Abstract
Growth characteristics have a complex inheritance pattern, and gene–environment interactions make predicting tree responses to environmental change difficult. In this study, we planted 44 Betula platyphylla families at three different sites (Mao er shan forestry center in Shangzhi, Jilin experiment forestry center in Jilin, Lang xiang forestry center in Langxiang) in northeastern China. Variation and stability of genotype–environment interactions of different families were analyzed using additive main effect and multiplicative interaction models. Variation analysis indicated significant differences between site × family interaction mean values for height, diameter at breast height, volume, and stem straight degree, suggesting that most genotypes responded differently according to location. The phenotypic coefficients of variation of different traits ranged from 12.84 % (stem straight degree in Langxiang) to 53.34 % (volume in Langxiang) and heritabilities of the different traits varied from 0.485 (diameter at breast height in Mao er shan) to 0.781 (height in Jilin). Correlation analysis showed a significantly positive association between tree height, diameter at breast height, and volume at the same and different sites, but stem straight degree showing a weaker correlation with other traits. Stability analysis indicated that some families had high tree heights but were sensitive to environmental conditions, whereas others had average tree heights but were resistant to environmental conditions. These results suggest that families should be bred in various habitats to assess growth under favorable and unfavorable environments. Under a selection ratio of 10 %, four families (family 1–7, 4–7, 3–12 and 4–13) were rated as superior families. The average height, diameter at breast height, volume, and stem straight degree of these four families were higher than average of all the families by 12.24, 16.82, 32.28 and 6.28 % in the four test sites, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Birch (Betula platyphylla) is one of the most extensively distributed broadleaf trees in the northern and southwestern forested areas of China (Zeng et al. 2003). Because of excellent wood quality, birch is widely used in papermaking, furniture making, and plywood production (Li et al. 1995). The native birch forest area in China has been gradually dwindling since the 1980s because of pests, disease, and environmental damage, and a large number of valuable genetic resources have been lost. Concurrently, demand for veneer plywood and papermaking fiber raw materials has increased rapidly and planted birch stands have not been able to meet production needs (Jiang et al. 2011). To address this, intensive birch seed orchards were created in 1999 at the Northeast Forestry University (Harbin, China) and many successful parents were established with the goal of obtaining superior seeds. Since then, seeds from intensive seed orchards have been distributed widely because birch plants grow quickly at the seedling stage (Yang et al. 2004). Beginning in 1991, Northeast Forestry University has developed a wel- established and widely promoted project for selection and cultivation of superior birch genotypes and intensive seed production.
Growth characteristics in woody plants have complex inheritance and are greatly influenced by various environmental conditions (Fang et al. 1999). The genotype × environment (G × E) interaction is defined as the differential response of genotypes to changing environmental conditions (Marron et al. 2010). It is essential to evaluate the importance of this interaction in developing breeding programs so that appropriate decisions can be made about testing and selection strategies (Codesido and Lopez 2009). The G × E interaction describes the situation in which a number of genotypes respond differently to various environments, so that the effects of genotypes and environments are not statistically additive (Lynch and Walsh 1998). In general, the environment can act on clonal (family) plant material in three different ways: (1) the G × E interaction is not significant; (2) the G × E interaction is significant owing to changes in differences among genotypes but not owing to changes in genotype ranking; and (3) the G × E interaction is significant owing to changes in genotype ranking from one environment to another (Nicolas et al. 2007). Only the last case causes problems for the breeder: for example, a genotype (family) selected for its growth vigor may not necessarily be vigorous if it grows in a different environment.
In this research, we focused on the growth traits of 44 12-year-old half-sib birch families at three different sites. Our study objectives were: (1) to compare the effects of G × E interactions on birch growth; (2) to compare the growth traits of different birch families; and (3) to determine which families exhibit the best growth traits under different environment conditions.
Materials and methods
Site description and materials
The materials used in this study included 44 families seeds (Table 1) of B.pubescens obtained in 2002 from Northeast Forestry University seed orchard. The next year, the seeds were sown and grown in artificial climate chambers of under a cycle of 1000 μmol m−2 s−1 light for 16 h from 0800 to 2400 hours and dark for 8 h. The temperature and humidity were set at 27 °C and 60 %, respectively. Experimental plantations were established in April 2004 at three sites [1, Mao er shan forestry center, Shangzhi, Heilongjiang Provence (MES). 2, Jilin Experiment Forestry Center, Jilin, Jilin Provence (JL). 3, Langxiang Forestry Center, Langxiang, Heilongjiang Provence (LX)] in China. Characteristics of the three sites are shown in Table 2. The experimental design consisted of six blocks per site. One-year-old seedlings of each family were planted using a randomized complete block design (Marron and Ceulemans 2006) in row plots containing 20 trees at a spacing of 3 m × 4 m.
Statistical analysis
Tree height (H), diameter at breath height (DBH) and stem straight degree (SSD) of all living, unbroken plants in the three sites were measured after leaf fall in 2014. Volume (V) of each plant were calculated according to Eq. 1 (Zhao et al. 2013), SSD were estimated by quantification (Table 3) based on Zhao et al. (2014a, b, c) and SSD should be square root-transformed before calculation.
Statistical analysis was carried out using SPSS 19.0 and DPS 9.50 software. The significance of fixed effects was tested by analysis of variance (ANOVA) F tests. The linear model (Eq. 2) used for joint analysis of the five sites (Dhillon et al. 2012) was
where y ijkl is the performance of individual l of family k growing in block j of site i, μ is the overall mean, S i is the effect of site i (i = 1,…,3), B j(i) is the effect of block j within site i (j = 1,…,6), C k is the effect of family k (k = 1,…,44), CS ik is the interactive effect of family k and site i, BC j(i)k is the interactive effect of clone k and block j (within site i) and ε ijkl is the random error.
Variation among families within site was analyzed by ANOVA according to linear model Eq. 3 (Hansen and Roulund 1997).
where y ij is the performance of an individual of family i within block j, μ is the overall mean, α i is the family effect (i = 1,…,44), β j is the block effect (i = 1,…,6), αβ i(j) is the random effect of family i within block j and ε ij is the random error.
The coefficient of phenotypic variation (PCV) was calculated using Eq. 4 (Hai et al. 2008).
where \( \bar{X} \) and SD are respectively the phenotypic mean and standard deviation of the trait.
Broad-sense heritabilities (h 2) was calculated following Hansen and Roulund (1997) as
where \( \sigma_{\text{A}}^{2} \) is the additive genetic variance component between families, \( \sigma_{\text{b}}^{2} \) is the block variance and \( \sigma_{\text{e}}^{2} \) is the error variance component.
Phenotypic correlation r A(xy) of traits x and y in the same sites was calculated according to Bi et al. (2000) as
where \( \text{cov}_{\text{p}} (x,y) \) is the covariance between families, \( \sigma_{\text{p}} (x) \) is the variance component for trait x and \( \sigma_{\text{p}} (y) \) is the variance component for trait y.
B-type correlations of traits between environments x and y, r g(xy), was calculated (Burdon 1977) as
where \( \text{cov}_{g} (x,y) \) is covariance for groups between the trait as it is expressed in environments x and y, respectively, and \( \sigma_{{g_{\text{x}} }}^{2} \) and \( \sigma_{{g_{\text{y}} }}^{2} \) are the variances between groups in environments x and y, respectively.
An additive main effects and multiplicative interaction (AMMI) model of diameter at breath height (DBH) was calculated following Zobel et al. (1988), with the linear model
where μ is the grand mean, α i is the family mean deviation (or provenance effect) with i = 1,..0.44, β j is the site effect with j = 1,…3, λ k is the singular value for axis k with k = 1,…,n, ψ ik is the provenance eigenvector value for axis k, δ jk is the site eigenvector for axis k, ρ ij is the residual, ε ijr is the error with r = 1,…, 6 replications, ε ijr = Y ijr minus the mean μ ij , and ε ijr is distributed normally with mean zero and standard deviation σ. These parameters were calculated by DPS software and the output could supply the parameter principal component analysis (PCA) which can explain the stability of different families.
Comprehensive evaluation of different families were calculated following Liu et al. (2015a, b) as
where a i = X ij /X jmax, Qi value is comprehensive evaluate value of each family, X ij is the average value of one trait and X jmax is the max value of the trait and n is the number of the traits.
Results
ANOVA analyses of H, DBH, V, and SSD at different sites
Analysis of variance showed that all main effects (site and family) and their interaction (site × family) were significant (P < 0.001) differences (Table 4).
Variation parameters analyses of different traits in the same site
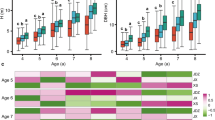
Significant differences (P < 0.01) were revealed among families (Table 5) in the same site. Heritability (h 2) of the different traits varied from 0.485 (DBH in MES) to 0.781 (H in JL). PCVs of different traits ranged from 12.84 % (SSD in LX) to 53.34 % (V in LX). PCV of H, DBH, and V at site LX showed higher values than the other two sites but were lower in SSD (Table 5).
Average each growth traits of all the families at different sites
The average H of all trees was highest at site JL (average temepreature was 3.1 °C and altitude was 360 m), with a mean value of 10.28 ± 1.23 m and a variation range of 8.33 ± 1.32 m (Family 1–2) to 11.89 ± 1.05 m (Family 1–7) (Table 6). The mean H at MES (average temepreature was 2.3 °C and altitude was 805 m) was 9.48 ± 1.44 m, ranging from 8.15 ± 1.28 (Family 1–2) to 11.00 ± 1.10 m (Family 4–13). The lowest mean H was 8.74 ± 1.60 m at site LX (average temperature was 0.36 °C and altitude was 588 m), with values ranging from 7.18 ± 2.93 m (Family 2–15) to 10.65 ± 0.71 m (Family 1–7). Average DBH of the 44 families was highest at site MES, with a mean DBH of 9.19 ± 1.85 cm and a variation range of 8.04 ± 2.58 (Family 4–3) to 10.35 ± 1.60 cm (Family 4–13) (Table 6). Averaged DBH of site JL was 9.13 ± 1.81 cm, ranging from 6.64 ± 2.47 cm (Family 1–13) to 11.12 ± 0.71 cm (Family 4–7). Similar to H, the lowest average DBH, 7.15 ± 1.90 cm, was also observed at site LX, where it ranged from 5.70 ± 3.05 (Family 1–3) to 9.44 ± 0.86 cm (Family –7). The highest average V of all plants was site JL, with a mean value of 0.033 ± 0.013 m3 and a variation range of 0.018 ± 0.010 m3 (Family 2–6) to 0.048 ± 0.007 m3 (Family 4–7) (Table 6). The mean V at MES was 0.032 ± 0.013 m3, ranging from 0.024 ± 0.011 (Family 1–2) to 0.040 ± 0.021 m3 (Family 3–12). The lowest mean V was 0.020 ± 0.010 m3 at site LX, with values ranging from 0.014 ± 0.008 m3 (Families 1–2 and 3–10) to 0.034 ± 0.006 m3 (Family 1–7). The average SSD of the 44 families was highest at site JL, with a mean SSD of 1.93 ± 0.24 and a variation range of 1.69 ± 0.42 (Family 3–7) to 2.18 ± 0.10 (Family 4–13) (Table 6). The mean SSD at site LX was 1.91 ± 0.22, ranging from 1.73 ± 0.19 (Family 4–8) to 2.15 ± 0.12 m (Family 4–13). The lowest average SSD, 1.88 ± 0.25, was observed at site MES, where it ranged from 1.71 ± 0.34 (Family 4–3) to 2.09 ± 0.21 cm (Family 1–8).
Average H, DBH, V, and SSD of different families at different sites
Families 4–13, 1–7, 3–12 and 4–7 showed higher average H values than other families, with mean H values of 10.95 ± 1.12, 10.88 ± 1.05, 10.65 ± 1.64 and 10.54 ± 1.37 m, respectively (Table 7). Families 1–2, 1–13 and 2–4 showed lower average H values than the other families, with mean H values of only 8.19 ± 1.28, 8.51 ± 2.13 and 8.73 ± 1.67 m, respectively. Families 4–13, 1–7, 3–12 and 4–7 also showed higher average DBH and V than other families, with DBH values of 9.95 ± 1.51, 10.00 ± 1.46, 9.68 ± 2.22, 9.99 ± 1.68 cm, respectively, and mean V values of 0.037 ± 0.012, 0.038 ± 0.012, 0.037 ± 0.021, 0.038 ± 0.013 m3, respectively. Families 1–8, 4–13, 2–11 and 4–7 showed higher average SSD values than other families, with mean SSD values of 2.05 ± 0.23, 2.04 ± 0.22, 2.03 ± 0.20 and 2.02 ± 0.20, respectively. Family 3-7 showed the lowest average SSD value (1.74 ± 0.27) (Table 7).
Correlation coefficients of different traits
The correlation coefficients of H, DBH, and V at the same site were significant (P < 0.01, Table 8). Within-site correlation coefficients of H, DBH and V ranged from 0.716 (H with DBH in JL) to 0.963 (DBH with V in MES). Correlation coefficients of SSD with other traits ranged from 0.198 (SSD with DBH in JL) to 0.730 (SSD with V in LX) in the same site. The B-type correlation coefficients among H at different sites ranged from 0.432 (JL with LX) to 0.700 (MER with LX). The correlation coefficients among DBH at different sites ranged from 0.402 (JL with LX) to 0.702 (JL with MES). The correlation coefficients among V at different sites ranged from 0.474 (JL with LX) to 0.706 (JL with MES) and among SSD at different sites ranged from 0.168 (MES with LX) to 0.498 (MES with JL), respectively (Table 8). The correlation coefficients of all the different traits at different sites varied from 0.002 to 0.704. The smallest (r = 0.002) correlation coefficient was found between DBH in JL with SSD in MES and the largest (r = 0.704) was DBH in MES with V in JL (Table 8).
AMMI analysis of different birch families at the three sites
All the three components, i.e., genotype (G), environment (E) and G × E interaction were highly significant (P < 0.01) (Table 9). The main effects, G and E, accounted for 27.33 % and 58.72 % of the total variation, respectively, while the G × E interaction accounted for 13.95 % of the total variation. The G × E interaction was further partitioned into principal component analysis (PCA1 and PCA2). The PCA1 component accounted for 71.14 % of the total variation in the G × E data (P = 0.125), whereas PCA2 and PCA3 were not significant (Table 9).
Biplot of genotype-environment interaction
A biplot generated from the AMMI analysis captured 100 % of the treatment sum of squares. Because the PCA1 scores of the genotypes were close to zero and had little interaction across environments, the overall mean rankings were very reliable. In contrast, the rankings of genotypes with large PCA1 scores (with either positive or negative overall rankings) were less reliable. Genotypes and locations combined with PCA1 scores of the same sign produced positive specific interaction effects, whereas combinations of positive signs had negative specific interactions. From the biplot (Figs. 1, 2), we observed that the families in which PCA1 values were negative had positive interactions with site LX and those with positive PCA1 values had positive interactions with sites JL and MES. The mean DBH at site LX was obviously the lowest and its G × E interaction was lower than at sites JL and MES. Site MES values were close to the horizontal ordinate with the largest mean DBH.
DBH biplot of genotype by environment interactions of different birch family growth in different sites. Abscissa average DBH of all the birch plant growth in the same site; ordinate PCA1 value of different sites. Site MES and JL showed higher average DBH value than LX and MES also showed more stability than the other two sites
DBH biplot of G × E interactions of different birch families. Abscissa DBH value and ordinate PCA1 value of different families. Families 4–7, 4–13, 1–7 and 3–12 were higher than other families in different sites. Most of the families were stable in different sites, and families 1–6, 1–5, 2–5, 3–16, 3–10 et al. had low stability than other families
When examining values above the horizontal ordinate on the biplot, we observed that the PCA1 values of families 3–10, 3–16, 3–7, 2–15 and 2–5 were distant from the horizontal ordinate, suggesting that these were the most unstable of all the studied families (Fig. 2). PCA1 values of the other families were more closely to the horizontal ordinate. Families 1–8 and 3–12 had lower PCA1 values (0.0060 and 0.0025, respectively) than other families, suggesting that these two families were more stable than the other families. Below the horizontal ordinate, the most unstable family was family 1–6, with a distance from the horizontal ordinate of 0.6274 and a mean DBH of 8.57 cm. The most stable family was family 2–7, which had an IPCA1 value of 0.0065 and a mean DBH of 8.28 cm. By observing the overall biplot, we determined that families 4–7, 4–13, 1–7 and 3–12 were fast growing and stable at all three sites.
Superior families selected
Based on the average H, DBH, V, and SSD at the three sites, families 1–7 and 4–13 showed the highest Qi value (1.99), family 4–7 and 3–12 just slightly lower with Qi value of 1.98 and 1.97, respectively. Family 1–13 showed the lowest Qi value of only 1.70 (Table 10). When the selected ratio was 10 %, families 1–7, 4–7, 3–12 and 4–13 were indicated as the superior families. The average H, DBH, V, and SSD of the top four families across the three sites were 10.76 m, 9.91 cm, 0.038 m3 and 2.02, respectively, which were higher than the average of all the families by 12.24, 16.82, 32.28 and 6.28 %, respectively.
Discussion
Variance among sites and familys
Understanding genetic variation within populations, families or clones is very important to understand and efficiently use genetic resources in breeding research (Safavi et al. 2010). ANOVA analysis is the most important method for estimating the extent of variability in a breeding population. In this study, the variation of H, DBH, V, and SSD showed significant differences (P < 0.01) among all variance sources, which indicated that estimations and selections of superior families were effective.
G × E interaction effects
Site effects reflect the response of trees to the combined effects of edaphic and local and regional climatic conditions (Pliura et al. 2007). When assessing the suitability of a plant for a particular environment, it is important to examine the interactions between the plant’s genotype and the environment (Yu and Pulkkien 2003). When such interactions are strong, tree breeders must decide whether to select for stability of performance and accept a slower rate of improvement, or to develop populations specifically adapted to each environment with the view of maximizing gain (Namkoong et al. 1988). In this study, the three sites represent different climate types in northeast China. The environmental conditions were different at the three sites, especially altitude and annual average temperature. The remarkable differences in H, DBH, V, and SSD among the families planted at the different sites indicated that the environment strongly influenced plant growth and development. This is especially significant because birch is one of the most important, quickly regenerating species in high altitude natural forests, and the ability to adapt to altitude impacts birch growth and development (Zhao et al. 2014a). At site LX, with an average temperature 0.36 °C and altitude of 588 m, plants had the lowest average H, DBH and V. However at site JL, where the temperature and altitude were 3.1 °C and 360 m, the trees had the highest average H and V, respectively. At site MES, where average temperature and altitude were 2.3 °C and 805 m, respectively, the trees showed the highest average DBH and moderate H and V.
PCV and h2 of different growth traits for the 44 birch families in different sites
The extent of variability in the breeding population was estimated by measuring different population parameters, including the phenotypic coefficient of variation and heritability (Liu et al. 2015a). In this research, PCV of different traits ranged from 12.84 to 53.34 %, which showed a wide range of genotype performance across different environments (Frew 2003). Trait V showed higher PCV values (42.64–53.34 %) than traits H, DBH, and SSD, but SSD showed lower PCV values (12.84–14.27 %) than other traits at different sites. These results are in agreement with those of Pliura et al. (2007), who found that the PCV values of H and DBH ranged from 10.05 to 31.60 % for poplar clones at different sites. Heritability magnitude indicated the reliability that the genotype could be recognized by its phenotype expression; heritability estimates are needed to assess the potential genetic gain that can be realized through selection (Montes et al. 2008). Heritability of the different traits varied from 0.485 to 0.781, which indicated that a great deal of variation in these families was heritable. Our heritability result is generally in agreement with the studies of Kien et al. (2008) in Eucalyptus urophylla and Zhao et al. (2014a) in poplar. High PCV among families indicated significant to excellent family selection, as marked differences between families could lead to considerable genetic gain. Furthermore, high repeatability estimates indicate that the selection for these traits will be effective and less influenced by environmental effects (Maniee et al. 2009).
Correlation coefficients
Although H and DBH are clearly related, the relationship between these parameters is rather complex (Sumida et al. 2013). The correlation coefficients among H, DBH, and V were significant at P < 0.01 for both the same and different sites, indicating that the selection for birch families was practical and predicting that the yield would be feasible. However, there were large differences among the coefficients of SSD with H, DBH, and V at the different sites. The coefficients of SSD with other traits were higher at LX (0.690–0.730) than at the other two sites. This indicates that superior families selected according to different traits could result in different effects according to the different sites. At LX, excellent families selected according to their H, DBH, and V might improve the SSD trait, which is beneficial for superior selections.
AMMI analysis
Different sites represent different environments that vary with longitude, latitude, altitude, temperature, and rainfall (Yu and Pulkkien 2003). The same genotype might present different phenotypic characteristics at different sites because of phenotypic plasticity. The AMMI model is a useful tool that combines ANOVA and principal components analysis and then exports the relationship among sites and among genotypes (Balestre et al. 2009). The model analysis generates a graphic representation of the major effects of the interaction (PCA1) of both genotypes and environments concurrently (Kempton 1984). In an AMMI biplot, genotypes that are distributed near the origin have minimal interactions with the environment, but those away from the origin are more sensitive to interactions with environment (Misra et al. 2009). In our study, sites MES and JL were more favorable for birch growth because of their high DBH and low PCA1 values (Fig. 1), which may occur because these sites have a higher annual average temperature than LX. Different families also showed different DBH values and stabilities (Fig. 2). Indeed, plants exhibiting wide adaptation perform well in nearly all environments (Annicchiarico 2002). Families 4–7, 3–12, 4–13 and 1–7 exhibited superior performance at all three sites, which indicated that these families exhibited excellent characteristics regardless of environmental conditions. Although family 3–12 showed lower DBH than families 4–7, 4–13 and 1–7, this family showed the lowest PCA1 among the four families, which indicated that family 3–12 had the highest stability of the four families. The DBH of families 3–10, 3–16, and 1–6 showed higher PCA1 absolute values than other families, indicating that these families were strongly influenced by the environment, displaying ideal growth under appropriate conditions but poor growth in other habitats. Yield stability data are useful for concurrently selecting high-yielding and stable genotypes (Kang and Pham 1991).
Superior families evaluated and selected
There are many different methods for selecting superior families according to different selected breeding targets. When the breeding goal is to encourage superior growth traits, as in wood production, H, DBH, V, and SSD are the most important traits. When comprehensively evaluating the families according to different traits, families 4–7, 1–7, 3–12 and 4–13 showed higher Qi values than other families; this result was also supported by our AMMI analysis. The average H, DBH, V, and SSD of the four families were higher than the average of all the families by 12.24, 16.82, 32.28 and 6.28 % in four sites, respectively.
Conclusion
In conclusion, our results demonstrate that birch families express G × E interactions for different growth traits. Superior families should be evaluated and selected separately for different sites. Families 1–7, 4–7, 3–12 and 4–13 were selected as superior families using comprehensive evaluation methods, but our analyses were based on a small number of testing sites and a growth period of 12 years. The results of the study could be improved by using more experiment sites and longer experiment durations. The methods employed herein and the implications of G × E interaction for calculating the gain of genetically improved birch should be applicable to further studies on other species in different environments, such as poplar and pine, which can supplement basic basis for stable and high yield materials selection.
References
Annicchiarico P (2002) Genotype × environment interactions: challenges and opportunities for plant breeding and cultivar recommendations. Washington (DC): Food and Agriculture Org. p 101
Balestre M, Pinho R, Soouza J, Oliverira R (2009) Genotypic stability and adaptability in tropical maize based on AMMI and GGE biplot analysis. Genet Mol Res 8:1311–1322
Bi CX, Guo JZ, Wang HW, Shu QY (2000) The correlation and path analysis on the quantity characters of Chinese pine. J Northwest For Univ 15(2):7–12
Burdon R (1977) Genetic correlation as concept for studying genotype-environment interactions in forest breeding. Silvae Genet 26:168–175
Codesido V, Lopez F (2009) Implication of genotype × site interaction on Pinus radiata breeding in Galicia. New Forest 37:17–34
Dhillon G, Singh A, Sidhu DS, Brar HS (2012) Variation among poplar clones for growth and crown traits under field conditions at two sites of north-western India. J For Res 24:61–67
Fang SZ, Xu XZ, Lu SX, Tang LZ (1999) Growth dynamics and biomass production in short-rotation poplar plantations: 6-year results for three clones at four spacings. Biomass Bioenergy 17:415–425
Frew M (2003) Yield stability in common bean (Phaseolus vulgaris L.) genotypes. Euphytica 130:147–153
Hai PH, Jansson G, Harwood C, Hannrup B, Thinh HH (2008) Genetic variation in growth, stem straightness and branch thickness in clonal trials of Acacia auriculiformis at three contrasting sites in Vietnam. For Ecol Manag 255:156–167
Hansen J, Roulund H (1997) Genetic parameters for spiral grain, stem form, pilodyn and growth in 13 years old clones of Sitka Spruce (Picea sitchensis (Bong.) Carr.). Silvae Genet 46:107–113
Jiang J, Yang G, Zhu ZB, Yang YL, Yang SZ (2011) Family selection from intensive seed orchard of Betula Platyphylla. J Northeast For Univ 39:1–4
Kang M, Pham H (1991) Simultaneous selection for high yielding and stable crop genotypes. Agron J 83:161–165
Kempton R (1984) The use of the biplots in interpreting variety by environment interactions. J Agric Sci 103:123–135
Kien ND, Jansson G, Harwood C, Almqvist C, Thinh HH (2008) Genetic variation in wood basic density and pilodyn penetration and their relationships with growth, stem straightness and branch size for Eucalyptus urophylla S. T. Blake in Northern Vietnam. NZ J For Sci 38:160–175
Li P, Fang G, Sun C (1995) Wood characteristics of pulpwood. Chem Indus For Prod 15:13–18
Liu DK, Liu MR, Li ZX, Wang GY, Li Y, Zheng M, Liu GF, Zhao XY (2015a) Variation analysis of growth traits of transgenic Populous simonii × P.nigra clones carrying TaLEA Gene. Bull Bot Res 35:540–546
Liu MR, Yin SP, Si DJ, Shao LT, Li Y, Zheng M, Wang FW, Li SC, Liu GF, Zhao XY (2015b) Variation and genetic stability analyses of transgenic TaLEA poplar clones from four different sites in China. Euphytica. doi:10.1007/s10681-015-1471-7
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Mass. Sinauer, Sunderland
Maniee MD, Kahrizi MD, Mohammadi R (2009) Genetic variability of some morphophysiological traits in durum wheat (Triticum turgidum var. durum). J Appl Sci 9:1383–1387
Marron N, Ceulemans R (2006) Genetic variation of leaf traits related to productivity in a Populus deltoides × Populus nigra family. Can J For Res 36:390–400
Marron N, Ricciotti L, Bastien C, Beritognolo L, Gaudet M, Paolucci I, Fabbrini F, Salani F, Dillen SY, Ceulemans R, Pinel M, Taylor G, Mugnozza GS, Sabatti M (2010) Plasticity of growth and biomass production of an intraspecific Populus alba family grown at three sites across Europe during three growing seasons. Can J For Res 40:1887–1903
Misra RC, Das S, Patnaik MC (2009) AMMI model analysis of stability and adaptability of late duration finger millet (Eleusine coracana) genotypes. World Appl Sci J 6:1650–1654
Montes CS, Hernandez RE, Beaulieu J, Weber JC (2008) Genetic variation in wood color and its correlations with tree growth and wood density of Calycophyllum spruceanum at an early age in the Peruvian Amazon. New Forest 35:57–73
Namkoong G, Kang HC, Brouard JS (1988) Tree breeding: principles and strategies. Springer, New York, p 180
Nicolas M, Sophie Y, Reinhart C (2007) Evaluation of leaf traits for indirect selection of high yielding poplar hybrids. Environ Exp Bot 61:103–116
Pliura A, Zhang YS, Mackay J, Bousquet J (2007) Genotypic variation in wood density and growth traits of poplar hybrids at four clonal trials. For Ecol Manag 238:92–106
Safavi SA, Pourdad SA, Mohmmad T, Mahmoud K (2010) Assessment of genetic variation among safflower (Carthamus tinctorius L.) accessions using agro-morphological traits nand molecular markers. J Food Agric Environ 8:616–625
Sumida A, Miyaura T, Hitoshi T (2013) Relationships of tree height and diameter at breast height revisited: analyses of stem growth using 20-year data of an even-aged Chamaecyparis obtuse stand. Tree Physiol 33:106–118
Yang CP, Liu GF, Wei ZG, Wu YL, Zhou YM (2004) Study on intensive breeding technique of accelerating Betula platyphylla flowering and seeding early. Sci Silvae Sin 40:14–17
Yu QB, Pulkkien P (2003) Genotype-environment interaction and stability in growth of aspen hybrid clones. For Ecol Manag 173:25–35
Zeng J, Zou YP, Bai JY, Zheng HS (2003) RAPD analysis of genetic variation in natural populations of Betula alnoides from Guangxi, China. Euphytica 134:33–41
Zhao XY, Li Y, Zhao L, Wu RL, Zhang ZY (2013) Analysis and evaluation of growth and adaptive performance of white poplar hybrid clones in different sites. J Beijing For Univ 35:7–14
Zhao XY, Bian XY, Li ZX, Wang XW, Yang CP, Liu GF, Jiang J, Kentbayev Y, Kentbayeva B, Yang CP (2014a) Genetic stability analysis of introduced Betula pendula, Bentula kirghisorum, and Betula pubescens families in saline-alkali soil of northeastern china. Scand J For Res 29:619–639
Zhao XY, Bian XY, Liu MR, Li ZX, Li Y, Zheng M, Teng WH, Jiang J, Liu GF (2014b) Analysis of genetic effects on complete diallel cross test of Betula platyphylla. Euphytica 200:221–229
Zhao XY, Hou W, Zheng HQ, Zhang ZY (2014c) Analyses of genotypic variation in white poplar clones at four sites in China. Silvae Genet 62:187–195
Zobel RW, Madison JW, Gauch HG (1988) Statistical analysis of a yield trial. Agron J 80:388–393
Acknowledgment
This work was supported by the grants of the National Science and Technology Pillar Program of China (No. 2012BAD21B02).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xiyang Zhao and Hui Xia have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhao, X., Xia, H., Wang, X. et al. Variance and stability analyses of growth characters in half-sib Betula platyphylla families at three different sites in China. Euphytica 208, 173–186 (2016). https://doi.org/10.1007/s10681-015-1617-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1617-7