Abstract
Brown planthopper (BPH) is a destructive insect pest of rice in Asia. Identification and the incorporation of new BPH resistance genes into modern rice cultivars are important breeding strategies to control the damage caused by new biotypes of BPH. In this study, a major resistance gene, Bph18(t), has been identified in an introgression line (IR65482-7-216-1-2) that has inherited the gene from the wild species Oryza australiensis. Genetic analysis revealed the dominant nature of the Bph18(t) gene and identified it as non-allelic to another gene, Bph10 that was earlier introgressed from O. australiensis. After linkage analysis using MapMaker followed by single-locus ANOVA on quantitatively expressed resistance levels of the progenies from an F2 mapping population identified with marker allele types, the Bph18(t) gene was initially located on the subterminal region of the long arm of chromosome 12 flanked by the SSR marker RM463 and the STS marker S15552. The corresponding physical region was identified in the Nipponbare genome pseudomolecule 3 through electronic chromosome landing (e-landing), in which 15 BAC clones covered 1.612 Mb. Eleven DNA markers tagging the BAC clones were used to construct a high-resolution genetic map of the target region. The Bph18(t) locus was further localized within a 0.843-Mb physical interval that includes three BAC clones between the markers R10289S and RM6869 by means of single-locus ANOVA of resistance levels of mapping population and marker-gene association analysis on 86 susceptible F2 progenies based on six time-point phenotyping. Using gene annotation information of TIGR, a putative resistance gene was identified in the BAC clone OSJNBa0028L05 and the sequence information was used to generate STS marker 7312.T4A. The marker allele of 1,078 bp completely co-segregated with the BPH resistance phenotype. STS marker 7312.T4A was validated using BC2F2 progenies derived from two temperate japonica backgrounds. Some 97 resistant BC2F2 individuals out of 433 screened completely co-segregated with the resistance-specific marker allele (1,078 bp) in either homozygous or heterozygous state. This further confirmed a major gene-controlled resistance to the BPH biotype of Korea. Identification of Bph18(t) enlarges the BPH resistance gene pool to help develop improved rice cultivars, and the PCR marker (7312.T4A) for the Bph18(t) gene should be readily applicable for marker-assisted selection (MAS).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brown planthopper (BPH), Nilaparvata lugens (Homoptera: Delphacidae), is one of the most destructive phloem-sap-sucking insect pests of tropical and temperate rice in Asia. It transmits viral diseases such as ragged stunt virus (RSV) and grassy stunt virus (GSV) into tropical indica rice cultivars (Khush and Brar 1991). Host-plant resistance is the most important breeding strategy to control BPH damage in contrast to the use of pesticides. In Korea, BPH does not overwinter but migrates to the peninsula during the rice-cropping season through summer winds and typhoons from the southern parts of mainland China. Biotype 1 is the common biotype, which has caused heavy losses to rice production in recent years (Jeon et al. 1999).
The genetics of BPH resistance is well studied and 17 monogenically controlled resistance genes have been reported until now (Yang et al. 2004). The genes conferring resistance to South and Southeast Asian biotypes are mostly dominant in nature and two dominant genes have been introduced into indica rice cultivars (Sharma et al. 2003). Of the eight known recessive genes, bph2 and bph4 are linked to the dominant genes Bph1 and Bph3, respectively, but are independent of each other (Kawaguchi et al. 2001). Two recessive genes, bph11 and bph12, confer resistance to the BPH biotype of Japan. The resistance genes Bph1, bph2, Bph9, and Bph10 are located on chromosome 12; Bph3 and bph12 on chromosome 4; bph4 on chromosome 6; Bph6 on chromosome 11; and bph11 and Bph13 on chromosome 3 (Hirabayashi et al. 1999; Ishii et al. 1994; Jena et al. 2003b; Renganayaki et al. 2002; Sharma et al. 2003). Quantitative trait loci (QTLs) for BPH resistance have also been identified and major QTLs conferring resistance to BPH biotypes 1 and 2 have been reported (Alam and Cohen 1998; Soundararajan et al. 2004; Xu et al. 2002). However, two dominant genes, Bph14 and Bph15 previously named as Qbp1 and Qbp2, conferring strong resistance to the BPH biotype of China have been mapped to the long arm of chromosome 3 and the short arm of chromosome 4, respectively (Ren et al. 2004; Yang et al. 2004).
The biotypes of BPH are widespread in South and Southeast Asia. Biotypes 1 and 2 are distributed in Southeast Asia and biotype 3 is a laboratory biotype produced in the Philippines. Biotype 4 is the most destructive biotype of South Asia and is distributed over the Indian subcontinent (Heinrichs 1986). The sources of resistance to BPH have been identified mostly in landraces as well as in accessions of wild Oryza species (Brar and Khush 1997; Ishii et al. 1994; Yang et al. 2004). Nonetheless, the resistance genes are not durable; resistance breaks down because of the biotype change. The source of BPH resistance genes in temperate japonica rice germplasm is very limited because of narrow genetic diversity. It is imperative to identify BPH resistance genes from alternate sources and incorporate them into japonica rice cultivars.
Several introgression lines having genes for BPH resistance from wild Oryza species have been developed at the International Rice Research Institute (IRRI) (Brar and Khush 1997; Jena et al. 1991). Of the seven BPH resistance genes of wild species origin, Bph10 is from O. australiensis; bph11, bph12, Bph13, Bph14, and Bph15 are from O. Officinalis; and Bph12 is from O. latifolia (Ishii et al. 1994; Ranganayaki et al. 2002; Ren et al. 2004; Yang et al. 2004). These genes have been tagged with molecular markers.
This paper reports on the identification and fine mapping of a new BPH resistance gene, Bph18(t), in the introgression line IR65482-7-216-1-2 derived from O. australiensis and marker-assisted selection (MAS) with the resistance gene linked to markers using the Korean biotype of BPH. From a breeder’s point of view, this paper also discusses practical ways to determine the target locus of the BPH resistance gene as well as tag it with tightly linked DNA markers to ensure high efficiency in producing resistant progenies with less impact on other important agronomic traits.
Materials and methods
Plant materials
Eleven introgression lines with genes from wild species of Oryza (3, 4, and 4 lines of O. longistaminata, O. minuta, and O. australiensis, respectively) along with the original recurrent parent, IR31917-45-3-2, and four temperate rice cultivars, Jinbubyeo, Junambyeo, Taebaekbyeo, and Andabyeo, were initially screened with the Korean biotype of BPH. The elite japonica cultivars Jinbubyeo and Junambyeo are susceptible to BPH and two tongil-type cultivars, Taebakbyeo and Andabyeo, were used as susceptible and resistant checks, respectively. Seeds of introgression lines and IR31917-45-3-2 were obtained from the Plant Breeding, Genetics and Biotechnology Division and O. australiensis (Acc. No. 100882) was obtained from the Genetic Resources Center of the IRRI, Los Baños, Philippines. Seeds of the Korean rice cultivars were obtained from the Genetics and Breeding Division of the National Institute of Crop Science (NICS), Rural Development Administration (RDA), Republic of Korea.
Population development and DNA extraction
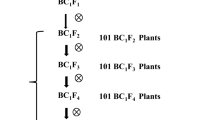
The introgression line IR65482-7-216-1-2 derived from an interspecific cross between IR31917-45-3-2 and O. australiensis (Acc. No. 100882) expressed strong resistance to the BPH biotype of Korea. The cross combinations between IR65482-7-216-1-2 and other lines are described in Table 1. In brief, progenies from a cross between Jinbubyeo and IR65482-7-216-1-2 were the base materials for conducting genetic evaluation of donor-derived resistance loci and developing markers tightly linked to the target locus for MAS application. The F2 mapping population was also used to collect data on important agronomic traits, such as fertility, days to heading, plant height, and number of tillers. Resistant seedlings of 18 BC2F2 families from crosses of Jinbubyeo × IR65482-7-216-1-2 and Junambyeo × IR65482-7-216-1-2 were used to evaluate the validity of the most tightly linked marker for the new resistance gene. Reciprocal crosses between the introgression lines (IR65482-4-136-2-2 carrying the Bph10 gene and IR65482-7-216-1-2) were made to generate F2 seeds for an allelism test of the resistance genes. Total genomic DNAs from young leaves of parental lines, mapping populations, and progenies after the BPH bioassay were prepared according to Murray and Thompson (1980), with minor modifications.
Bioassay for BPH resistance
A pure BPH population was developed from a single colony of BPH and was grown on the susceptible variety Taebaekbyeo in a cage in the temperature-controlled greenhouse facility of the Genetics and Breeding Division of NICS, RDA. The bioassay was done with a modified bulk seedling test following the method of Pathak et al. (1969). Seedlings at the three-leaf stage were infested with second- or third-instar nymphs at a density of 10–12 nymphs per seedling. Seedlings of F3 families of the F2 mapping population (Jinbubyeo × IR65482-7-216-1-2) were planted in a row with three replications. Other F2 and BC2F2 progenies (see Table 1) were planted into randomly selected rows. Evaluations were based on the degree of susceptibility of the S check and the insect bioassay was done once the S check was dead. Genotypes of BPH response (RR: homozygous resistant, RS: segregating heterozygous, SS: homozygous susceptible) of 94 F2 individuals were determined by assaying the phenotypes (R or S) of the corresponding F3 progenies. Seedling survival was considered in a quantitative manner. A complete resistance score (0–1) was given to progenies expressing 91–100% seedling survival, a resistance score of 2–3 was for progenies with 76–90% seedling survival, a score of 5–7 was for progenies with 11–75% survival, and a score of 8–9 was for progenies with complete susceptibility and 0–10% survival. Bioassays of 245 F2 progenies were conducted by six-time point collection of susceptible progenies to determine the most putative DNA marker linked to the BPH resistance gene. The japonica variety, Taebaekbyeo was used as the susceptible check and Andabyeo was used as the resistant check.
Linkage map construction and fine mapping of BPH resistance gene
Based on the recently developed high-resolution rice linkage map with SSR and STS markers (McCouch et al. 2002), 186 SSR and STS primer sets were tested on the parental lines Jinbubyeo and IR65482-7-216-1-2. Four restriction enzymes (AluI, HaeIII, HinfI, and RsaI: New England Biolabs, Beverly, MA, USA) were applied on the monomorphic STS-PCR products to find latent polymorphism. A total of 48 SSR and 10 STS primer sets displaying polymorphism were selected as anchor markers after careful comparisons with previous reports in terms of band sizes of amplified products. The F2 mapping population of 94 individuals was genotyped at the loci tagged by the preselected anchor markers. MAPMAKER/EXP 3.0 (Lincoln et al. 1992) was used to create linkage maps from the 58 markers spanning the 12 rice chromosomes. The Haldane mapping function (Haldane 1919) was used to convert recombination frequencies to map distances in centiMorgans (cM).
The intervening physical region of 1.612 Mb equivalent on the Nipponbare genome between markers RM463 and S15552 on chromosome 12 was identified using information from Gramene (http://www.gramene.org/markers/; Ware et al. 2002) and TIGR rice pseudomolecule 3 (http://www.tigr.org/tdb/e2k1/osa1/). Representative SSR and STS markers tagging the 15 BAC clones composing the 1.612 Mb virtual contig were screened on IR31917-45-3-2, IR65482-7-216-1-2, and Jinbubyeo for integration and a polymorphism test. The progenies of the F2 mapping population as well as BPH-susceptible seedlings of the additional 245 F2 progenies derived from the Jinbubyeo × IR65482-7-216-1-2 cross were further genotyped with positive markers to narrow down the putative locus of the resistance gene. The most tightly linked marker, 7312.T4A, with primer sequences F:5′ ACGGCGGTGAGCATTGG 3′ and R:5′ TACAGCGAAAAGCATAAAGAGTC 3′, was identified in the BAC clone OSJNBa0028L05 (GeneBank acc. AL935072). This marker was also used for a MAS test of the BPH resistance gene in BC2F2 progenies.
Statistical analysis
Chi-square analysis was used to analyze F3 progeny-row segregation for the BPH resistance gene and F2 segregation for the allelism test of reciprocal crosses between Bph10 and Bph18(t) genes. The percentage of phenotypic variation (R 2) explained was obtained from PROC GLM of the SAS statistical package (SAS Institute 2000) to estimate the relative contribution of particular loci on rice chromosome 12 for BPH resistance as well as agronomic traits recorded from 94 progenies of the F2 mapping population by single-locus ANOVA. For the F-test, markers having a P value of less than 0.01, which was approximately equivalent to an LOD of 2.5 in MARKER/QTL (Version 3.0b), were declared as significant empirically. The additive and dominance effects, and degree of dominance, were then estimated for the declared loci.
Results
BPH bioassay and genetic analysis
The introgression line IR65482-7-216-1-2 expressed strong resistance to the Korean biotype of BPH. However, four introgression lines derived from O. minuta, three introgression lines derived from O. australiensis, and three introgression lines derived from O. longistaminata were either susceptible or moderately resistant to BPH. The Korean japonica rice cultivars Jinbubyeo and Junambyeo were completely susceptible to BPH. The F1 plants of Jinbubyeo × IR65482-7-216-1-2 and Junambyeo × IR65482-7-216-1-2 showed complete resistance to BPH, indicating that BPH resistance in IR65482-7-216-1-2 is controlled by a dominant gene(s). Owing to the low fertility of some F2 plants, F3 progenies of 86 F2 individuals derived from an F2 mapping population of Jinbubyeo × IR65482-7-216-1-2 were screened for BPH reaction based on percent survival scores. F2 segregation for BPH resistance showed resistance or susceptibility ranging from complete susceptibility (15 plants) to segregating (47 plants) to resistance (24 plants). The F2 segregation showed a 1:2:1 segregation ratio (χ2=2.15) that indicated the presence of a major dominant gene conferring resistance to BPH.
Linkage map construction and localization of a resistance gene
Of the 186 SSR and STS markers used in the analysis of parental polymorphism, only 58 markers (48 SSR and 10 STS) detected polymorphism between Jinbubyeo and IR65482-7-216-1-2. These markers were distributed over the entire genome and PCR analysis of the mapping population using the anchor markers showed segregation of marker alleles (Fig. 1). The mapping population was successively genotyped with all 58 anchor markers, and a linkage map was constructed with an average marker density of one marker per 30 cM (Table 2).
Segregation pattern of anchor markers in an F2 mapping population with RM463 (a) and S15552 (b, cleaved by AluI). Sequencing gel electrophoresis (5% polyacrylamide, 6 M urea, 1× TBE) was applied for RM463 followed by standard silver staining visualization. For S15552, a natural polyacrylamide gel (5% polyacrylamide, 0.5X TBE) was used and stained by ethidium bromide. a Jinbubyeo, b IR65482-7-216-1-2
The genotypes of the F2 individuals of the mapping population for BPH resistance were inferred based on the phenotypes of the corresponding F3 progeny-rows using seedling survival ratings with ranges of 0–10%, 11–90%, and 91–100% for homozygous susceptible, segregating heterozygous, and homozygous resistant, respectively. All anchor markers on the framework map (linkage map skeleton) were tested for linkage relationships and a BPH resistance gene was localized on the long arm of chromosome 12. Two anchor markers, RM463 and S15552, on the long arm of chromosome 12 could not be separated from the putative BPH resistance-gene locus (Fig. 2a) even under extreme linkage criteria (minimum LOD score of 10 and maximum distance of 20 cM). Compared to the interval between RM463 and S15552 (15.9 cM), however, placing the putative resistance-gene locus near the two anchor markers tended to exaggerate the accumulative map length over a region of 20 cM (data not shown). ANOVA on the three subgroups divided into the genotypes of each locus with respect to the BPH bioassays percent survival for each individual in the subgroups also indicated that the 15.9-cM region delimited by RM463 and S15552 was strongly associated with BPH resistance (Table 3). This new BPH resistance-gene locus is named as Bph18(t).
a Linkage map skeleton of rice chromosome 12 showing ten polymorphic anchor markers and the putative R-gene location obtained by association analysis between marker genotypes of F2 progenies. Quantitative phenotyping of F3 lines (single-locus ANOVA) is indicated by two R- or S-associated markers (in boldface; RM463 and S15552). The open rectangle indicates the centromere region. b High-resolution map of Bph18(t) locus showing 15 Nipponbare BAC clones corresponding to the 15.9 cM interval delimited by RM463 and S15552. The BAC clones, which were not tested with any DNA marker, are indicated in italics. The negative markers for the integration events of O. australiensis segmented into IR31917-45-3-2 (progenitor line) are indicated in italics. The R- or S-associated DNA markers, polymorphic between Jinbubyeo and IR65482-7-216-1-2, are underlined. The PCR primer set for the 7312.T4A (in boldface) marker tagging the Bph18(t) gene originated from Nipponbare BAC clone OSJNBa0028L05. The intervals between tested loci were estimated by using 94 F2 mapping populations and corresponding physical distances (cM and Mb) were calculated based on the genome sequence information of Nipponbare chromosome 12
Minor QTL for BPH resistance affecting Bph18(t)
Single-locus ANOVA results suggested that the genetic effect of at least two more loci may be compounded with that of the Bph18(t) locus (R 2=0.47) during bioassay of the mapping population, one of them was on the short arm of chromosome 5 (tagged by RM13; R 2=0.14) and the other was on the end of chromosome 12 (tagged by C901; R 2=0.17; Table 3). In both cases, the allele type of IR65482-7-216-1-2 was favorable for increasing BPH resistance, and it is believed that the co-segregation of those loci with Bph18(t) has affected the genotypes of the F2 individuals to some extent.
Allelic relationship between Bph18(t) and Bph10 genes
The introgression line IR65482-4-136-2-2 of O. australiensis carries the Bph10 gene for BPH resistance. The F2 progenies derived from reciprocal crosses between IR65482-7-216-1-2 and IR65482-4-136-2-2 segregated for BPH resistance and showed a good fit to a 15:1 ratio (Table 4). Furthermore, the subcentromeric region of chromosome 12 (Fig. 2a), on which the Bph10 gene is located, did not respond significantly to linkage as well as single-locus ANOVA (Table 3). These results indicated that the BPH resistance gene derived from IR65482-7-216-1-2 and initially tagged by RM463 and S15552 is not in an allelic phase with the Bph10 gene derived from IR65482-4-136-2-2 (see also Tables 3, 5). Therefore, this new BPH resistance gene was nominated as Bph18(t).
High-resolution mapping of the Bph18(t) gene through e-landing
Electronic chromosome landing (e-landing) was opted to localize the Bph18(t) gene on the recently elucidated genome sequence of Nipponbare. Through a database search, the intervening physical regions between the DNA markers RM463 and S15552 were identified, in which the 1.612 Mb virtual contig is composed of 15 overlapping BAC clones (Fig. 2b). Seven SSR and four STS markers identified from the 15 BAC clones were selected and were surveyed for polymorphism between Jinbubyeo and IR65482-7-216-1-2 and the progenitor line IR31917-45-3-2 (Fig. 2b). Seven DNA markers (RM6869, RM6217, R10289S, RM3726, R2708, RM7376, and RM3331) were positive for polymorphism between Jinbubyeo and IR65482-7-216-1-2 and segregated for marker alleles in F2 progenies as well as between IR65482-7-216-1-2 and IR31917-45-3-2 (the putative integration events of O. australiensis segments). Those seven markers were further tested on the F2 mapping population as well as susceptible seedlings derived from additional screening of F2 progenies. Some 86 susceptible seedlings were collected at six time points and genotyped with the seven DNA markers along with RM101 and S10704 to test the centromeric region (Fig. 2a, b). However, the segregating allele types tagged by RM6217 did not exhibit faithful banding patterns and were not included in further analysis. As a posteriori judgment on the genotypes of the susceptible F2 seedlings, it was realized that the high degree of R or S association rapidly broke down across all the dispersed loci tested, including the centromere region tagging markers from the collection of the 12th day. Therefore, the genotypes from the 49 susceptible F2 seedlings collected until the ninth day after infestation were included to identify the most putative location of the Bph18(t) gene (Table 5). The relative ratios for detection of homozygous alleles of IR65482-7-216-1-2 at all tested loci were compared taking into account conflicting evidence; this data suggests that the most likely position for Bph18(t) is near the STS marker R10289S on BAC clone OSJNBb0076G11 (Table 5; Fig. 2b).
The TIGR annotated gene contents of each BAC clone were surveyed. A putative resistance gene locus with the temporary identifier 7312.m00152 as annotated by TIGR was identified in a 155.6-kb BAC clone OSJNBb0028L05 (Acc. AL935702) that is the successive BAC clone to OSJNBb0076G11 containing the locus R10289S. The 7312.m00152 locus has two exons split by one intron, and a primer set for STS marker 7312.T4A was designed from the first exon with a predicted PCR product size of 1033 bp on the Nipponbare genome.
Evidence of O. australiensis chromosome segment integration
The STS primer set 7312.T4A amplified a single strong band having the predicted size of 1033 bp for Jinbubyeo and Junambyeo. Meanwhile, the 1078 bp PCR products were identified for both IR65482-7-216-1-2 and O. australiensis Acc. 100882 (Fig. 3). This positive result supported the integration of O. australiensis DNA segment(s) into IR31917-45-3-2, which occurred near the 7312.T4A locus.
Introgression test of O. australiensis segment at the locus tagged by the marker 7312.T4A. Tested lines are A Jinbubyeo, D Junambyeo, B IR65482-7-216-1-2, C IR31917-45-3-2, and E O. australiensis (accession # 100882). The 7312.T4A primer set amplified two different allele types (1,033 and 1,078 bp) among Jinbubyeo, Junambyeo, IR65482-7-216-1-2, and IR31917-45-3-2 (a). The HinfI-digested PCR products revealed the 1,078-bp band of IR65482-7-216-1-2 as a different allele type from that of IR31917-45-3-2 (b). The 7312.T4A primer set also generated 1,078 bp in O. australiensis that was of the same band size identified in IR65482-7-216-1-2 (c). Natural polyacrylaimde gel electrophoresis was used to resolve the PCR products and cleaved fragments by HinfI. The gels were stained by ethidium bromide. M 2-Log DNA ladder (NEB)
MAS test for the BPH resistance gene linked to marker 7312.T4A
The individual genotyping of 86 susceptible F2 seedlings for the 7312.T4A locus exhibited the strongest R or S association in the BPH bioassay. The directions of the crossover events between two adjacent loci supported that 7312.T4A is the marker most tightly linked to the Bph18(t) gene (Table 5). The highest F value was detected at the 7312.T4A locus (P=6.8E−10, R 2=0.47) during single-locus ANOVA of all DNA markers on the long arm of chromosome 12 for BPH resistance levels of the F2 mapping population (Table 3). Related to the important agronomic traits tested simultaneously, two putative minor QTLs were detected on the long arm of chromosome 12 for plant height (RM463) and panicle number (C60772). Those putative QTL alleles of IR65482-7-216-1-2, however, are separate from the 7312.T4A locus allowing them to be eliminated among progenies through background selection if their allele effects are considered as deleterious linkage drags (see the marker intervals in Table 3).
The PCR marker 7312.T4A was further validated as a complete R- or S-associated DNA marker by genotyping the resistant BC2F2 progenies having different genetic backgrounds of Jinbubyeo and Junambyeo. Some 97 resistant BC2F2 individuals were randomly selected from the resistant seedlings rescued after insect bioassay of 433 BC2F2 progenies (see Table 1). Amplification of the 7312.T4A locus in BC2F2-resistant progenies did not detect any homozygous susceptible marker allele of Jinbubyeo or Junambyeo (Fig. 4). The resistant plants were from two genetic backgrounds but revealed a resistance-specific marker allele in the resistant plants in either the homozygous or heterozygous state, thus confirming the dominant nature of BPH resistance to the biotype of Korea.
PCR amplification of 45 and 52 BC2F2-resistant plants from JinbubyeoX IR65482-7-216-1-2 (a) and JunambyeoX IR65482-7-216-1-2 (b), respectively, with the primer set of 7312.T4A as a complete R or S marker-associated with the Bph18(t) gene. Progenies marked with a closed circle are homozygous for the IR65482-7-216-1-2 (lanes B and D) allele and asterisks indicate heterozygous progenies. Lanes A Jinbubyeo and Junambyeo. Note: None of the progenies was homozygous for the Jinbubyeo or Junambyeo allele. PCR products were cleaved by HinfI and gels were stained by ethidium bromide
Discussion
BPH is a major biotic stress in rice production in most Asian countries, including Korea and Japan. In Korea, BPH resistance genes such as Bph1 and bph2 were introduced into Korean japonica cultivars by conventional breeding methods during the 1980s. However, because of changes in BPH biotype and infestation patterns, varieties with the Bph1 gene for resistance have become susceptible. It is also reported that biotype changes occurred because of the immigration of new biotypes of BPH from neighboring China by summer wind since BPH never overwinters in Korea (Choi et al. 1979; Yeo et al. 1998). Therefore, identification of a new source of BPH resistance genes followed by cost-effective incorporation into japonica cultivars are important breeding strategies in temperate rice.
The main finding of this study is the identification of a new BPH resistance gene originally transferred from the EE genome wild species O. australiensis (Jena et al. 1991) and its molecular mapping on chromosome 12. Previously, the Bph10 gene from O. australiensis conferring resistance to the Philippine biotype of BPH was identified in breeding line IR65482-4-136-2-2 at IRRI, and the gene was mapped on chromosome 12 (Ishii et al. 1994). Even though the source of the resistance genes is the same, genetic analysis has revealed the non-allelic nature of the two genes. The two genes segregated as two independent dominant genes, suggesting that several restricted homoeologous recombinations might have occurred between the EE genome of O. australiensis and the AA genome of O. sativa.
Of the 17 major BPH resistance genes reported to date, nearly half of them are derived from four distantly related wild species of Oryza (Yang et al. 2004). Twelve of the 17 resistance genes have been tagged by different DNA markers. Nevertheless, most of the markers, except for the Bph1, bph2, and Bph15 genes, are not tightly linked to resistance genes and are difficult to use as markers in practical breeding for BPH resistance. In this context, accurate phenotyping, fine mapping of resistance-gene loci, and development of reliable PCR markers would be of great value in MAS application.
This study has identified a major resistance gene, Bph18(t), on the subterminal region of the long arm of chromosome 12 using six time-point BPH bioassays with quantitative phenotyping and has used Nipponbare genome sequence information for precise mapping of the resistance gene (Sasaki 2005). The location of the Bph18(t) gene locus is quite interesting as this study has identified BAC clones in the region flanked by RM463 and S15552. PCR markers corresponding to the BAC clones have been used to analyze susceptible plants derived from time-point BPH bioassays. This approach has enabled one to localize the R gene in a 0.843-Mb genomic region flanked by markers RM6869 and R10289S. Further using Nipponbare genome sequence information and gene annotation system from Gramene and TIGR one could identify a BAC clone, OSJNBa0028L05, located near the BAC clone OSJNBb0076G11. BAC clone OSJNBa0028L05 contains 26 ORF with a range of genes, including different retro-elements. In the study, PCR primers were designed for STS marker 7312.T4A that was linked to a putative resistance protein sequence. The PCR analysis of F2 progenies with the marker corresponding to the first exon completely co-segregated with the resistance phenotype and also did not reveal a resistance-specific marker band in the plants with a susceptible phenotype. Thus, it is predicted that the STS marker 7312.T4A could be the location of the BPH resistance gene inherited from the wild species O. australiensis. This approach of resistance gene mapping has shown an advantage over the standard methods of gene tagging, thus saving time and resources. The absence of polymorphism for four BAC clone-derived DNA markers flanking RM463 and S15552 within the physical distance of 1.612 Mb may be attributed to the high degree of synteny between the chromosome segments derived from O. australiensis and O. sativa. BPH resistance genes such as Bph1, bph2, Bph9, and Bph10 are also located on the long arm of chromosome 12. However, they have been tagged to different classes of DNA markers such as RFLP and AFLP without using prior sequence information, therefore making their application in breeding for BPH resistance less reliable. The presence of five BPH resistance genes on the long arm of chromosome 12 indicates clustering of resistance gene loci similar to the clustering of blast resistance genes in rice (Khush and Brar 2001). Further studies are needed to better understand the interaction between different R genes for BPH resistance on chromosome 12.
Another important finding of this study is the MAS application of the PCR marker linked to the Bph18(t) resistance gene. Identification of a tightly linked DNA marker is a prerequisite for MAS application in rice improvement (Jena et al. 2003a). In this study, BC2F2 progenies segregating for BPH resistance were derived from two crosses that involved japonica rice cultivars Jinbubyeo and Junambyeo. MAS application of the R-gene-linked marker 7312.T4A on randomly selected R plants from both crosses accurately amplified the R-gene-specific marker allele, thus demonstrating the potential of fine mapping the resistance gene.
None of the BPH resistance genes have been cloned in crop plants. To date, the only example of a cloned resistance gene to a pest is the Mi gene conferring resistance to nematodes and to potato aphids that belongs to the NBS-LRR class of resistance genes (Vos et al. 1998; Ronald 1998). The gene Bph18(t) identified in this study is of wild species origin, and it could be a novel resistance gene that should be cloned. Fortunately, BAC libraries of 12 wild species of Oryza belonging to 12 genomes have been completed (Wing 2005). These BAC libraries include accession number 100882 of O. australiensis, the source of Bph18(t). It would be ideal to clone the new BPH resistance gene through selection of the BAC clone corresponding to the segment carrying the gene.
References
Alam SN, Cohen MB (1998) Detection and analysis of QTLs for resistance to the brown planthopper, Nilaparvata lugens, in a doubled-haploid rice population. Theor Appl Genet 97:1370–1379
Brar DS, Khush GS (1997) Alien introgression in rice. Plant Mol Biol 35:35–47
Choi SY, Heu MH, Lee JO (1979) Varietal resistance to brown planthopper in Korea: In: Brown planthopper: threat to rice production in Asia. IRRI, Los Baños, pp 219–232
Haldane JBS (1919) The combination of linkage values and the calculation of distances between the loci of linked factors. J Genet 8:299–309
Heinrichs EA (1986) Perspectives and directions for the continued development of insect resistant rice varieties. Agric Ecosyst Environ 18: 9–36
Hirabayashi H, Kaji R, Angeles ER, Ogawa T, Brar DS, Khush GS (1999) RFLP analysis of a new gene for resistance to brown planthopper derived from O. officinalis on rice chromosome 4 (abstract in Japanese). Breed Res 1(Suppl 1):48
Ishii T, Brar DS, Multani DS, Khush GS (1994) Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, Oryza sativa. Genome 37:217–221
Jena KK, Multani DS, Khush GS (1991) Monosomic alien addition lines of Oryza australiensis and alien gene transfer. Rice Genet II:728
Jena KK, Moon HP, Mackill DJ (2003a) Marker-assisted selection: a new paradigm in plant breeding. Korean J Breed 35:133–140
Jena KK, Pasalu IC, Rao YK, Varalaxmi Y, Krishnaiah K, Khush GS, Kochert G (2003b) Molecular tagging of a gene for resistance to brown planthopper in rice (Oryza sativa L.). Euphytica 129:81–88
Jeon YH, Ahn SN, Choi HC, Hahn TR, Moon HP (1999) Identification of a RAPD marker linked to a brown planthopper resistance gene. Euphytica 107:23–28
Kawaguchi M, Murata K, Ishii T, Takumi S, Mori N, Nakamura C (2001) Assignment of a brown planthopper (Nilaparvata lugens Stal) resistance gene bph4 to rice chromosome 6. Breed Sci 51:13–18
Khush GS, Brar DS (2001) Rice genetics from Mendel to functional genomics. Rice Genet IV:3–25
Khush GS, Brar DS (1991) Genetics of resistance to insects in crop plants. Adv Agron 45:223–274
Lincoln S, Daly M, Lander ES (1992) Constructing genetic maps with MAPMAKER/EXP 3.0, 3rd edn. Whitehead Institute Technical Report, Cambridge
McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton M, Fu BY, Maghirang R, Li ZK, Xing YZ, Zhang QF, Kono I, Yano M, Fjellstom R, Declerck G, Scheider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Res 9:199–207
Murray MG, Thompson WF (1980) Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res 8:4321–4325
Pathak MD, Cheng CH, Fortuno ME (1969) Resistance to Nephotettix impictiveps and Nilaparvata lugens in varieties of rice. Nature 223:502–504
Ren X, Weng QM, ZhuLL, He GC (2004) Dynamic mapping of quantitative trait loci for resistance to brown planthopper in rice. Cereal Res Commun 32:31–38
Renganayaki K, Friz AK, Sadasivam S, Pammi S, Harrington SE, McCouch SR, Kumar SM, Reddy AS (2002) Mapping and progress toward map-based cloning of brown planthopper biotype-4 resistance gene introgressed from Oryza officinalis into cultivated rice, O. sativa Crop Sci 42:2112–2117
Ronald PC (1998) Resistance gene evolution. Curr Opin Plant Biol 1:294–298
SAS Institute (2000) SAS language and procedure: Usage, Release & 01. SAS Inst., Cary
Sasaki T (2005) The completion of the rice genome sequence by IRGSP. Plant Animal Genome XIII:73
Sharma PN, Ketipearachchi Y, Murata K, Torii A, Takumi S, Mori N, Nakamura C (2003) RFLP/AFLP mapping of a brown planthopper (Nilaparvata lugens Stal) resistance gene Bph1 in rice. Euphytica 129(1):109–117
Soundararajan RP, Kadirvel P, Gunathilagraj K, Maheswara M (2004) Mapping of quantitative trait loci associated with resistance to brown planthopper in rice by means of a doubled haploid population. Crop Sci 44:2214–2220
Vos P, Simmon G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, Fierens-Onstenk J, Both MD, Peleman J, Liharska T, Hontelez J, Zabeau M (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotech 16:1365–1369
Ware DH, Jaiswal P, Ni J, Yap IV, Pan X, Clark KY, Teytelman L, Schmidt SC, Zhao W, Chang K, Cartinhour S, Stein LD, McCouch SR (2002) Gramene, a tool for grass genomics. Plant Physiol 130:1606–1613
Wing R (2005) Development of BAC fingerprint/sequence-tagged–connector based physical maps to understand and sequence plant genomes. Plant Animal Genome XIII:51
Xu XF, Mei HW, Luo LJ, Cheng XN, Li ZK (2002) RFLP-facilitated investigation of the quantitative resistance of rice to brown planthopper (Nilaparvata lugens). Theor Appl Genet 104:248–253
Yang HY, You AQ, Yang ZF, Zhang F, He RF, Zhu LL, He GG (2004) High resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.). Theor Appl Genet 110:182–191
Yeo US, Jin YD, Kim HY, Park NB, Lim SJ, Hwang HG, Kim SC, Shon JK (1998) Linkage between shikimic dehydrogenase isomerase and a brown planthopper (Nilaparvata lugens) resistance gene in a japonica rice Milyang 4. Korean J. Breed 30:162–167
Acknowledgements
We are grateful to the Rural Development Administration (RDA), Suwon, Korea, for financial support of this study. We are thankful to Bill Hardy, IRRI editor, for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Q. Zhang
K. K. Jena and J. U. Jeung contributed equally to this study.
Rights and permissions
About this article
Cite this article
Jena, K.K., Jeung, J.U., Lee, J.H. et al. High-resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18(t), and marker-assisted selection for BPH resistance in rice (Oryza sativa L.). Theor Appl Genet 112, 288–297 (2006). https://doi.org/10.1007/s00122-005-0127-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0127-8