Abstract
Brown planthopper (BPH) has emerged as one of the most devastating pests of rice in several Asian countries. Cultivation of planthopper resistant varieties is an ecologically acceptable strategy. A population of 255 F2:3 families from the cross Taichung Native 1/ARC10550 was used to map BPH resistance with 106 polymorphic simple sequence repeat markers. The inheritance pattern of different traits suggested that the resistance in ARC10550 is controlled by quantitative traits instead of a single recessive gene. The quantitative trait loci (QTLs) for BPH resistance were analysed for nine phenotypic traits. Several of these phenotypic traits recorded high degree of positive or negative correlations between them, suggesting dependence or redundancy of the tests. QTL analysis revealed that five major loci were associated with resistance, one for damage score (qBphDs6) on chromosome 6, two for nymphal preference at 48 and 72 h (qBphNp(48h)-1 and qBphNp(72h)-12) on chromosome 1 and 12 and two for days to wilt (qBphDw(30)-3 and qBphDw(30)-8) on chromosome 3 and 8 explaining the phenotypic variance of 24.23, 8.69, 7.66, 4.55 and 10.48% respectively. These QTLs indicated the negative additive effects suggesting that the resistant alleles identified were from ARC10550 donor parent. These QTLs jointly explained 55.6% of the phenotypic variance for BPH resistance in this population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The brown planthopper [Nilaparvata lugens (Stål); BPH] is one of the most destructive pests of rice. It can suck sap from the phloem of rice plant, as well as transmit several kinds of viruses (Cha et al. 2008). The severe BPH infestation can cause extensive damage to the rice plants resulting complete crop loss and referred as “hopper burn” phenomenon (Sogawa 1982). Since past two decades, the infestations of planthoppers (mainly BPH) have intensified across Asia, resulted in heavy yield losses (Normile 2008). Conventionally, BPH is controlled mainly by chemical pesticides, which are expensive and environmentally unsafe. Moreover, pesticides would kill BPH along with the predators, and over-use of pesticides prompts the insect to evolve resistance, which in turn leads to pest resurgence. Therefore, cultivation of BPH-resistant varieties is the most economical and environmentally safe.
The differential reaction of the resistance sources indicated that BPH has four different biotypes. Biotypes 1 and 2 are distributed in Southeast Asia and biotype 3 is a laboratory biotype produced in IRRI, Philippines. Biotype 4 is the most destructive biotype of South Asia and is distributed over the Indian subcontinent (Heinrichs 1986). So far 30 major genes for resistance to BPH have been identified from cultivated and wild species of rice (http://www.shigen.nig.ac.jp/rice/oryzabase_submission/gene_nomenclature/). Many of the BPH resistance genes identified against biotype 1 and 2 are not effective against biotype 4 (Ram et al. 2010; Deen et al. 2010). Among the major genes identified, most of them have been mapped with molecular markers and some of the genes are presently being used extensively in the practical rice breeding programs using MAS (Qiu et al. 2012).
Besides the major genes, several quantitative trait loci (QTLs) associated with BPH resistance have been identified in the cultivated and wild species of rice. Su et al. (2002) reported three QTLs on chromosomes 2, 10, and 12, associated with BPH resistance from the population derived from Nipponbare × Kasalath. The QTLs (Qbph-2, Qbph-4, Qbph-7, and Qbph-9) accounting for 5.64–12.77% of the phenotypic variance for BPH resistance were identified by Liu et al. (2009) from the landrace Yagyaw. Traditionally, the resistance mechanisms are seedling resistance (MSST), antixenosis (non-preference) involving a reduction in colonization or oviposition, antibiosis involving a reduction in insect survival, growth rate, or reproduction after ingestion of host tissue and tolerance involving production of a crop of high quality and yield despite insect infestation (Alam and Cohen 1998a). The previous studies have shown that these resistance mechanisms were conferred by the major BPH resistance genes such as Bph14 appeared to confer antibiosis only in transgenic lines (Du et al. 2009). While, Qiu et al. (2012) found that Bph6 and Bph12 conferred both antixenotic and antibiotic effects, respectively in near isogenic lines. In addition, several polygenes or QTLs have been identified to be associated with seedling resistance, antixenosis, antibiosis, or tolerance in rice cultivars. Soundararajan et al. (2004) reported that the QTLs on chromosome 7 and 2 were associated with seedling resistance and antibiosis, respectively, while QTLs on chromosomes 1, 6, and 7 showed association with tolerance.
It is well known that pyramiding of major BPH resistance genes can significantly improve resistance levels (Myint et al. 2012; Qiu et al. 2012; Wang et al. 2013; Hu et al. 2015) but the spectrum and durability of resistance over the biotypes have not been studied. Most of the donors showing durable resistance over the years and biotypes have one or two major genes along with several QTLs. Hence identification and use of the QTLs along with major genes will help in developing durable resistance. Cohen et al. (1997) detected that the cultivar IR64 carrying the major gene Bph1 and other minor QTLs directed the cultivar to display a higher and durable resistance to the BPH in the field. The land race ARC10550 has been reported to be resistant to BPH biotype 4 having the major (bph5) recessive resistance gene (Khush et al. 1985). In the present study we analysed the QTLs associated with resistance to BPH in ARC10550.
Materials and methods
Plant material
The parents used in the present study include the ARC10550 (acc. 12507), a traditional tall, late duration indica land race with bold grains and resistant to biotype 4 collected from Assam province of India and Taichung Native1 (TN1), a semi dwarf, medium duration indica variety with short bold grains and susceptible to all the biotypes of BPH. The seeds of both the parents have been collected from International Rice Brown Planthopper Nursery, IRRI, Philippines. The mapping population consisted of 255 F2:3 families derived from the single F1 plant developed from the cross between BPH susceptible parent TN1 and resistant parent ARC10550. The confirmed F1 plant was selfed by bagging all the panicles at the anthesis with butter paper bag. At maturity more than 350 F2 seeds were obtained. The F2 plants were planted in field and 255 F2 plants were randomly selected for genetic study. Leaf samples from each individual plant were collected for DNA isolation and genotyping with polymorphic markers. At the flowering each plant was again bagged to produce F3 seeds. At maturity the seeds of individual plants were harvested separately as F3 seeds. The F2:3 families along with the parents TN1 and ARC10550 were used for phenotyping for different traits associated with BPH resistance. The PTB33 was also used as resistant check in BPH screening experiments along with resistant (ARC10550) and susceptible (TN1) parents.
Insect population
At the Indian Institute of Rice Research, Hyderabad, India, BPH biotype 4 population is being reared under controlled greenhouse conditions on the susceptible rice variety TN1 (Kalode et al. 1975). Freshly hatched nymphs or adults of specified age were utilized for various screening tests. Necessary precautions were also taken to keep the BPH culture away from predators and other natural enemies to prevent contamination.
Greenhouse evaluation for BPH resistance
Phenotyping experiments were conducted in the green-house at 25 ± 5 °C with 50 ± 10% relative humidity (RH) under natural light/dark conditions. The F2:3 families along with parents TN1 and ARC10550, and resistant check PTB33 were evaluated for different resistance traits following standard protocols. For seedling reaction in MSST, the seed boxes were prepared one day before by filling with well puddled soils mixed with FYM and fertilizer. The sprouted seeds of F3 populations were sown in tray of each seed box containing 20 test lines with 15 seedlings each, besides two border rows of susceptible check (TN1) and one row of resistant check PTB-33 in the middle. Ten days after sowing, the seedling in 3–4 leaf stage were infested with 6–8 first and second instars nymphs per seedling. The damage scores were recorded in 0–9 scale following the standard evaluation system (SES) of rice (SES 1996) in each of F3 plant of each line when more than 90% plants of the susceptible check TN1 were dead. A total of 255 F3 lines derived from TN1/ARC10550 cross were screened for damage score. All the F3 progenies were replicated thrice for screening with 15 seedlings in each replication (Kalode et al. 1975).
Recommended protocols for other tests such as nymphal preference at 24, 48, and 72 h, nymphal survival, nymphal duration, honeydew area on plants 30 and 60 days after sowing (DAS) and days to wilt 30 DAS were adopted according to Heinrichs et al. (1985), Renganayaki et al. (2000), Myint et al. (2012).
Genotyping
A set of 662 simple sequence repeat (SSR) markers uniformly spread over 12 rice chromosomes were screened for parental polymorphism survey between TN1 and ARC10550, which resulted in identification of 106 polymorphic markers between both the parents (Fig. 1). DNA was isolated from the leaf samples of 255 F2 plants along with the parents using the modified method of Zheng et al. (1995) and then used for polymerase chain reaction (PCR) for amplification following the protocol of Chen et al. (1997). The entire mapping population was genotyped with 106 polymorphic markers, and alleles were scored on agarose gel (Fig. 2). The original sources and motifs for all SSR markers used in the present study are available at http://www.gramene.org. PCR amplification of SSRs was performed in 10 µl reaction volume containing template DNA (20–25 ng), 250 µM each of dNTPs, PCR buffer (1×), 0.6 U/µl of Taq DNA polymerase (Genei, Bangalore, India), and ≥0.2 µl of both forward and reverse primers. PCR amplifications were performed in 96-well plates on a thermal cycler (Eppendorf, Hamburg, Germany) using the following PCR conditions: hot start at 94 °C for 5 min followed by denaturing at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min for 35 amplification cycles, with final extension at 72 °C for 7 min. The PCR products were resolved on 3% agarose gel (SeaKem, Rockland, USA) stained with ethidium bromide (0.5 mg/ml; Sigma-Aldrich, St. Louis, USA) in 0.59 Tris/borate/ethylene diamine tetra acetic acid buffer at 100–150 V for 2 h using a submarine electrophoresis unit (Genei, Bangalore, India) and photographed under ultraviolet light. The size of the amplified fragments was calculated using Alpha-ease software (Alpha Innotech, San Leandro, USA) with a 100-bp ladder (MBI Fermentas, Vilnius, Lithuania) as size reference standard.
Data analysis
Phenotypic data for each of the tests recorded for the 255 F2:3 families were subjected to goodness-of-fit analysis with expected Mendelian ratios for simple inherited traits. The F2:3 families with mean value similar to the resistant parent ARC10550 were treated as resistant, and the rest as susceptible. The frequency distribution of F2:3 families across levels of phenotypic values was plotted, and data were analyzed for normal distribution using the SAS software, mean, range, standard deviation, standard error, paired t test, χ 2 test, correlation analysis, and the probability estimates for null hypothesis rejection using these tests for all the phenotypic values of BPH resistance were obtained using MS Excel software.
Resistance against BPH was also investigated by QTL analysis. Genotypic data assembled for all the polymorphic makers among all the 255 F2 plants were subjected to linkage analysis using Join Map version 4.0 (Van Ooijen 2006). Map distances were calculated using the Kosambi (1944) mapping function. Placement of markers into different linkage groups was performed with “LOD groupings” and “Create group using the mapping tree” commands. Mean χ 2 contributions or average contributions to the goodness of fit of each locus were also checked to determine the best fitting position for markers in genetic maps. Markers showing negative map distances or a large jump in mean χ 2 value were discarded. Final maps were drawn with the help of Map Chart version 2.2 (Voorrips 2002). QTL analysis of F2:3 families was performed using a composite interval mapping (CIM) method (Zeng 1994) in Windows QTL Cartographer version 2.5 (Wang et al. 2007). A permutation number of 1000 was applied for each trait in QTL analysis, and a LOD threshold of 2.5 was adjusted for identification of significant QTLs. The relative contribution of each QTL towards phenotype was calculated as the percentage of phenotypic variance explained (PVE, %). The percentages of variation explained by a QTL and the additive effect were also estimated using the software.
Results
Inheritance of BPH resistance
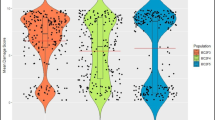
The significant differences between TN1 (susceptible) and ARC10550 (resistant) were observed for all the phenotypic traits under study (Table 1). However, ARC10550 differed from the standard resistant check PTB33 in terms of damage score, nymphal survival, feeding rate and nymphal preference. Mean and range of all nine phenotypic traits for the F3 families are also presented in Table 1 and the frequency distribution (Fig. 3) indicated that the traits were inherited quantitatively. Transgressive segregants in F2:3 families were observed for the traits damage score, days to wilt, nymphal preference, nymphal survival, nymphal duration, and honeydew area on plants 30 and 60 DAS, suggesting multiple genes with additive effects influencing the trait (Fig. 3). However, the normal distribution of values for each traits recorded in the 255 F2:3 families confirms that the traits are inherited quantitatively (Fig. 3; Table 1). Data pertaining to the traits listed in Table 1 did not show significant fitness to simple Mendelian ratio indicating that the BPH resistance is governed by polygenes in ARC10550.
Correlation among traits
Significant positive correlations were observed among the nine traits analyzed indicating their interdependence (Table 2). The seedling resistance in terms of damage scores was positively correlated with the traits feeding rate at 30 DAS (0.909), 60 DAS (0.903), nymphal survival (0.783) and negatively correlated with nymphal duration (−0.488), days to wilt (−0.418), While the correlation between damage score and nymphal preference 24 h (−0.126), nymphal preference 48 h (−0.104) and nymphal preference 72 h (−0.098) were not significant. The nympal survival (%) was positively correlated to feeding rate 30 days (0.759) and 60 days (0.764). Nympal duration had positive correlation with days to wilt (0.236).
QTL identification
Of the 662 SSR markers tested for polymorphism between TN1 and ARC10550, 106 (20%) were found to be polymorphic and distributed in all the chromosomes. All the 255 F2 plants were genotyped with the polymorphic markers. Thus, the molecular linkage map was constructed with 106 polymorphic markers to identify QTLs conferring BPH resistance. The map covered 1532.2 cM on all 12 chromosomes, with average interval of 14.45 cM. QTL analysis using Windows QTL Cartographer version 2.5 with a LOD threshold of 2.5 and significance level of 0.01 detected 5 QTLs for BPH resistance qBphDs-6, qBphNp(48h)-1, qBphNp(72h)-12, qBphDw(30)-3 and qBphDw(30)-8 explaining the phenotypic variance, of 24.23, 8.69, 7.66, 4.55 and 10.48% with LOD value 2.56, 2.75, 3.57, 4.21 and 4.19 as well as an additive effect −3.915, −0.720, −0.714, −0.509 and −0.864 on chromosome 1, 3, 6, 8 and 12, respectively (Table 3; Figs. 4, 5). The negative additive effects of the QTLs indicated the resistant alleles are from donor parent ARC10550.
Linkage map showing chromosomal locations of QTLs detected for resistance to BPH in TN1/ARC10550 F2:3 population. Marker names are listed on the right hand side of the chromosome with the distances (in cM) indicated on the left. The solid bars indicates the locations of the different loci for BPH resistance, designated as qBphNp(48h)-1, qBphDw(30)-3, qBphDs6, qBphDw(30)-8 and qBphNp(72h)-12
a–e Composite interval mapping, showing QTLs graph, LOD score is on the Y-axis and relative location of marker name and distribution in cM on the X-axis and below is R2 value. a Damage score-QTL (qbphDS-6-1) flanking marker between RM19697–RM5855 with LOD 2.6 and PVE 24.23%; b days to wilt (30 days)-QTL (qbphDW) flanking marker between RM547–RM22741 with LOD 4.2 and PVE 10.48%; c days to wilt (30 days)-QTL qbphDW(30)-3 flanking marker between RM7197–RM6987 with LOD 4.2 and PWE 4.55%; d nymphal preference (48 h)-QTL (qbphNP-48h-1) flanking marker between RM11704–RM1068 with LOD 2.8 and PVE 8.69%; e nymphal preference (72 h) QTL (qbphNP-72h-12) flanking marker between RM27971–RM28024 with LOD 3.6 and PVE 7.66%
Discussion
The brown planthopper, historically was recognized as minor pest of rice, emerged as a major pest in the tropical Asia during green revolution (Heninrichs and Mochida 1984; Gallagher et al. 1994). Host plant resistance has been recognized as a viable alternative to chemical control methods to reduce the yield losses by this pest.
The brown planthopper has assumed great importance in rice in India, all efforts are being made to identify resistant donors/varieties, to understand the genetics of host plant resistance and utilize it breeding programs. Currently, more than 30 genes for resistance to BPH have been reported from indica and wild rice species (Wu et al. 2014). Fourteen of these genes have been fine mapped to specific regions on chromosomes 3, 4, 6 and 12. Bph14 and Bph26 (or Bph2) were recently cloned and found to encode coiled-coil, nucleotide-binding, and leucine-rich repeat (CC-NB-LRR) proteins (Du et al. 2009; Tamura et al. 2014). Most of the major genes identified and mapped were against BPH biotype 1, 2 and 3 hence majority of them are ineffective against Biotype 4 (Ram et al. 2010; Deen et al. 2010). Krishna et al. (1984) reported that 18 of the 114 land races from Assam, India were resistant to BPH biotype 4 prevalent in India and ARC10550 was further identified as possessing major recessive gene (bph5) by Khush et al. (1985).
The mapping population was developed by crossing TN1 with ARC10550. The F1 was tested for its hybridity using three markers from each chromosome. The panicles of the single F1 plant were bagged before anthesis and harvested at maturity and used for producing F2 population. All the plants in F2 population were selfed to develop F3 progenies. The F2 population was used for genotyping while F3 population was used for phenotyping to construct linkage map for BPH resistance. Similar populations have been also used earlier by Sun et al. (2007), Li et al. (2011), Santhanalakshmi et al. (2010) and Qiu et al. (2011) for mapping of gene/QTLs associated with resistance to BPH in rice.
Three mechanism or modalities of plant resistance to insects are generally recognized: Antixenosis, a quality that repels or disturbs the insects by causing a reduction in colonization or oviposition; antibiosis a quality that reduces insect survival, growth rate or reproduction following the ingestion of host tissue; and tolerance, the capacity to produce a crop of high quality and yield despite insect infestation (Kennedy et al. 1987). These properties can be quantified by making relative comparisons among genotypes. Antixenosis, antibiosis and tolerance have been detected in various combinations in rice germplasm conferring resistance to BPH (Heinrichs et al. 1985; Kennedy et al. 1987; Cohen et al. 1997; Alam and Cohen 1998a, b).
In the present study a series of screening techniques were applied for phenotyping of the traits associated with gene/QTLs for BPH resistance in TN1 × ARC10550 mapping population. The parents ARC10550 and TN1 showed differences for the nine traits under study viz, damage scores to measure seedling resistance, days of nymphal survival at 30 DAS, nymphal duration before converting adults at 30 DAS, days to wilt at 30 DAS (tolerance), feeding rate at 30 DAS (antibiosis), feeding rate at 60 DAS (antibiosis) and Nymphal preference at 24, 48 and 72 h after infestation, respectively (antixenosis) (Table 1). Substantial variations in F3 population for the nine traits during screening indicated that the traits were genetically inherited. The results of honeydew excretions, survival rate of nymphs and emergence rate of adults indicated that the antibiosis was one of the mechanisms involved in the BPH resistance in ARC10550. The lower feeding rates were measured in terms of less area of honey dew spread, indicating that the tolerance in ARC10550 was associated with antibiosis. The range of honeydew spread in the F3 population 345–754 and 254–637 mm2 at 30 and 60 days old plants infested with BPH, suggested that tolerance in ARC10550 is due to antibiosis that reduces the feeding and growth rate of the insect, similar results were also reported by Panda and Heinrichs (1983). In the case of Bph14, mainly resistance appears to be due to antibiosis which reduces the feeding and growth rate of BPH insect (Du et al. 2009). More recently, Qiu et al. (2010) reported that Bph6 exerted antixenotic and antibiosis effects while conferring BPH resistance. Sonalikar et al. (2011), reported that BPH tolerance in Dagad Deshi is mainly due to antibiosis. However the mechanisms of resistance in most of the genes are still unknown. Therefore it is necessary to identify the level of antibiosis and tolerance in donors carrying BPH resistance gene/QTLs which will help in improving durable resistance in rice varieties (Paguia et al. 1980; Alam and Cohen 1998b; Huang et al. 2001; Hao et al. 2008; Myint et al. 2009a, b). The feeding rate was assessed in terms of the amount of honeydew excreted. The amount of honeydew excreted is directly proportional to the amount of sap sucked by BPH. Alagar et al. (2008) and Sonalikar et al. (2011) studied the feeding behaviour of BPH on few selected rice genotypes and indicated that feeding marks and feeding rate could be the reliable parameters to evaluate the resistance nature of the genotypes against insect pests. It is known that the quantitative resistance to BPH may result from the cumulative effect of different mechanisms, such as non-preference or antixenosis, antibiosis and tolerance (Alam and Cohen 1998a) and result in the durable resistance (Ren et al. 2004; Murata et al. 2001; Xu et al. 2002).
The seedling bulk test is widely used to assess the BPH resistance in the rice germplasm because it is a rapid and efficient screening method. The seedling bulk test is actually the cumulative expression of most of the mechanisms (antibiosis, antixenosis and tolerance) associated with resistance and hence being widely used by most of the workers (Shi et al. 2003; Jena et al. 2006; Li et al. 2006; Qiu et al. 2010, 2011). The correlation analysis also suggested the association of resistance in terms of damage scores with feeding rate, nymphal survival, nymphal duration and days to wilt (Table 2).
The frequency distribution of the plants in the F3 population (Table 1; Fig. 1) for different traits showed a fit to normal distribution indicating that many loci are involved in the tolerance but not a major gene bph5 which was reported earlier (Khush et al. 1985). Also transgressive segregation in both directions was observed for most of the traits. This indicated that resistance is under polygenic control and several loci related to deferent traits are involved in the expression of the traits might be contributed from both the parents.
In the present study we identified two major QTLs one each for damage score and days to wilt and three minor QTLs, two for Nymphal preference and one for days to wilt. The major QTLs for damage score explaining phenotypic variance of 24.23% was identified in the on chromosomes 6 (Table 3). Similar type of trait enhancing QTLs for seedling resistance was also identified earlier on chromosome # 1, 2, 4, 6 and 8 in DH lines of IR64 × Azucena (Alam and Cohen 1998a, b) and on chromosome #2 (Sun et al. 2006) in a cross involving Col 5.T/02428 against mixed population of biotype 1 and 2. Likewise Ren et al. (2004) have reported the QTLs for BPH resistance on chromosome # 2 against biotype 1 using standard seed box screening technique in the RILs derived from the cross MH 63 × B5. QTLs associated with seedling resistance were also reported earlier on chromosome 3 and 4 (Sun et al. 2005; Huang et al. 2001; Soundararajan et al. 2004; Heinrichs et al. 1985; Renganayaki et al. 2000; Myint et al. 2012; Qiu et al. 2012; Wang et al. 2013; Hu et al. 2015).
The QTL qBphDS6 accounted for 24.23% of phenotypic variance noted in SSST as damage score between RM19697 and RM5855 with LOD 2.56 on short arm of chromosome 6. Yang et al. (2012) also reported the presence of bph20(t) on the short arm of chromosome 6 about 2.7 cM to the upper marker RM435 and 2.5 cM to lower marker RM540 and in a 2.5 cM region flanked by two new SSR markers BYL7 and BYL8. Xu et al. (2002) identified two QTLs for damage score on chromosome 6 with flanking RFLP marker G294d–G294a and RZ2-C having a LOD of 3.39 and 6.82 and a phenotypic variance of 4.3 and 6.3% respectively. Fujita et al. (2013) noted a cluster of BPH resistance genes (cluster C) on chromosome 6 which includes Bph3, bph4, qBph6(t), Bph25(t) and Ovc.
Two QTLs were identified for days to wilt in ARC10550 located on chromosome 3 and 8 explaining 4.55 and 10.48% phenotypic variance, respectively which were different from those detected by earlier researchers (Soundararajan et al. 2004; Alam and Cohen 1998a). Soundararajan et al. (2004) revealed that two QTLs reported one each for days to wilt 30 and 60 DAS were mapped on chromosome # 6 and 7 respectively in a doubled haploid population derived from IR64 × Azucena when tested against biotype similar to biotype 4.
A QTL qBphDW(30)-3 was identified on long arm of chromosome 3 with flanking marker RM7197 and RM6987 with a LOD of 4.21 and phenotypic variance of 4.55% and at a physical position of 34.7 Mb. Wang et al. (2013) also reported a QTL for SBPH resistance, designated qSBPH3 which was mapped between RM22 and RM545 on chromosomes 3 with LOD scores of 2.54 and phenotypic variance 7.7%. Alam and Cohen (1998a, b) reported six QTLs for BPH on chromosome 3 between RG191 and RZ67. Duan et al. (2010) identified a QTL on chromosome 3 between R2170 and C1135 SBPH (small brown planthopper) resistance.
Another major QTL qBphDW(30)-8 for days to wilt was identified on short arm chromosome 8 between RM547 and RM22741 with a LOD of 4.1 and showing with 10.48%. phenotypic variance. Alam and Cohen (1998a, b) also reported a QTLs for BPH damage score (field) and antixenosis on chromosome 8. Hou et al. (2011) reported one QTL on short arm chromosome 8, which was located at RM2655–RM3572 with LOD values of 3.15, explaining 14.9% of the phenotypic variation. The presumed view of the QTLs additive effect indicated that the gene effect increasing resistance to BPH originated from the parent WB01, there after, they were named temporarily as bph23(t). Tuyen et al. (2012) also reported QTLs on SSST method chromosome 8 in SBPH resistance. The QTL for resistance index (bulk test) and resistance index (independent test) were reported on chromosome 8 flanking RFLP marker C1073a–G187 with LOD 2.10 and phenotypic variance 3.5% by Xu et al. (2002).
Two QTLs were identified for nymphal preference (48 h) and nimphal preference (72 h). One QTL for nymphal preference (48 h) was mapped on long arm chromosomes 1 explaining 8.69% phenotypic variance between RM11704 and RM1068 with LOD 2.7. Another QTL qBphNP(72) was identified for Nymphal preference (72 h) on long arm chromosomes 12 explaining 7.66% phenotypic variance between RM27971 and RM28024 with LOD 3.57. Alam and Cohen (1998a, b) reported QTLs for BPH on chromosome 1 for damage score (seed box) and antixenosis. Xu et al. (2002) reported QTL between RFLP marker R210-RZ382 with LOD 4.51 and phenotypic variance 13.7 on resistance index (bulk test) and resistance index (independent test) while Sonalikar et al. (2011) reported QTLs for feeding rate linked with HvSSR-1-46. Hu et al. (2015) reported that trait RS2 (phenotypes of BPH resistance scores 10 and 12 DAI) qBph12 on chromosome 12 between RM235 and RM17 with phenotypic variance 4.93% and LOD 4.27. Fujita et al. (2013) have summed up that (cluster A) chromosome 12 long arm (Chr12L) as the Bph-resistance region (RR). Since this region has eight genes (Bph1, bph2, Bph9, Bph10, Bph16(t), Bph18(t), Bph21(t) and Bph26(t)). All of eight genes are associated with BPH resistance and have been identified from either O. sativa or wild rice species. The low proportion of genetic variation explained by the QTLs may be due to two factors, first, BPH resistance is a very complex trait and difficult to measure as the screening parameters vary with the conditions of plant growth, the insects and the environments in which the test has been carried out. Second, may be the use of F2.3 mapping population which might have overestimated the experimental error because of genetic heterogeneity within each of the F3 families.
Breakdown of resistance to BPH was reported in the varieties carrying single major resistance gene (Medina et al. 1996). In India, the BPH biotype 4 is prevalent and it is very virulent hence many of the major resistant genes identified against other biotype of BPH are ineffective, (Ram et al. 2010; Deen et al. 2010). The donor like PTB33 with major genes along with some QTLs are stable over the years and locations for resistance to all biotype of BPH. Similar observations were also reported by Alam and Cohen (1998a, b) in case of IR64 which showed durable resistance to BPH due to the involvement of quantitative trait loci and Bph1 a major gene. QTLs for BPH resistance in rice have been reported by several researchers (Alam and Cohen 1998a, b; Murata et al. 2001; Xu et al. 2002) but use of those QTLs in improving BPH resistance have not been used so for using MAS. The reported QTLs with major effects need to be validated and fine mapped for using in marker aided breeding along with major resistance genes having additive effects to develop effective and durable resistance against BPH. The two major QTLs identified in the present study for damage score (qBphDS-6) and for days to wilt (qBphDW(30)-8) are presently in the processes of validation and fine mapping for use in marker aided breeding programme.
Conclusion
The present results indicated that bph 5 gene in ARC10550 as reported earlier (Khush et al. 1985) is not a single recessive gene but it could be controlled with multiple loci for the resistance to BPH. We identified five QTLs qBphDs6, qBphNp1, qBphNp12, qBphDw3 and qBphDw8 associated BPH (biotype 4) resistance in ARC10550. The two major QTLs qBphDs6 for damage score and qBphDw8 for days to wilt are important for further investigation and use in breeding programme. The positive correlations between the traits related to different mechanism is important for pyramiding the traits. It is known that the quantitative resistance to BPH may result from different mechanisms; such as non-preference or antixenosis, antibiosis and tolerance would help in marker aided pyramiding of major resistance gene along with some QTLs could provide effective and suitable control to BPH. We are in the processes of fine mapping of both the major QTLs which can be further used in marker aided breeding along with major genes to improve resistance and durability.
References
Alagar M, Suresh S, Saravanan PA (2008) Feeding behaviour of Nilaparvata lugens (Stål.) on selected rice genotypes. Ann Plant Prot Sci 16(1):55–60
Alam SN, Cohen MB (1998a) Detection and analysis of QTLs for resistance to the brown planthopper, Nilaparvata lugens, in a double-haploid rice population. Theor Appl Genet 97:1370–1379
Alam SN, Cohen MB (1998b) Durability of brown planthopper, Nilaparvata lugens, resistance in rice variety IR64 in greenhouse selection studies. Entomol Exp Appl 89:71–78
Cha YS, Ji H, Yun DW, Ahn BO, Lee MC, Suh SC, Lee CS, Ahn EK, Jeon YH, Jin ID, Sohn JK, Koh HJ, Eun MY (2008) Fine mapping of the rice Bph1 gene, which confers resistance to the brown planthopper [Nilaparvata lugens (Stål)], and development of STS markers for marker-assisted selection. Mol Cells 26:146–151
Chen D, Wang GL, Ronald PC (1997) Location of the rice blast resistance locus Pi 5 (t) in Moroberekan by AFLP bulk segregant analysis. Rice Genet Newsl 14:95–97
Cohen MB, Alam SN, Medina EB, Bernal CC (1997) Brown planthopper, Nilaparvata lugens, resistance in rice cultivar IR64: mechanism and role in successful N. lugens management in Central Luzon, Philippines. Entomol Exp Appl 85:221–229
Deen R, Ramesh K, Gautam SK, Rao YK, Lakshmi VJ, Viraktamath BC, Brar DS, Ram T (2010) Identification of new gene for BPH resistance introgressed from O. rufipogon. Rice Genet Newsl 25:70–72
Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, He G (2009) Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci USA 106(52):22163–22168
Duan CX, Su N, Cheng Z, Lei CL, Wang JL, Zhai HQ, Wan JM (2010) QTL analysis for the resistance to small brown planthopper, Laodelphax striatellus (Fallen) in rice using backcross inbred lines. Plant Breed 129:63–67
Fujita Daisuke, Kohli Ajay, Horgan Finbarr G (2013) Rice resistance to planthoppers and leafhoppers. Crit Rev Plant Sci 32(3):162–191. doi:10.1080/07352689.2012.735986
Gallagher KD, Kenmore PE, Sogawa K (1994) Judicial use of insecticides deter planthopper outbreaks and extend the life of resistant varieties in Southeast Asian rice. In: Denno RF, Perfect JT (eds) Planthoppers: their ecology and management. Chapman and Hall, New York, pp 599–614
Hao P, Liu C, Wang Y, Chen R, Tang M, Du B, Zhu L, He G (2008) Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism of host resistance. Plant Physiol 146:1810–1820
Heinrichs EA (1986) Perspectives and directions for the continued development of insect resistant rice varieties. Agric Ecosyst Environ 18(1):9–36
Heinrichs EA, Medrano FG, Rapusas HR (1985) Genetic evaluation for insect resistance in rice. International Rice Research Institute, Los Baños, pp 1–356
Heninrichs EA, Mochida O (1984) From secondary to major pest status: the case of insecticide-induced rice brown planthopper, Nilaparvata lugens resurgence. Prot Ecol 7:201–218
Hou LY, Ping YU, Qun XU, Yuan XP, Yu HY, Wang YP, Wang CH, Guo WA, Tang SX, Peng ST, Wei XH (2011) Genetic analysis and preliminary mapping of two recessive resistance genes to brown planthopper, Nilaparvata lugens Stål in rice. Rice Sci 18(3):238–242
Hu J, Xiao C, Cheng MX, Gao GJ, Zhang QL, He YQ (2015) A new finely mapped Oryza australiensis-derived QTL in rice confers resistance to brown planthopper. Gene 561(1):132–137. doi:10.1016/j.gene.2015.02.026
Huang Z, He G, Shu L, Li X, Zhang Q (2001) Identification and mapping of two brown planthopper resistance genes in rice. Theor Appl Genet 102:929–934
Standard evaluation system for rice (1996) Genetic Resources Centre, 4th edn. International Rice Research Institute, Los Baños, pp 29–30
Jena KK, Jeung JU, Lee JH, Choi HC, Brar DS (2006) High resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18(t) and marker assisted selection for BPH resistance in rice (Oryza sativa L.). Theor Appl Genet 112:288–297
Kalode MB, Kasi PR, Viswanathan Seshu DV (1975) Standard test to characterize host plant resistance to brown planthopper in rice. Indian J Plant Prot 3(2):204–206
Kar S, Verulkar SB, Verma S, Pali V (2011) Identification of QTLs for brown planthopper (Nilaparvata lugens Stål) resistance in RIL mapping population of rice (Oryza sativa L.). Plant Arch 11(2):687–692
Kennedy GG, Gould F, Deponti OMB, Stinner RE (1987) Ecological, agricultural, genetic and commercial considerations in the deployment of insect-resistant germplasm. Environ Entomol 16:327–338
Khush GS, Karim ANMR, Angeles ER (1985) Genetics of resistance of rice cultivar ARC10550 to Bangladesh brown planthopper biotype. J Genet 64:121–125
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Krishna TS, Sheshu DV, Kalode MB (1984) Inheritance of resistance to whitebacked planthopper in rice. Indian J Genet 44(2):329–335
Li R, Li L, Wei S, Wei Y, Chen Y, Bai D, Yang L, Huang E, Lu W, Zhang X, Li X, Yang X, Wei Y (2006) The evaluation and utilization of new genes for brown planthopper resistance in common wild rice (Oryza rufipogon Griff.). Mol Plant Breed 4:365–371
Liu Y, Su C, Jiang L, He J, Wu H, Peng C, Wan J (2009) The distribution and identification of brown planthopper resistance genes in rice. Hereditas 146(2):67–73
Medina EB, Bernal CC, Cohen MB (1996) Role of host plant resistance in successful control of brown planthopper in Central Luzon, Philippinenes. IRRN 21(2–3):53
Murata K, Fujiwara M, Murai H, Takumi S, Mori N, Nakamura C (2001) Mapping of a brown planthopper (Nilaparvata lugens Stål) resistance gene Bph9 on the long arm of chromosome 12. Cereal Res Commun 29:245–250
Myint KKM, Yasui H, Takagi M, Matsumura M (2009a) Virulence of long-term laboratory populations of the brown planthopper, Nilaparvata lugens (Stål), and whitebacked planthopper, Sogatella furcifera (Horvath; Homoptera: Delphacidae), on rice differential varieties. Appl Entomol Zool 44:149–153
Myint KKM, Matsumura M, Takagi M, Yasui H (2009b) Demographic parameters of long-term laboratory strains of the brown planthopper, Nilaparvata lugens Stål (Homoptera: Delphacidae) on resistance genes bph20(t) and Bph21(t) in rice. J Fac Agric Kyushu Univ 54:159–164
Myint KKM, Fujita D, Matsumura M, Sonoda T, Yoshimura A, Yasui H (2012) Mapping and pyramiding of two major genes for resistance to the brown planthopper (Nilaparvata lugens Stål) in the rice cultivar ADR52. Theor Appl Genet 124:495–504
Normile D (2008) Agricultural research reinventing rice to feed the world. Science 321:330–333
Paguia P, Pathak MD, Heinrichs EA (1980) Honeydew excretion measurement techniques for determining differential feeding activity of biotypes of Nilaparvata lugens on rice varieties. J Econ Entomol 73:35–40
Panda N, Heinrichs EA (1983) Levels of tolerance and antibiosis in rice varieties having moderate resistance to the brown planthopper, Nilaparvata lugens (Stål) (Hemiptera: Delphacidae). Environ Entomol 12:1204–1214
Qiu YF, Guo JP, Jing SL, Zhu LL, He GC (2010) High-resolution mapping of the brown planthopper resistance gene Bph6 in rice and characterizing its resistance in the 9311 and Nipponbare near isogenic backgrounds. Theor Appl Genet 121:1601–1611
Qiu YF, Guo JP, Jing SL, Tang M, Zhu LL, He GC (2011) Identification of antibiosis and tolerance in rice varieties carrying brown planthopper resistance genes. Entomol Exp Appl 141:224–231
Qiu YF, Guo JP, Jing SL, Zhu LL, He GC (2012) Development and characterization of japonica rice lines carrying the brown planthopper resistance genes Bph12 and Bph6. Theor Appl Genet 124(3):485–494
Ram T, Deen R, Ramesh K, Gautam SK, Rao YK, Brar DS (2010) Identification of new genes for brown planthopper resistance in rice introgressed from O. glaberrima and O. minuta. Rice Genet Newsl 25:67–69
Ren X, Wang X, Yuan H, Weng Q, Zhu L, He G (2004) Mapping quantitative trait loci and expressed sequence tags related to brown planthopper resistance in rice. Plant Breed 123:342–348
Renganayaki K, Fritz AK, Sadasivam S, Pammi S, Harrington S, McCouch SR, Kumar SM, Reddy AS (2000) Mapping and progress toward map-based cloning of brown planthopper biotype-4 resistance gene introgressed from Oryza officinalis into cultivated rice O. sativa. Crop Sci 42:2112–2117
Santhanalakshmi S, Saikumar S, Rao S, Sai Harini A, Khera P, Shashidhar HE, Kadirvel P (2010) Mapping genetic locus linked to brown planthopper resistance in rice (Oryza sativa L.). Int J Plant Breed Genet 4(1):13–22
Shi ZY, Ren X, Weng QM, Li XH, He CG (2003) Construction of a genomic library from a brown planthopper resistant rice line spanning the Qvb one locus. Plant Sci 165:879–885
Sogawa K (1982) The rice brown planthopper: feeding physiology and host plant interactions. Ann Rev Entomol 27:49–73
Soundararajan RP, Kadirvel P, Gunathilagaraj K, Maheswaran M (2004) Mapping of quantitative trait loci associated with resistance to brown planthopper in rice by means of a doubled haploid population. Crop Sci 44:2214–2220
Su CC, Cheng XN, Zhai HQ, Wan JM (2002) Detection and analysis of QTL for resistance to brown planthopper, Nilaparvata lugens (Stål), in rice (Oryza sativa L.), using backcross inbred lines. Acta Genet Sin 29:332–338
Sun L, Su C, Wang C, Zai H, Wan J (2005) Mapping of a major resistance gene to brown planthopper in the rice cultivar Rathu Heenati. Breed Sci 55:391–396
Sun LH, Wang CM, Su CC, Liu YQ, Zhai HQ, Wan J (2006) Mapping and marker-assisted selection of a brown planthopper resistance gene bph2 in rice (Oryza sativa L.). Acta Genet Sin 33:717–723
Sun L, Liu Y, Jiang L, Su C, Wang G, Zhai H, Wan J (2007) Identification of quantitative trait loci associated with resistance to brown planthopper in the indica rice cultivar Col. 5 Thailand. Hereditas 144(2):48–52
Tamura Y, Hottori M, Yoshioka H, Yoshioka M, Takahashi A, Sentoku N, Yasui H (2014) Map-based cloning and characterization of a brown planthopper resistance gene Bph26 from Oryza sativa L. ssp. indica cultivar ADR52. Sci Rep 4:5872–5879
Tuyen L, Liu Y, Jiang L, Wang BX, Wang Q, Than TTH, Wan JM (2012) Identification of quantitative trait loci associated with small brown planthopper, Laodelphax striatellus (Fallen) resistance in rice (Oryza sativa L.). Hereditas 149:16–23
Van Ooijen JW (2006) JoinMap 4.0 software for the calculation of genetic linkage maps in experimental populations. Kyzama B V, Wageningen
Voorrips R (2002) Map Chart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh. User manual, pp 1–87. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Wang Q, Liu Y, Hu J, Zhang Y, Xie K, Wang B, Tuyen LQ, Song Z, Wu H, Liu Y, Jiang L, Liu S, Cheng X, Wang C, Zhai H, Wan J (2013) Detection of quantitative trait loci (QTLs) for resistances to small brown planthopper and rice stripe virus in rice using recombinant inbred lines. Int J Mol Sci 14:8406–8421. doi:10.3390/ijms14048406
Wu H, Liu Y, He J, Liu Y, Jiang L, Liu L, Wang C, Cheng X, Wan J (2014) Fine mapping of brown planthopper (Nilaparvata lugens Stål) resis-tancegene Bph28(t) in rice (Oryza sativa L.). Mol Breed 33:909–918
Xu XF, Mei HW, Luo LJ, Cheng XN, Li ZK (2002) RFLP facilitated investigation of the quantitative resistance of rice to brown planthopper (Nilaparvata lugens). Theor Appl Genet 104:248–253
Yang L, Li RB, Li YR, Huang FK, Chen YZ, Huang SS, Huang LF, Liu C, Ma ZF, Huang DH, Jiang JJ (2012) Genetic mapping of bph20(t) and bph21(t) loci conferring brown planthopper resistance to Nilaparvata lugens Stål in rice (Oryza sativa L.). Euphytica 183:161–171
Zeng Z (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zheng KL, Subudhi PK, Domingo J, Magpantay G (1995) Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genet Newsl 12:255–258
Acknowledgements
We thank the Director, Indian Institute of Rice Research, Hyderabad for the facilities and encouragement. This work was partly supported by a grant from the Department of Biotechnology (DBT F.No. BT/AB/FG-2 (PH-II) Sep-2009), Government of India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Deen, R., Ramesh, K., Padmavathi, G. et al. Mapping of brown planthopper [Nilaparvata lugens (Stål)] resistance gene (bph5) in rice (Oryza sativa L.). Euphytica 213, 35 (2017). https://doi.org/10.1007/s10681-016-1786-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-016-1786-z