Abstract
Retrotransposon-based molecular markers were applied for the first time within the genera Xanthosoma and Colocasia to assess intraspecific variability among 27 accessions of cocoyam (Xanthosoma sagittifolium) and taro (Colocasia esulenta). Retrotransposons were isolated and sequenced; long terminal repeat (LTR) primers were designed to obtain inter-retrotransposon amplified polymorphism (IRAP) fingerprints. A set of six chosen LTR primers yielded 433 reproducible bands across 20 X. sagittifolium samples. Out of the 433 bands, 400 fragments (92 %) were polymorphic. In seven C. esculenta accessions, the six primers amplified a total of 354 reproducible, informative data points, of which 285 (80.5 %) were polymorphic. Cluster analysis placed all the accessions in two groups according to their species. The accessions of X. sagittifolium were further divided into two subgroups corresponding to their ploidy level. Moreover, the genetic variability accessed by IRAP markers allowed separation of X. sagittifolium and C. esculenta accessions according to their type and botanical variety respectively. The results suggest that retrotransposon activity continued after Xanthosoma speciation. The data and approach provides a basis for better germplasm management, future systematic studies and genetic improvement, as well as for exploration of the role of retrotransposons in cocoyam and taro polyploid formation and genome dynamics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The monocotyledonous Araceae family comprises about a hundred genera, among which five are cultivated as food crops: Cyrtosperma, Amorphophallus, Alocasia, Colocasia, and Xanthosoma (Purseglove 1972). The common edible aroids are found in the genera Xanthosoma and Colocasia, with Xanthosoma sagittifolium and Colocasia esculenta being the most important species. X. sagittifolium is an annual crop grown for its edible, corms, cormels, and leaves in the tropics and sub-tropics. It originated in the tropical Americas and spread later to Southeast Asia, the Pacific Islands, and Africa. Presently, X. sagittifolium is widely cultivated in subtropical and tropical regions including Central and South America, Africa, Asia, and the South Pacific islands. Consequently, X. sagittifolium is also referred to as tannia, new cocoyam, cocoyam, macabo, and other names depending on the place of cultivation.
X. sagittifolium is a rich source of carbohydrates, essential amino acids, vitamins, and minerals for the human diet (Sefa-Dedeh and Agyir-Sackey 2004). C. esculenta, or taro, is intercropped with cocoyam. The two species are phenotypically close enough to be interchangeable for preparation of many traditional West African dishes. In addition to its high nutritional value, cocoyam is progressively becoming an important cash crop, particularly for poor resource-rural populations. It is among the six most important root and tuber crops worldwide and has an annual production of about 10 million tons (FAOSTAT 2014). Despite its widespread production and importance, improved breeding and processing of cocoyam could greatly increase its utilization and value (Owusu-Darko et al. 2014).
Cocoyam root rot disease (CRRD), caused by Pythium myriotylum, has emerged as the main production constraint in most cocoyam growing areas worldwide (Pacumbaba et al. 1992; Perneel et al. 2006). Recommended cultural practices provide only short-term control of CRRD (Adiobo et al. 2007) and strategies to control it with biological agents are still under examination (Perneel et al. 2007). C. esculenta suffers great damage due to the taro leaf blight caused by Phytophthora colocasiae (Singh et al. 2012), but resistant varieties are available. The use of resistant varieties could be an approach to sustainable disease control, but to date no acceptable disease-resistant cocoyam variety has been bred. Furthermore, little is known about cocoyam plant genetics.
Generally poor knowledge of the genetic variation within the existing cocoyam germplasm has constrained development of improved varieties. Until recently, the diversity of cocoyams, e.g. in Cameroon, has been assessed only with phenotypic descriptors and cytological features (Ngouo et al. 1989), resulting in the identification of three cocoyam types (White, 2n = 26; Red, 2n = 26; Yellow, 4n = 52), which were later characterized by morphology and with biochemical markers (Mbouobda et al. 2007; Onokpise et al. 1999). Efforts have been made to develop PCR-based molecular markers as well. Use of RAPD markers revealed either low (Schnell et al. 1999) or high (Offei et al. 2004) genetic variation, and no correlations were made to plant morphology; mitochondrial and chloroplast-specific restriction fragment polymorphisms revealed limited species-level variability (Brown and Asemota 2009). Microsatellite markers were recently introduced (Cathebras et al. 2014), giving very variable degrees of heterozygosity, which were observed at levels ranging from 0.00 to 0.97. Cathebras et al. (2014) were able to transfer the markers to other species of Xanthosoma and Caladium at success rates from 23.5 to 100 %. Likewise, studies using AFLP were used only to determine the intergeneric relationships between Caladium, Xanthosoma, and other genera of Araceae (Loh et al. 2000). We sought a highly polymorphic marker system for cocoyam, which would be suited to low- and medium-throughput analyses, and a simple laboratory setup.
Retrotransposons have been established in many plant species as highly effective marker systems due to their ubiquity, abundance, dispersion, and genomic dynamism (Kalendar and Schulman 2006; Kalendar et al. 2011a; Schulman et al. 2012).The long terminal repeat (LTR) retrotransposons propagate within the genome through a “copy and paste” mechanism, whereby reverse-transcribed daughter copies of the retrotransposon transcripts are inserted into new locations (Schulman 2013). The LTRs, conserved within retrotransposon families, form joints with the genomic DNA at the retrotransposon insertion points and provide a means of detecting the insertions.
The retrotransposon integration joints can be detected by PCR amplification between one primer matching the LTR and another matching a nearby motif in the genome, and the polymorphic insertion sites detected by standard agarose gel electrophoresis of the PCR products (Schulman et al. 2012). For example, the inter-retrotransposon amplified polymorphism (IRAP) method amplifies segments between two nearby LTRs; retrotransposon microsatellite amplified polymorphism (REMAP) amplifies segments between an LTR and a microsatellite (Kalendar et al. 1999; Schulman et al. 2012). Retrotransposons and LTRs can be found and annotated in sequenced genomes bioinformatically. However, in uncharacterized genomes such as that of cocoyam, the LTR sequences for the development of marker methods must be isolated either by laborious walking to the LTR ends from conserved domains in retrotransposon polyproteins (Pearce et al. 1999) or by the more recently developed and rapid method known as iPBS, which is based on a highly conserved motif adjacent to LTRs (Kalendar et al. 2010).
The aim of this work was therefore to develop retrotransposon-based markers for cocoyam using the iPBS method. The LTRs isolated by iPBS were converted into IRAP primers and used as molecular markers for identifying cultivars and determining genetic relationships in X. sagittifolium. The transferability of the markers to the closely related araceae species, C. esculenta was also tested.
Materials and methods
Plant materials and DNA isolation
X. sagittifolium and taro are not native to the African continent. Much of the cocoyam cultivated in Cameroon and the rest of West Africa are landraces derived from earlier introductions especially by the Portuguese. Passport data and ultimate geographic origins of the materials are not available. The genotypes used in the experiments are listed in Table 1. Twenty different accessions of X. sagittifolium and seven accessions of C. esculenta were obtained in Cameroon from farms or from the Institute of Agricultural Research for Development (IRAD), Ekona Regional Centre, and used for the study. Accession W305 is an artificial hybrid derived from crosses between White and Red accessions of X. sagittifolium. Accessions RO1054 and RO2063 were selections of the ROTREP cocoyam breeding program of Cameroon.
Following previous tests in the field for root rot susceptibility, tolerance, or resistance, these two accessions were rated as tolerant (Nyochembeng et al. 2007). Total genomic DNA was extracted from leaf samples of these genotypes using a modification of the CTAB extraction protocol (http://primerdigital.com/dna.html) with RNAse A treatment. The DNA samples were diluted in 1 × TE buffer and the DNA quality was checked spectrophotometrically with a Nanodrop apparatus (Thermo Fisher Scientific Inc.) as well as electrophoretically.
Isolation of retrotransposon fragments
The iPBS method (Kalendar et al. 2008, 2010) was used to obtain retrotransposon fragments containing LTR regions from X. sagittifolium. The method is based on PCR amplification between the primer binding sites (PBS) that serve in minus-strand cDNA synthesis, which are highly conserved in virtually all retrotransposons and located adjacent to the 5′ LTR (Kalendar et al. 2010). The PCR amplification was performed with one or two primers complementary to particular PBS types. A total of 96 PCR fragments were isolated and cloned into the vector pGEM-T (Promega, USA), then sequenced using an ABI3700 (Applied Biosystems, USA) capillary sequencer.
Retrotransposon segments were recognized by a combination of approaches: alignment of the sequenced PCR fragments to identify conserved repetitive segments; comparison of these segments to LTRs and internal regions of annotated retrotransposons; identification of the diagnostic structural features of retrotransposons, including the inverted terminal repeats of LTRs containing the terminal nucleotides 5′ TG…CA 3′ and the 3′ terminus of the 5′ LTR being positioned one to five nucleotides away from the PBS. Primers were designed with the FastPCR software (Kalendar et al. 2011b), to match the conserved segments of retrotransposon LTRs in various orientations.
IRAP reactions
IRAP analysis was carried out according to Kalendar and Schulman (2006). PCR reactions for IRAP were performed in 25 µl containing 25 ng genomic DNA, 1 × DreamTaq buffer, 0.3 µM each primer, 0.2 mM each dNTP, and 1 U DreamTaq DNA polymerase (Thermo Scientific). Amplifications were performed in a Mastercycler Gradient (Eppendorf AG, Germany) in 0.2 ml tubes or in 96-well plates. After an initial denaturation step at 95 °C for 3 min, thermocycling was performed for 32 cycles at 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 90 s, with a final extension at 72 °C for 5 min.
Each primer was tested individually in PCR reactions using a genomic DNA mixture composed of equal amounts from each of the accessions. The PCR products were separated by electrophoresis at 60 V for 8 h in a 1.4 % agarose gel (RESolute Wide Range, BIOzym) with 0.5 × TBE electrophoresis buffer. The Thermo Scientific GeneRuler DNA Ladder Mix, 100-10,000 bp, #SM0332, was used as a standard. Gels were stained with EtBr and scanned using a FLA-5100 imaging system (Fuji Photo Film GmbH) at a resolution of 50 µm.
IRAP fingerprint analyses
From the IRAP profiles, all distinct bands were scored as present (1) or absent (0) at each band position for each primer in the 27 samples. Each IRAP band was treated as a single locus. The presence or absence of a fragment of a given length was recorded in binary code. The sets that contained missing values were removed from the raw scored data sets. Monomorphic bands were scored and removed from the data set before analysis.
The 50 IRAP primers were tested for the number, distinctness, robustness, and ease of scoring of the fingerprint bands (Table 2). Six primers generating reproducible and robust patterns were retained for subsequent analysis. The discriminatory power of these six primers was evaluated by means of the number of bands generated and by the following two parameters. The polymorphic information content (PIC) for each primer was calculated using the online program PICcalc (Nagy et al. 2012). The effective multiplex ratio (the number of polymorphic products from a single amplification reaction) was calculated according to Powell et al. (1996).
Several software programs were then used to analyze the data. Genetic similarity was evaluated using PopGene software version 1.32 (Yeh and Boyle 1997) for the Shannon index (H’j; Shannon and Weaver 1949), which is defined for multilocus markers as: H′j = −∑ pi log pi, where pi is the frequency of the ith fragment in the sample. In addition, the genetic similarity index of Jaccard (1908) was used to calculate genetic similarity, employing the software GenStat Discovery 4 (2012) and FAMD (Schlüter and Harris 2006). Given two genotypes, A and B, the Jaccard similarity index J is defined as: J = M 11/M 01 + M 10 + M 11, where M 11 is the total number of bands both having a value of 1, M 01 is the total number of bands whose values are 0 in A and 1 in B, and M 10 is the total number of bands whose values are 1 in A and 0 in B. The average Jaccard indices were calculated separately for data obtained on X. sagittifolium and C. esculenta accessions.
In order to compare levels of diversity detected by different primers, the various parameters were calculated for each primer separately. One-way ANOVA and Tukey’s test were performed on genetic similarity indices. Principal component analysis (PCA) using the GenAlex 6.5 program (Peakall and Smouse 2006, 2012) was carried out to define the relationships among X. sagittifolium types (White, Red and Yellow) and among C. esculenta varieties (C. esculenta var. esculenta and C. esculenta var. antiquorum), based on the data from all six chosen primers. A pairwise genetic distance matrix was calculated, then converted to a covariance matrix and used to perform the PCA.
The ability of IRAP markers to reveal genetic relationships among all X. sagittifolium and C. esculenta accessions and between X. sagittifolium and C. esculenta accessions was evaluated phylogenetically by Neighbor-Joining (NJ). An algorithm was constructed using PAUP software (Swofford 1998). Support for the tree was determined by performing 1000 bootstrap operations on the data set generated by distance analysis. To study the partition of IRAP genetic variation into among- and within-population variance components, an analysis of molecular variance (AMOVA) was conducted with the program GenAlex 6.5 (Peakall and Smouse 2006, 2012). The genetic distances among groups were analyzed with the Phi statistic (Φst). To test the significance of the genetic distance among groups, AMOVA was used with 999 permutations. The accessions of X. sagittifolium were defined as diploid (White and Red types) or tetraploid (Yellow type).
Results
LTR isolation
Retroviruses and retrotransposons prime minus-strand reverse transcription, an essential step of replication, with specific tRNAs of the host cell (Schulman 2013). The PBS site is complementary to the tRNA primer and adjacent to the 5′ LTR. The iPBS method exploits primers matching common PBS motifs as a means of amplifying segments of two LTR retrotransposons in close proximity to one another. To screen the cocoyam genome for retrotransposons, a set of 83 PBS primers were used in iPBS. Most produced multiple bands, expected for abundant and clustered retrotransposons. A total of 96 iPBS PCR reaction products, ranging from 400 to 2000 bp long, were cloned, sequenced, and aligned, identifying clusters of highly similar fragments. A total of 93 clusters were identified, which have features typical for LTRs (Benachenhou et al. 2013; Schulman 2013); 22 LTRs were deposited in Genbank (accessions GU810535, KF813039-KF813057). Primers were designed to match these (Table 2), positioned close to the predicted ends of the LTRs and oriented outwards from the LTRs.
Initial screening of primers
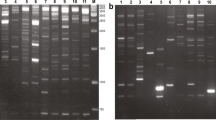
The IRAP protocol was used to screen the 50 primers for their ability to produce polymorphic patterns across 27 accessions of X. sagittifolium and C. esculenta. The majority of primers amplified bands which were easy to score on standard agarose gels stained with ethidium bromide. The PCR reactions produced variable numbers of amplified fragments depending on the primer tested (Table 2). Only four primers (2969, 2973, 2987 and 2995) produced no bands. Altogether 18 primers gave superior results, with many strong and equally amplified bands. Six of these (2952, 2964, 2971, 2977, 2978, 2979) were selected for evaluation of genetic diversity in the accessions and for further analysis.
Genetic diversity in X. sagittifolium accessions
The IRAP protocol was used to study genetic variability among 20 accessions of White, Red, and Yellow cocoyam using the six chosen primers singly in PCR reactions (Table 3; Fig. 1). A total of 433 reproducible bands, ranging from 400 bp (primer 2950) to 3000 bp (primer 2950) were detected using the six selected primers. The number of bands per primer varied from 60 (primer 2952) to 86 (primer 2977). Of the 433 bands, 400 fragments (92 %) were polymorphic across the whole set of samples (Table 3). The primers displayed between 82 % (primer 2952) and 97 % (primer 2977) polymorphism across the samples analyzed. Based on the Jaccard index, a genetic similarity matrix was constructed using the IRAP data to assess the genetic relatedness among the 20 X. sagittifolium accessions. The minimum genetic similarity (0.23) was between R5 and Y4 and the greatest (0.85) between R1 and R3. Genetic similarity coefficients indicating the extent of relatedness between each pair of accessions are presented in Supplementary Table S1 and ANOVA for the data of all six primers taken together are found in Table S2.
Agarose gel electrophoresis of IRAP reactions using the primer 2971for X. sagittifolium accessions. Lanes, left to right: 1, size ladder (bp); 2, W1; 3, R6; 4, W3; 5, W4; 6, Y5; 7, not analyzed; 8, W7; 9, RO1054; 10, RO2063; 11, RO31; 12, W305; 13, Jinika; 14, R1; 15, R2; 16, R3; 17, R4; 18, R5; 19 IB; 20, IB1; 21, IB2; 22, AS; 23, AS1; 24, AS2; 25, Nejet; 26, Y1; 27, Y2; 28, Y3; 29, Y4
Genetic diversity in diploid and tetraploid X. sagittifolium accessions
In a different analysis, IRAP was used to study the relationship between ploidy level and cocoyam type using the six primers. Four White diploids (W1, W3, W4 and W7), five Red diploids (R1, R2, R3, R4 and R6), and five yellow tetraploids (Y1, Y2, Y3, Y4 and Y5) were examined. The selection of these accessions was based on their having a known ploidy level and on their grouping in the dendrogram. Within the X. sagittifolium accessions analyzed, the number of bands scored was higher for tetraploids than for diploids: 278 for the White diploids, 301 in Red diploids, and 367 for Tetraploids. In parallel, the number of bands for individual primers varied from 35 (primer 2979, Red diploids) to 74 (primer 2977, tetraploids). Primer 2977 used with tetraploid accessions showed the highest number of polymorphic (56) and private (23) bands (Table 4). The total number of polymorphic loci showed a pattern similar to the total scored, from 101 (White diploids) to 147 (Red diploids) and 259 (Tetraploids), as did the number of private bands exclusively present in accessions of only one of the genome groups, respectively 9, 14, and 72 for White, Red, and Tetraploids, with respectively three, four, and twelve being monomorphic.
Genetic variability based on IRAP analysis in C. esculenta
The IRAP method was used to study genetic variability among different accessions of C. esculenta using the same primers developed for X. sagittifolium. The PCR reactions produced a high number of bands for all IRAP primers. As shown in Table 3, the six primers amplified a total of 354 reproducible and informative data points, of which 285 (81 %) were polymorphic, in the set of seven C. esculenta accessions. Between 47 (primer 2979) and 70 (primers 2971 and 2977) bands, varying from 300 to 3000 bp, were produced per primer. These products were from 58 % (primer 2964) to 90 % (primer 2971) polymorphic. The IB and Nejet accessions showed the lowest genetic Jaccard index (0.10), whereas the highest (0.63) was observed between IB and AS. Genetic similarity coefficients indicating the extent of relatedness between each pair of C. esculenta accessions are presented in Supplementary Table S3.
X. sagittifolium versus C. esculenta variability
Overall, also C. esculenta genotypes exhibited a high level of genetic diversity, even though their fingerprints contained generally fewer bands than those of X. sagittifolium accessions for all six primers. The Shannon index was on average 0.494 (range 0.458–0.519) for X. sagittifolium and 0.508 (range 0.469–0.536) for C. esculenta genotypes (Table 3). The Jaccard index shows a similar trend (Table 3), but there is no significant difference for these indices between the two groups. The lowest PIC scores (respectively 0.364 and 0.387) were obtained respectively with primer 2978 for X. sagittifolium accessions and primer 2971 for C. esculenta accessions. The remaining PIC values for the six primers were almost similar and ranged between 0.372 and 0.374 for the two groups of accessions (Table 5). The effective multiplex ratio (EMR) values (Table 5) for X. sagittifolium accessions ranged from 46.81 to 80.10 (respectively primers 2952 and 2977), whereas those for C. esculenta ranged from 37.53 to 56.7 (primers 2979 and 2971).
Multivariate analyses
Phylogenetic relationships among the 27 accessions of cocoyam and taro were estimated by NJ according to Nei’s genetic distance matrices. In the resulting dendogram (Fig. 2), the two species each produce a major cluster. The accessions of X. sagittifolium were further divided into two well-supported clusters that are consistent with the cytogenetic genome groups, the tetraploids (4n = 52) and the diploids (2n = 26). The diploid cocoyam accessions were further clustered into almost type-specific subclusters resolving the White and Red cocoyams. However, three accessions, two Red (RO2063 and R5) and one White (RO1054), did not cluster with their types. Interestingly, accessions W305, an artificial hybrid, and Jinika, apparently a natural hybrid, clustered with the white types. The second major cluster, comprising C. esculenta accessions, resolved two subgroups representing two subspecies, Nejet for C. esculenta var. esculenta and. the six remaining accessions for C. esculenta var. antiquorum.
When the combined IRAP data of the six primers was subjected to PCA analysis (Supplementary Fig. S1), the tetraploid Yellow X. sagittifolium type was separated from the White and Red accessions by the first three principal components (47.85 % of the diversity). Most of the Yellow accessions were well resolved by Coordinate 3 alone. The hybrids W305 and Jinika, respectively White and Red, were closer to the Red accessions by Coordinate 1 and intermediate between White and the Red accessions by Coordinate 2. For the taro accessions (Supplementary Fig. S2), the first two components accounted for 58.41 % of the variation and PCA separated accession Nejet (C. esculenta var. esculenta) from the accessions belonging to the antiquorum subspecies. The accessions are widely scattered, confirming the large variability among them.
Analyses of molecular variance (AMOVA) revealed that a highly significant genetic differentiation occurred in the X. sagittifolium types (Supplementary Table S4). At two hierarchical levels, most of the genetic variation (67 %) could be attributed to differences within the populations (i.e., within White, Red, and Yellow types) and only 33 % to differences among the populations. Pairwise Φst values (Supplementary Table S5) were obtained based on the AMOVA analysis and showed that the greatest genetic distances are between White and Yellow types (0.439) and the smallest between White and Red types (0.189), with the distance between Red and Yellow types being intermediate (0.368).
Discussion
Most recent advances in the fields of genomics and molecular genetics have been aimed at those crops, particularly the major cereals, that sustain the economy and ensure food security primarily in the developed world. However, crops of similar potential importance to the developing world are still understudied. X. sagittifolium (cocoyam) and C. esculenta (taro) are important West African food crops. Currently, molecular markers have been only limitedly applied to cocoyam, the genetic variation of which is poorly characterized; both cocoyam and taro are classified as orphan crops by the African Orphan Crops Consortium (http://www.mars.com/global/african-orphan-crops.aspx). No acceptable disease-resistant cocoyam variety has been bred. In this work, we have taken advantage of the ubiquity and abundance of LTR retrotransposons in plant genomes and their role in genomic diversification to develop and apply retrotransposon marker systems for cocoyam and taro, to our knowledge for the first time.
Cocoyam genotyping by retrotransposon markers
Using the iPBS method (Kalendar et al. 2010), we cloned and sequenced 92 PCR fragments, from 400 to 2000 bp, representing the first identified retrotransposon segments from Xanthosoma. When converted into IRAP primers, the majority displayed sufficiently high levels of polymorphism to be generally applicable as molecular markers. The 6 most informative primers were applied to 20 X. sagittifolium accessions and yielded a high number of bands. By comparison, RAPD markers used for Xanthosoma spp. genotypes (Schnell et al. 1999; Offei et al. 2004) showed low numbers of amplified bands. The abundant IRAP and iPBS bands may indicate that retrotransposons are abundant in the Xanthosoma genome or they are predominantly clustered, or both. The X. sagittifolium genome has been reported at 8592 Mbp (Ghosh et al. 2001). Even if, for the sake of argument, we assume a relatively high gene number of 40,000 of average length 5000 bp, the total gene space would comprise only 200 Mbp of the genome. The remaining 98 % would therefore comprise repetitious sequences, the great majority of which being retrotransposons. The abundance of iPBS and IRAP bands amplified from the X. sagittifolium is consistent with this perspective.
Impact on X. sagittifolium characterization and taxonomy
We investigated whether the three cocoyam groups defined by morphological and cytological features (Ngouo et al. 1989; Onokpise et al. 1999) can also be differentiated by the retrotransposon genomic insertion patterns revealed by IRAP markers. Indeed, a set of six IRAP markers separated the Yellow tetraploid accessions from the diploid White and Red ones. The hybrid accessions displayed an intermediate position in PCA based on their IRAP banding patterns. Moreover, IRAP markers distinguished individual accessions, which could not be achieved by RAPD markers (Offei et al. 2004).
A broader problem related to germplasm and biodiversity conservation of cocoyam is the unclear taxonomic position of cultivated Xanthosoma. The tendency has been to classify most edible, tuber bearing members of the genus as X. sagittifolium (Giacometti and Leon 1994). However, using leaf shape, pigmentation, and other vegetative characteristics, Wilson (1984) identified several species, including X. violaceum, X. caracu, and X. atrovirens. Bown (2000) divides the genus into two main species based on the color of the corm, cormels, and leaves and on the shape of the cormels. The results from IRAP analyses reported here support the splitting of cultivated Xanthosoma into several species. Further insight into the genetic diversity of cocoyams therefore would be gained from analysis of all species belonging to Xanthosoma by retrotransposon-based markers. A well-defined and established systematics of Xanthosoma would enable better management of germplasm collections and facilitate clonal selection.
Tetraploidy and IRAP polymorphism
The tetraploid accessions on average showed more polymorphic IRAP bands than did the diploids. The greater number of bands in the tetraploids may be due to their having more retrotransposon insertion loci. The increased polymorphism may be explained in several ways. The “genome shock” (McClintock 1984) of polyploidization has been reported to activate the transcription (Kashkush et al. 2003) and propagation (Kraitshtein et al. 2010; Parisod et al. 2010) of retrotransposons. Alternatively, the redundancy of the gene complement in polyploids, especially autopolyploids, could reduce the selective pressure against retrotransposon insertions into genes and thereby lead both to loss of fewer retrotransposon insertions by purifying selection and to fixation of more (Hazzouri et al. 2008). A third explanation is that the increased IRAP polymorphism of tetraploids reflects an increase in overall genetic diversity in polyploids (Wendel 2000) in general and cocoyam in particular (Ngouo et al. 1989). Last, shrinkage of genomes subsequent to polyploidization (Grover et al. 2008) may lead to differential loss of retrotransposon insertions in various populations and increased polymorphism as detected by marker methods. All four scenarios are consistent with the results obtained.
Implications for cocoyam breeding
The IRAP method resolved cocoyam accessions well. The two hybrids could also be distinguished from the parental sets and grouped together. Furthermore, IRAP markers enabled detection of accessions with similar physiological traits. For example, RO1054 and RO2063, which are tolerant to disease (Nyochembeng et al. 2007), clustered together in the dendogram even though they are morphologically different. This result also indicated that the morphological system may be useful for morphotype management but not appropriate for defining genetic diversity in cocoyam.
One of the most important problems in X. sagittifolium production is the susceptibility to CRRD caused by the fungus P. myriotylum. In response, efforts are being made to broaden the genetic base of breeding populations to allow for selection of resistant lines. However, acceptable cocoyam varieties resistant to CRRD are not yet available, despite considerable advances including the creation of hybrids and mutants, the introduction of polyploids, and study of interactions between cocoyam and P. myriotylum (Boudjeko et al. 2005; Nyochembeng et al. 2007). Because intervarietal hybridization is a potential means to genetically improve X. sagittifolium for major agronomic traits and for resistance to CRRD, IRAP markers are an important molecular tool that may assist cocoyam breeders when applied to map QTLs or major genes in mapping crosses or for marker-assisted selection. The resistance of Yellow cocoyam to disease has been attributed partly to its tetraploid nature; therefore, it has been proposed that inducing polyploidy in White and Red diploid types will improve their resistance (Tambong et al. 1998; Doungous et al. 2011). IRAP markers developed in this work can assist cocoyam breeders selecting desired genotypes following polyploidy induction treatments.
Suitability of X. sagittifolium IRAP markers in C. esculenta
Here, we assessed the usefulness of X. sagittifolium-derived retrotransposon markers to investigate genetic variability and relationships among seven C. esculenta accessions, and observed a high level of polymorphism (80.5 %). This is the first report to our knowledge of genetic relationships in taro using retrotransposon markers. The X. sagittifolium and C. esculenta accessions that were examined were well separated into two distinct groups by IRAP markers, as others had been earlier by RAPD (Brown and Asemota 2009). However, both RAPD (Irwin et al. 1998) and AFLP methods (Quero-Garcia et al. 2004) failed in some cases to resolve or reflect geographical origin or morphological characters in taro. Moreover, the IRAP markers proved to be powerful enough to differentiate even the two botanical varieties (esculenta and antiquorum), although RAPD markers could not (Irwin et al. 1998). Confirmation of the utility of IRAP on taro will require, however, a wider set of material to be examined.
The genetic separation of X. sagittifolium from C. esculenta accessions supports the suitability of IRAP for differentiating between closely related taxa of Araceae. In contrast, the AFLP fingerprint patterns based on primers effective for C. esculenta were too dissimilar to those of X. sagittifolium and other closely related species to include them in parallel diversity analyses (Kreike et al. 2004). In previous studies, IRAP has been used successfully for assessing genetic identities and relationships of closely related species (Antonius-Kemola et al. 2006) as well as in the studies of genetic diversity in Helianthus (Vukich et al. 2009) and Linum (Smykal et al. 2011). Despite the use of IRAP primers derived from X. sagittifolium retrotransposons, the number and polymorphism of IRAP products in C. esculenta was high for all six primers. This suggests that both species may share active and conserved families of retrotransposons as described previously for the grasses (Vicient et al. 2001) and confirms the inter-generic transferability of retrotransposon markers previously reported in Spartina (Baumel et al. 2002) and Crocus (Alavi-Kia et al. 2008).
Conclusion
We demonstrate that the IRAP marker system provides a simple and useful approach for studying genetic diversity of cocoyam and taro. The high percentage of polymorphism and the large number of bands obtained per assay using a single primer shows that IRAP is the most informative marker system so far developed for X. sagittifolium. It was possible to identify cocoyam accessions including hybrids, diploid, and tetraploid plants and accessions that are susceptible or tolerant to the CRRD based on the marker pattern generated by six IRAP primer sets. Furthermore, the primers could also be used for C. esculenta and were successful in identifying different C. esculenta subspecies. The results obtained in this study could be very useful in the future for defining germplasm conservation and designing cocoyam and taro improvement programs for the development of disease resistant varieties.
Abbreviations
- IRAP:
-
Inter-retrotransposon amplified polymorphism
- TEs:
-
Transposable elements
- LTR:
-
Long terminal repeat
- PBS:
-
Primer-binding site
- CRRD:
-
Cocoyam root rot disease
References
Adiobo A, Doungous O, Perneel M, Zok S, Höfte M (2007) Variation of Pythium-induced cocoyam root rot severity in response to soil type. Soil Biol Biochem 39:2915–2925
Alavi-Kia SS, Mohammadi SA, Aharizad S, Moghaddam M (2008) Analysis of genetic diversity and phylogenetic relationships in Crocus genus of Iran using inter-retrotransposon amplified polymorphism. Biotechnol Biotechnol Equip 22:795–800
Antonius-Kemola K, Kalendar R, Schulman AH (2006) TRIM retrotransposons occur in apple and are polymorphic between varieties but not sports. Theor Appl Genet 112:999–1008
Baumel A, Ainouche M, Kalendar R, Schulman AH (2002) Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Mol Biol Evol 19:1218–1227
Benachenhou F, Sperber GO, Bongcam-Rudloff E, Andersson G, Boeke JD, Blomberg J (2013) Conserved structure and inferred evolutionary history of long terminal repeats (LTRs). Mob DNA 4:5
Boudjeko T, Omokolo ND, Driouich A, Balangé AP (2005) Peroxydase and pectin methylesterase activities in cocoyam (Xanthosoma sagittifolium L. Schott) roots upon Pythium myriotylum inoculation. J Phytopathol 153:409–416
Bown D (2000) Aroids: plants of the Arum family, 2nd edn. Timber Press, Portland
Brown VM, Asemota H (2009) PCR-based characterization of dasheen (Colocasia sp.) and cocoyam (Xanthosoma sp.). J Biotech Res 1:28–40
Cathebras C, Traore R, Malapa R, Risterucci AM, Chaïr H (2014) Characterization of microsatellites in Xanthosoma sagittifolium (Araceae) and cross-amplification in related species. Appl Plant Sci 2:1400027
Doungous O, Sama AE, Adiobo A, Zok S (2011) Determination of ploidy level by flow cytometry and autopolyploid induction in cocoyam (Xanthosoma sagittifolium). Afr J Biotechnol 10:16491–16494
FAOstat (2014) Food and Agriculture Organization of the United Nations. Rome, Italy. http://faostat3.fao.org/home/E. Accessed 30 Oct 2014
GenStat Discovery 4th edn 4 (2012)VSN International Ltd., Hemel Hempstead
Ghosh P, Mukherjee S, Sharma AK (2001) Cytophotometric estimation of in situ DNA content in several species of Araceae. Cytobios 105:177–183
Giacometti DC, Leon J (1994) Tannia, yautia (Xanthosoma sagittifolium). In: Hernando Bermejo JE, Leon J (eds) Neglected Crops: 1492 from a different perspective. FAO Plant Production and Protection Service, Rome
Grover CE, Yu Y, Wing RA, Paterson AH, Wendel JF (2008) A phylogenetic analysis of indel dynamics in the cotton genus. Mol Biol Evol 25:1415–1428
Hazzouri KM, Mohajer A, Dejak SI, Otto SP, Wright SI (2008) Contrasting patterns of transposable-element insertion polymorphism and nucleotide diversity in autotetraploid and allotetraploid Arabidopsis species. Genetics 179:581–592
Irwin SV, Kaufuis P, Banks K, de la Pena R, Cho JJ (1998) Molecular characterization of taro (Colocasia esculenta) using RAPD markers. Euphytica 99:183–199
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Kalendar R, Schulman AH (2006) IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nature Protoc 1:2478–2484
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman AH (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH (2008) Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA 105:5833–5838
Kalendar R, Antonius K, Smykal P, Schulman AH (2010) iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor Appl Genet 121:1419–1430
Kalendar R, Flavell A, Ellis THN, Sjakste T, Moisy C, Schulman AH (2011a) Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 106:520–530
Kalendar R, Lee D, Schulman AH (2011b) Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. Genomics 98:137–144
Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33:102–106
Kraitshtein Z, Yaakov B, Khasdan V, Kashkush K (2010) Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186:801–812
Kreike CM, Van Eck HJ, Lebot V (2004) Genetic diversity of taro, Colocasia esculenta (L.) Schott, in Southeast Asia and the Pacific. Theor Appl Genet 109:761–768
Loh JP, Kiew R, Hay A, Kee A, Gan LH, Gan YY (2000) Intergeneric and interspecific relationships in Araceae tribe Caladieae and development of molecular markers using amplified fragment length polymorphism (AFLP). Ann Bot 85:371–378
Mbouobda HD, Boudjeko T, Djocgoue PF, Tsafack TJJ, Omokolo DN (2007) Morphological characterization and agronomic evaluation of cocoyam (Xanthosoma sagittifolium L. Schott) germplasm in Cameroon. J Biol Sci 7:27–33
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Nagy S, Poczai P, Cernák I, Gorji AM, Taller GHJ (2012) PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem Genet 50:670–672
Ngouo LV, Nzietchueng S, Valet G (1989) Organisation inflorentielle et florale, fertilité et germination des pollens chez trois Xanthosoma spp. cultivés au Cameroun. Agro Afr 1:95–104
Nyochembeng LM, Beyl CA, Pacumbaba RP (2007) Peroxydase activity, isozyme patterns and electrolyte leakage in roots of cocoyam infected with Pythium myriotylum. J Phytopathology 115:454–461
Offei SK, Asante IK, Danquah EY (2004) Genetic structure of seventy cocoyam (Xanthosoma sagittifolium, Linn, Schott) accessions in Ghana based on RAPD. Hereditas 140:123–128
Onokpise OU, Wutoh JG, Ndzana X, Tambong JT, Meboka MM, Sama AE, Nyochembeng L, Agueguia A, Nzietchueng S, Wilson JG, Bursn M (1999) Evaluation of macabo cocoyam germplasm in Cameroon. In: Janick J (ed) Perspectives on News Crops and New Uses. ASHA Press, Alexandra, pp 394–396
Owusu-Darko PG, Paterson A, Omenyo EL (2014) Cocoyam (corms and cormels)—An underexploited food and feed resource. J Agric Chem Environ 3:22–29
Pacumbaba RP, Wutoh JG, Sama AE, Tambong JT, Nyochembeng LM (1992) Isolation and pathogenicity of rhizosphere fungi of cocoyam in relation to cocoyam root rot disease. J Phytopathol 135:265–273
Parisod C, Alix K, Just J, Petit M, Sarilar V, Mhiri C, Ainouche M, Chalhoub B, Grandbastien MA (2010) Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol 186:37–45
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlex 6.5: genetic analysis in Excel. Population genetic software for teaching and research -an update. Bioinformatics 28:2537–2539
Pearce SR, Stuart-Rogers C, Knox MR, Kumar A, Ellis THN, Flavell AJ (1999) Rapid isolation of plant Ty1-copia group retrotransposon LTR sequences for molecular marker studies. Plant J 19:711–717
Perneel M, Tambong JT, Adiobo A, Floren C, Saborio F, Levesque A, Höfte M (2006) Intraspecific variability of Pythium myriotylum isolates from cocoyam and the host crops. Mycol Res 110:583–593
Perneel M, Heyrman J, Adiobo A, De Maeyer K, Raaijimakers JM, De Vos P, Höfte M (2007) Characterization of CMR5c and CMR12a, novel fluorescent Pseudomonas strains from the cocoyam rhizosphere with biocontrol activity. J Appl Microbiol 103:1007–1020
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–248
Purseglove JW (1972) Tropical crops: monocotyledons I. Longman Ltd., London
Quero-Garcia J, Noyer JL, Perrier X, Marchand JL, Lebot V (2004) A germplasm stratification of taro (Colocasia esculenta) based on agro-morphological descriptors, validation by AFLP markers. Euphytica 137:387–395
Schlüter PM, Harris SA (2006) Analysis of multilocus fingerprinting data sets containing missing data. Mol Ecol Notes 6:569–572
Schnell RJ, Goenaga R, Olano CT (1999) Genetic similarity among cocoyam cultivars based on randomly amplified polymorphic DNA (RAPD) analysis. Sci Hortic 80:267–276
Schulman AH (2013) Retrotransposon replication in plants. Curr Opin Virol 3:604–614
Schulman A, Flavell A, Paux E, Ellis THN (2012) The application of LTR retrotransposons as molecular markers in plants. Methods Mol Biol 859:115–153
Sefa-Dedeh S, Agyir-Sackey KE (2004) Chemical composition and effect of processing on oxalate content of cocoyam Xanthosoma sagittifolium and Colocasia esculenta cormels. Food Chem 85:479–487
Shannon CE, Weaver W (1949) The mathematical theory of communication. The University of Illinois Press, Urbana
Singh D, Jackson G, Hunter D, Fullerton R, Lebot V, Taylor M, Iosefa T, Okpul T, Tyson J (2012) Taro leaf blight—a threat to food security. Agriculture 2:182–203
Smykal P, Bacova-kerteszova N, Kalendar R, Corander J, Schulman AH, Pavelek M (2011) Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-based markers. Theor Appl Genet 122:1385–1397
Swofford DL (1998) PAUP. Phylogenetic analysis using parsimony (and other methods). Version 4. Sinauer Associates, Sunderland
Tambong JT, Sapra VT, Garton S (1998) In vitro induction of tetraploids in colchicine-treated cocoyam plantlets. Euphytica 104:191–197
Vicient CMM, Jääskeläinen M, Kalendar R, Schulman AH (2001) Active retrotransposons are a common feature of grass genomes. Plant Physiol 125:1283–1292
Vukich M, Schulman AH, Giordani T, Natali L, Kalendar R, Cavallini A (2009) Genetic diversity in sunflower (Helianthus annus L.) and the Helianthus genus as assessed by retrotransposon-based molecular markers. Theor Appl Genet 119:1027–1038
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Wilson JE (1984) Cocoyam. In: Goldsworthy PR, Fisher NM (eds) The physiology of tropical field crops. Wiley, London, pp 589–605
Yeh FC, Boyle TBJ (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157
Acknowledgments
Anne-Mari Narvanto and Ursula Lönnqvist are thanked for their valuable technical assistance. D.O. was supported by an 8 month-mobility fellowship (Decision TM-09-6259) and a travel grant issued by the Finnish Centre for International Mobility (CIMO) and the Kirkhouse Trust respectively. R.K. was supported by a grant from the Academy of Finland (Decision 134079).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest. The experiments comply with the current laws of the countries in which they were performed.
Additional information
Data deposition The sequences reported here have been deposited in the GenBank database (accession nos.GU810535, KF813039–KF813057).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Doungous, O., Kalendar, R., Adiobo, A. et al. Retrotransposon molecular markers resolve cocoyam (Xanthosoma sagittifolium) and taro (Colocasia esculenta) by type and variety. Euphytica 206, 541–554 (2015). https://doi.org/10.1007/s10681-015-1537-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1537-6