https://doi.org/10.34133/2020/4216373
Mikolov T, Sutskever I, Chen K, et al (2013) Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Syst. https://doi.org/10.48550/arXiv.1310.4546
Milioto A, Lottes P, Stachniss C (2018) Real-Time Semantic Segmentation of Crop and Weed for Precision Agriculture Robots Leveraging Background Knowledge in CNNs. In: 2018 IEEE International Conference on Robotics and Automation (ICRA). IEEE, pp 2229–2235
Minervini M, Scharr H, Tsaftaris SA (2015) Image analysis: the new bottleneck in plant phenotyping. IEEE Signal Process Mag 32:126–131. https://doi.org/10.1109/MSP.2015.2405111
Minervini M, Fischbach A, Scharr H et al (2016) Finely-grained annotated datasets for image-based plant phenotyping. Pattern Recogn Lett 81:80–89. https://doi.org/10.1016/j.patrec.2015.10.013
Misra T, Arora A, Marwaha S et al (2020) SpikeSegNet-a deep learning approach utilizing encoder-decoder network with hourglass for spike segmentation and counting in wheat plant from visual imaging. Plant Methods 16:40. https://doi.org/10.1186/s13007-020-00582-9
Moghimi A, Yang C, Anderson JA (2020) Aerial hyperspectral imagery and deep neural networks for high-throughput yield phenotyping in wheat. Comput Electron Agric 172:105299. https://doi.org/10.1016/j.compag.2020.105299
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1419. https://doi.org/10.3389/fpls.2016.01419
Nie S, Gu L, Zheng Y, et al (2018) Deeply learned filter response functions for hyperspectral reconstruction. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. IEEE, pp 4767–4776
Nyine M, Uwimana B, Blavet N et al (2018) Genomic prediction in a multiploid crop: genotype by environment interaction and allele dosage effects on predictive ability in banana. Plant Genome. https://doi.org/10.3835/plantgenome2017.10.0090
Oquab M, Bottou L, Laptev I, Sivic J (2014) Learning and Transferring Mid-level Image Representations Using Convolutional Neural Networks. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition. IEEE, pp 1717–1724
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22:1345–1359. https://doi.org/10.1109/TKDE.2009.191
Partel V, Charan Kakarla S, Ampatzidis Y (2019) Development and evaluation of a low-cost and smart technology for precision weed management utilizing artificial intelligence. Comput Electron Agric 157:339–350. https://doi.org/10.1016/j.compag.2018.12.048
Pound MP, Atkinson JA, Townsend AJ et al (2017a) Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience 6:1–10. https://doi.org/10.1093/gigascience/gix083
Pound MP, Atkinson JA, Townsend AJ et al (2018) Erratum to: deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience. https://doi.org/10.1093/gigascience/giy042
Pound MP, Atkinson JA, Wells DM, et al (2017b) Deep Learning for Multi-task Plant Phenotyping. In: 2017b IEEE International Conference on Computer Vision Workshops (ICCVW). IEEE, pp 2055–2063
Ramcharan A, McCloskey P, Baranowski K et al (2019) A mobile-based deep learning model for cassava disease diagnosis. Front Plant Sci 10:272. https://doi.org/10.3389/fpls.2019.00272
Redmon J, Divvala S, Girshick R, Farhadi A (2016) YOLO You Only Look Once: Unified, Real-Time Object Detection. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, pp 779–788
Ren S, He K, Girshick R, Sun J (2017) Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell 39:1137–1149. https://doi.org/10.1109/TPAMI.2016.2577031
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds) Medical image computing and computer-assisted intervention (MICCAI). Springer International Publishing, Cham, pp 234–241
Sadeghi-Tehran P, Virlet N, Ampe EM et al (2019) DeepCount: in-field automatic quantification of wheat spikes using simple linear iterative clustering and deep convolutional neural networks. Front Plant Sci 10:1176. https://doi.org/10.3389/fpls.2019.01176
Sandhu KS, Lozada DN, Zhang Z et al (2020) Deep learning for predicting complex traits in spring wheat breeding program. Front Plant Sci 11:613325. https://doi.org/10.3389/fpls.2020.613325
Sandhu KS, Mihalyov PD, Lewien MJ et al (2021a) Combining genomic and phenomic information for predicting grain protein content and grain yield in spring wheat. Front Plant Sci 12:613300. https://doi.org/10.3389/fpls.2021.613300
Sandhu KS, Mihalyov PD, Lewien MJ et al (2021b) Genome-wide association studies and genomic selection for grain protein content stability in a nested association mapping population of spring wheat. BioRxiv. https://doi.org/10.1101/2021.04.15.440064
Sandhu KS, Patil SS, Pumphrey MO, Carter AH (2021c) Multi-trait machine and deep learning models for genomic selection using spectral information in a wheat breeding program. BioRxiv. https://doi.org/10.1101/2021.04.12.439532
Sa I, Ge Z, Dayoub F et al (2016) Deepfruits: a fruit detection system using deep neural networks. Sensors. https://doi.org/10.3390/s16081222
Sankaran S, Quirós JJ, Miklas PN (2019) Unmanned aerial system and satellite-based high resolution imagery for high-throughput phenotyping in dry bean. Comput Electron Agric 165:104965. https://doi.org/10.1016/j.compag.2019.104965
Schmidhuber J (2015) Deep learning in neural networks: an overview. Neural Netw 61:85–117. https://doi.org/10.1016/j.neunet.2014.09.003
Seide F, Agarwal A (2016) CNTK: Microsoft’s Open-Source Deep-Learning Toolkit. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ’16. ACM Press, New York, New York, USA, pp 2135–2135
Shi Z, Chen C, Xiong Z, et al (2018) HSCNN+: Advanced CNN-Based Hyperspectral Recovery from RGB Images. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). IEEE, pp 1052–10528
Signoroni A, Savardi M, Baronio A, Benini S (2019) Deep learning meets hyperspectral image analysis: a multidisciplinary review. J Imag 5:52. https://doi.org/10.3390/jimaging5050052
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556
Singh AK, Ganapathysubramanian B, Sarkar S, Singh A (2018) Deep learning for plant stress phenotyping: trends and future perspectives. Trends Plant Sci 23:883–898. https://doi.org/10.1016/j.tplants.2018.07.004
Sladojevic S, Arsenovic M, Anderla A et al (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Comput Intell Neurosci. https://doi.org/10.1155/2016/3289801
Stein M, Bargoti S, Underwood J (2016) Image based mango fruit detection, localisation and yield estimation using multiple view geometry. Sensors. https://doi.org/10.3390/s16111915
Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI Conference on Artificial Intelligence 31:
Szegedy C, Wei Liu, Yangqing Jia, et al (2015) Going deeper with convolutions. In: Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, pp 1–9
Taghavi Namin S, Esmaeilzadeh M, Najafi M et al (2018) Deep phenotyping: deep learning for temporal phenotype/genotype classification. Plant Methods 14:66. https://doi.org/10.1186/s13007-018-0333-4
Tapas A (2016) Transfer learning for image classification and plant phenotyping. Int J Adv Res Computer Eng Technol (IJARCET) 5:2664–2669
Team PC (2017) Pytorch: tensors and dynamic neural networks in Python with strong GPU acceleration. GitHub pub online: March 22, 2017.
Tsoulias N, Paraforos DS, Fountas S, Zude-Sasse M (2019) Estimating canopy parameters based on the stem position in apple trees using a 2D lidar. Agronomy 9:740. https://doi.org/10.3390/agronomy9110740
Ubbens J, Ceislak M, Prusinkiewicz P, Stavness I (2018) Synthetic arabidopsis rosettes for leaf counting. Figshare. https://doi.org/10.6084/m9.figshare.5450080.v5
Ubbens JR, Stavness I (2017) Deep plant phenomics: a deep learning platform for complex plant phenotyping tasks. Front Plant Sci 8:1190. https://doi.org/10.3389/fpls.2017.01190
Uzal LC, Grinblat GL, Namías R et al (2018) Seed-per-pod estimation for plant breeding using deep learning. Comput Electron Agric 150:196–204. https://doi.org/10.1016/j.compag.2018.04.024
Veit A, Wilber M, Belongie S (2016) Residual networks behave like ensembles of relatively shallow networks. arXiv preprint arXiv:160506431
Vincent P, Larochelle H, Bengio Y, Manzagol P-A (2008) Extracting and composing robust features with denoising autoencoders. In: Proceedings of the 25th international conference on Machine learning - ICML ’08. ACM Press, New York, New York, USA, pp 1096–1103
Voulodimos A, Doulamis N, Doulamis A, Protopapadakis E (2018) Deep learning for computer vision: a brief review. Comput Intell Neurosci 2018:7068349. https://doi.org/10.1155/2018/7068349
Waldmann P, Pfeiffer C, Mészáros G (2020) Sparse convolutional neural networks for genome-wide prediction. Front Genet 11:25. https://doi.org/10.3389/fgene.2020.00025
Wang C, Li X, Caragea D et al (2020a) Root anatomy based on root cross-section image analysis with deep learning. Comput Electron Agric 175:105549. https://doi.org/10.1016/j.compag.2020.105549
Wang X, Xuan H, Evers B et al (2019) High-throughput phenotyping with deep learning gives insight into the genetic architecture of flowering time in wheat. Gigascience. https://doi.org/10.1093/gigascience/giz120
Wang X, Zhao Y, Pourpanah F (2020b) Recent advances in deep learning. Int J Mach Learn & Cyber 11:747–750. https://doi.org/10.1007/s13042-020-01096-5
Wang Z, Hu M, Zhai G (2018) Application of deep learning architectures for accurate and rapid detection of internal mechanical damage of blueberry using hyperspectral transmittance data. Sensors https://doi.org/10.3390/s18041126
Ward D, Moghadam P (2020) Scalable learning for bridging the species gap in image-based plant phenotyping. Comput vis Image Underst 197–198:103009. https://doi.org/10.1016/j.cviu.2020.103009
Wu C, Zeng R, Pan J et al (2019) Plant phenotyping by deep-learning-based planner for multi-robots. IEEE Robot Autom Lett 4:3113–3120. https://doi.org/10.1109/LRA.2019.2924125
Xiong Z, Shi Z, Li H, et al (2017) HSCNN: CNN-Based hyperspectral image recovery from spectrally undersampled projections. In: 2017 IEEE International Conference on Computer Vision Workshops (ICCVW). IEEE, pp. 518–525
Yalcin H (2017) Plant phenology recognition using deep learning: Deep-Pheno. In: 2017 6th International Conference on Agro-Geoinformatics. IEEE, pp 1–5
Yamamoto K, Togami T, Yamaguchi N (2017) Super-resolution of plant disease images for the acceleration of image-based phenotyping and vigor diagnosis in agriculture. Sensors. https://doi.org/10.3390/s17112557
Yasrab R, Atkinson JA, Wells DM et al (2019) RootNav 2.0: Deep learning for automatic navigation of complex plant root architectures. Gigascience. https://doi.org/10.1093/gigascience/giz123
Yendrek CR, Tomaz T, Montes CM et al (2017) High-throughput phenotyping of maize leaf physiological and biochemical traits using hyperspectral reflectance. Plant Physiol 173:614–626. https://doi.org/10.1104/pp.16.01447
Yuan Y, Zheng X, Lu X (2017) Hyperspectral image superresolution by transfer learning. IEEE J Sel Top Appl Earth Observ Remote Sens 10:1963–1974. https://doi.org/10.1109/JSTARS.2017.2655112
Zhang J, He L, Karkee M et al (2018) Branch detection for apple trees trained in fruiting wall architecture using depth features and Regions-Convolutional Neural Network (R-CNN). Comput Electron Agric 155:386–393. https://doi.org/10.1016/j.compag.2018.10.029
Zhang X, Fu L, Karkee M et al (2019) Canopy segmentation using resnet for mechanical harvesting of apples. IFAC-PapersOnLine 52:300–305. https://doi.org/10.1016/j.ifacol.2019.12.550
Zhao Y, Li J, Zhao R et al (2020) Genome-wide association study reveals the genetic basis of cold tolerance in wheat. Mol Breed 40:36. https://doi.org/10.1007/s11032-020-01115-x
Zhou L, Zhang C, Taha MF et al (2020) Wheat kernel variety identification based on a large near-infrared spectral dataset and a novel deep learning-based feature selection method. Front Plant Sci 11:575810. https://doi.org/10.3389/fpls.2020.575810
Zhuang F, Qi Z, Duan K et al (2020) A comprehensive survey on transfer learning. Proc IEEE. https://doi.org/10.1109/JPROC.2020.3004555
Zhu N, Liu X, Liu Z et al (2018) Deep learning for smart agriculture: concepts, tools, applications, and opportunities. Int J Agri Biol Eng 11:21–28
Zhu W, Sun Z, Yang T et al (2020) Estimating leaf chlorophyll content of crops via optimal unmanned aerial vehicle hyperspectral data at multi-scales. Comput Electron Agric 178:105786. https://doi.org/10.1016/j.compag.2020.105786
Author information
Authors and Affiliations
Contributions
All authors contributed equally.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Arya, S., Sandhu, K.S., Singh, J. et al. Deep learning: as the new frontier in high-throughput plant phenotyping. Euphytica 218, 47 (2022). https://doi.org/10.1007/s10681-022-02992-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-02992-3