Abstract
Asian soybean rust (ASR), caused by the fungus Phakopsora pachyrhizi, is one of the most serious soybean diseases in South America and other tropical and sub-tropical areas. The soybean lines, ‘Xiao Jing Huang,’ ‘Himeshirazu,’ and ‘Iyodaizu B’ were previously identified for their resistance to ASR fungus, while the genetic basis of the resistance has yet to be known. In this study, we mapped the ASR resistance loci in these three lines using three independent mapping populations derived from crosses with an ASR susceptible variety, BRS184. In each population, resistance to ASR appeared to be primarily controlled by a single major gene. The resistance genes with largest effect from Xiao Jing Huang and Himeshirazu were genetically mapped on chromosome 18 corresponding to the same location of known resistant locus, Rpp1. On the other hand, quantitative trait locus analysis mapped the major ASR resistance locus of Iyodaizu B to the region of chromosome 16 where Rpp2 was previously mapped. Genetic mapping with DNA markers and disease reactions of seven candidates carrying Rpp1 to four Brazilian ASR isolates revealed a significant variation in their ASR resistance reaction, indicating that they share different resistance genes tightly linked to each other or different resistant alleles of a single Rpp1 gene. Therefore, these seven soybean lines could be clearly separated into at least two functional groups.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asian soybean rust (ASR) caused by biotrophic basidiomycete Phakopsora pachyrhizi (H. Sydow and Sydow) occurs in all major soybean-growing regions of the world. The disease is considered as one of the most serious economic threats for soybean growers (Goellner et al. 2010) and severe yield losses are common in South America when environmental conditions are conducive for disease development (Yorinori 2008). Several management tactics have been employed to control ASR and to minimize the impact of this disease. Chemical treatment with fungicide has been perceived as the first line of defense against the disease (Levy 2005). However, limited number of appropriate fungicide, specific application requirements, increased production costs, environmental pollution and development of fungicide resistant races are the main concerns of using fungicides (Schneider et al. 2008). Hence, an environment friendly, cost-effective and long-term management of the disease can be achieved through utilization of host genetic resistance to the disease (Ribeiro et al. 2007). A major impediment to breeding for ASR resistance is the existence of the high pathogenic variability in the pathogen population and the length of time that takes to incorporate effective and novel Rpp genes into new cultivars. Therefore, rational management of ASR in sustainable cropping systems should be based on integrated management strategy that includes host resistance and fungicide.
Genetic resistance to ASR has been identified. Six dominant genes (Rpp1 to Rpp6) controlling pathotype-specific resistance to ASR have so far been mapped at different loci (Hyten et al. 2007; Garcia et al. 2008; Silva et al. 2008; Hyten et al. 2009; Li et al. 2012). Incompatible interactions mediated by some of these genes have an immune phenotype, whereas resistances conferred by other R genes are characterized by limited fungal growth and sporulation and the formation of reddish-brown lesions. Compatible interactions typically are characterized by tan-colored lesions with fully sporulating uredinia. Partial or rate-reducing resistance to ASR has also been documented in soybean, but it has not been widely employed because of complexities in assessment. Although these major Rpp genes are now available for breeding, they rarely offer durable resistance to highly variable ASR pathogen (Oliveira et al. 2005). These genes are effective to specific P. pachyrhizi isolates, while they were ineffective when challenged with other isolates. This has limited the use of single genes for resistance in soybean especially in South America, where ASR populations are highly virulent and divergent (Yamanaka et al. 2011; Akamatsu et al. 2013). These commonly encountered problems associated with the ineffectiveness of the specific resistance genes and identification of the durable resistance against highly variable ASR population have led to the continuous search for new resistant gene as a breeding remedy for ASR. Development of ASR resistant cultivar has been an important aspect of genetic improvement programs in soybean in the current days and would be augmented by the identification of gene conditioning the ASR resistance in wide range of soybean varieties. However, very limited sources of soybean germplasm are known to be resistant to ASR. In order to identify new sources of resistance in soybean, Miles et al. (2006) evaluated the entire germplasm collection (16,000 accessions) of the United States Department of Agriculture (USDA) against a mixture of four P. pachyrhizi isolates. After two rounds of evaluation, only 850 accessions were identified with partial tolerance or resistance reactions to P. pachyrhizi, which correlates to less than 5 % of the USDA Soybean Germplasm Collection. Although specific P. pachyrhizi races are virulent on these single gene resistant sources, it may be beneficial to pyramid these known resistant genes into modern cultivars to create broad spectrum resistance to ASR.
Previously intensive screening of Chinese and Japanese soybean genetic resources had identified a Chinese land race, Xiao Jing Huang and Japanese cultivar, Himeshirazu with resistance to Brazilian ASR population (Yamanaka et al. 2008). Resistance to Japanese and Brazilian ASR population in Xiao Jing Huang was confirmed in subsequent experiments (Yamanaka et al. 2010, 2011). ASR resistance in Iyodaizu B was identified against the infection by Japanese ASR population (Benitez et al. 2008). Although these three lines do not possess a very high level of resistance against the tested ASR population, they could be beneficial for pyramiding broad spectrum ASR resistance. Therefore, the aims of the present study were to study the mode of inheritance and identify the resistance loci in Xiao Jing Huang, Himeshirazu, and Iyodaizu B.
Materials and methods
Plant materials
The parental soybean lines used in this study include an ASR susceptible Brazilian variety BRS184 and three ASR resistant Asian lines, Xiao Jing Huang (China, Chinese ID number, ZDD: 00880), Himeshirazu (Japan, Japanese ID number, JP: 67990), and Iyodaizu B (Japan, no national ID number). The resistant lines were crossed with susceptible Brazilian variety BRS184 to develop three F2 mapping populations; POP-A, POP-B, and POP-C. The mapping population POP-A was consisted of 90 F2 plants derived from the cross of Xiao Jing Huang × BRS184. The mapping population POP-B with a pooled collection of 120 F2 plants was developed from the cross of BRS184 × Himeshirazu while in POP-C, a population of 143 F2 plants was produced from the cross of Iyodaizu B × BRS184 (for POP-C).
A total of ten soybean lines were tested against four Brazilian ASR isolates to determine allelic variation within Rpp1 and Rpp2 loci. The candidate lines carrying Rpp1 included Komata (PI 200492), Huang Dou (PI 587880A), Bai Dou (PI 587886), Xiao Huang Dou (PI 587905), Zhao Ping Hei Dou (PI 594767A), Xiao Jing Huang (PI number not assigned) and Himeshirazu (PI 594177). The evaluated soybean lines which may carry Rpp2 were No. 3 (PI 230970), Hougyoku (PI 224270), and Iyodaizu B (PI number not assigned) (Table 1) The three mapping populations, the parentals and the other ten candidates carrying Rpp1 or Rpp2 were grown as described by Yamanaka et al. (2010).
Pathogen inoculation and resistance evaluation
The Japanese ASR isolate T1-2 (Yamanaka et al. 2013b; Hossain et al. 2014) was used for inoculation of POP-A, while isolate E1-4-12 was used for inoculating POP-B and POP-C. ASR isolate T1-2 was obtained from ASR population T1 via single-lesion isolation by Yamaoka et al. (2014). ASR population T1 was originally collected from soybean cultivar ‘Tachinagaha’ in Tsukuba, Japan on September 20, 2007 (Yamaoka et al. 2014). ASR isolate E1-4-12 was obtained from Japanese ASR isolate E1-4 (Yamaoka et al. 2014) by single-lesion isolation following the manual (Yamanaka et al. 2013a). E1-4 has previously been known to be either avirulent or have only weak pathogenicity on differential varieties carrying different ASR resistance genes (Rpp1 to Rpp6) (Yamaoka et al. 2014). Hence E1-4-12 was expected to be suitable for mapping weak ASR resistance genes in Himeshirazu, and Iyodaizu B.
When the plants of three mapping populations reached V3 to V4 growth stage (approximately 3 weeks after sowing), they were inoculated with the urediniospores of T1-2 and E1-4-12. The optimal spore concentration used for inoculation of soybean plants was 5 × 104 urediniospores/mL. Preservation, multiplication and adjustment of spore concentration were carried out according to the manual (Yamanaka et al. 2013a). The inoculation of parental lines and POP-A, POP-B and POP-C plants was done following the same technique as described by Hossain et al. (2014). Inoculated plants were kept overnight in the humid chamber and then transferred to the growth chamber under the condition described in the previous study (Yamanaka et al. 2013b). Two weeks after inoculation, three populations and their parental lines were evaluated for ASR reactions based on number of uredinia per lesion (NoU) and sporulation level (SL). Three infected leaflets were detached from each inoculated plant and examined the abaxial side of each leaflet microscopically following the same methods of previous study (Hossain et al. 2014). For determining NoU and SL, a maximum of 30 lesions in total, 10 lesions from each leaflet were observed. The SL was rated on a scale of 0–3 as described by Yamanaka et al. (2010) where 0 = none, 1 = little, 2 = moderate and 3 = abundant sporulation. In T1-2 inoculated POP-A, infection phenotypes with lesions with NoU and SL < 2.0 were classified as resistant (R) and those with lesions with NoU and SL ≥ 2.0 were classified as susceptible (S) (Hossain et al. 2014). The boundary value of 1.0 was applied for E1-4-12 infection to POP-B, since the infection with less virulent ASR isolate, E1-4-12 produced lower NoU and SL than T1-2 infection and made the gaps (0.0 < NoU < 1.1, 0.0 < SL < 1.53) between R and S phenotypes in the frequencies distributions (Fig. 1). In contrast, the gaps between R and S were not observed in POP-C even though frequency distributions of NoU and SL seemed to be bimodal (Fig. 1). Therefore, boundary values of NoU and SL for R and S phenotypes were not determined and quantitative trait locus (QTL) analysis was applied for genetic mapping in POP-C.
Frequency distributions of number of uredinia per lesion (NoU) (a, c and e) and sporulation level (SL) (b, d and f) in POP-A (Xiao Jing Huang × BRS184) (a and b), POP-B (BRS184 × Himeshirazu) (c and d) and POP-C (Iyodaizu B × BRS184) (e and f) against Japanese Asian soybean rust (ASR) isolates. ASR isolates, T1-2 and E1-4-12 were used for POP-A, and other two populations: POP-B and POP-C, respectively. Triangles indicate the values of respective resistant and susceptible parents. Estimated boundary values between resistant and susceptible phenotypes for NoU and SL in POP-A and POP-B are shown by arrows
In order to compare the ASR resistance among the ten selected Rpp1 or Rpp2 lines, we have used four Brazilian ASR isolates, BRP-2.1, BRP-2.5, BRP-2.6, and BRP-2.49 for inoculation. A detached-leaf method was used for this experiment using experimental condition as described in the previous study (Yamanaka et al. 2013b). Two weeks after inoculation, a total of 30 lesions from four infected leaflets of each line were evaluated by examining frequency of lesions with uredinia (%LU), NoU, and SL. Classification of infection types was done following the same criteria as used in our previous study (Yamanaka et al. 2013b).
SSR marker analysis
The same mapping strategy was adopted and followed in the present study as we did in our previous study (Hossain et al. 2014) for identifying the major ASR resistance loci in the three mapping population. Briefly, DNA was extracted from the parental lines as well as individual F2 plant. Simple sequence repeat (SSR) markers linked to six Rpp (1–6) loci were employed to investigate the linkage to unknown loci of ASR resistance in three lines. Although several SSR markers linked to each of the known ASR resistance genes (Rpp1–6) were tested; one was selected as a polymorphic marker for each gene. SSR marker Sat_064, Satt380, Sat_263, Satt288, Sat_280 and Satt324 linked to Rpp1, 2, 3, 4, 5, and 6, respectively were found polymorphic between mapping parents of POP-A and used for initial screening of a small subset (24 F2 plants) of this population. In POP-B, SSR marker Sat_064, Satt380, Sat_263, AF162283, Sat_275 and Satt324 were polymorphic between mapping parents and selected for initial screening of a small subset of this population. In POP-C, the selected polymorphic SSR markers for initial screening of its small subset population were Sat_064, Satt380, Satt460, AF162283, Sat_280 and Satt324. Once significant association between NoU and genotypes of marker linked to Rpp loci was found, full mapping population was tested for confirmation. Additional markers around the regions where the potential ASR resistance gene was mapped were chosen from SoyBase (Grant et al. 2010). All SSR markers used in this study were co-dominant for parents. PCR and subsequent electrophoresis were performed following the procedures described in the manual (Yamanaka et al. 2013a).
Genetic mapping and statistical analysis
Goodness-of-fit between observed and expected segregation ratios of ASR resistance phenotypes and SSR marker genotypes in F2 populations was tested using Chi square (χ2) analysis. ANOVA and regression analysis were applied to identify the significance of the association between ASR phenotype (NoU and SL) and nearest flanking markers and to estimate the genetic effects and the portion of the phenotypic variation explained by the resistance genes based on genotypes of flanking markers. MAPMAKER/EXP 3.0b (Lander et al. 1987) was used to determine the order of loci for SSR markers and ASR resistance in each mapping population. The Kosambi mapping function was used to convert recombination values into map distances. The minimum logarithm of the odds (LOD) score and the maximum genetic distance for linkage map construction were adjusted to 3.0 and 37.2 cM, respectively. In POP-C, genomic region associated with NoU and SL was identified by quantitative trait loci (QTLs) analysis using Windows QTL Cartographer v2.5 (Wang et al. 2007). The interval mapping with the walk speed of 0.5 cM was applied for the QTL analysis. The estimated order of markers and distances between markers determined by MAPMAKER were used for QTL analysis. The LOD score threshold for declaring a putative locus as significant was determined by permutation testing using 1,000 permutations and significant at the 0.01 level. The threshold level of the LOD score was set at 1.78 for NoU and 1.75 for SL. The loci for NoU and SL were defined as the peaks of maximum LOD score. The nearest flanking marker was chosen according to the distance between peak of LOD score and markers.
In order to compare the linkage map around Rpp1 obtained from this study with those of previous studies more precisely, the linkage maps from two independent F2 populations derived from BRS184 × PI 594767A and BRS184 × PI 587905, respectively (Hossain et al. 2014) were re-constructed by the progeny test using F3 populations. In both populations, fifteen or fewer (0–15) F2:3 progeny from each resistant F2 plant was cultivated and evaluated for their resistance to T1–2, as it was done in F2 (Hossain et al. 2014). Firstly, we started this evaluation with a few F3 plants, and then the numbers of evaluated plants were enlarged according to their phenotype. If a susceptible phenotype was observed among F2:3 progeny, the Rpp1 genotype of the F2 plant was considered to have been heterozygous. If all the F2:3 progeny were resistant, the F2 plant was considered to have been either homozygous for the resistance allele or heterozygous. These genotype data for resistance loci were used for reconstruction of previous genetic maps.
Results
Evaluation of Asian soybean rust resistance
Significant differences were observed in NoU and SL between the susceptible parent BRS184 and three resistant parents, Xiao Jing Huang, Himeshirazu, and Iyodaizu B (Fig. 1). The susceptible parent BRS184 showed the highest susceptibility among the four varieties to both T1-2 and E1-4-12. However, the susceptibility was relatively higher to T1-2 (NoU = 3.13, SL = 3.00) than E1-4-12 infections (NoU = 1.62, SL = 1.97) (Fig. 1). Inoculations of Himeshirazu and Iyodaizu B with E1-4-12 isolate gave rise to strong resistance or almost immune reaction, showing imperfect formation of RB lesions with no uredinia production. The mean values of both NoU and SL in these varieties were therefore, scored as zero. On the other hand, Xiao Jing Huang produced RB lesions with limited uredinia and spore production (NoU = 1.13, SL = 1.43) after T1–2 infection.
Phenotypic analysis of the POP-A showed a segregation of 66 plants with resistant phenotype and 24 plants with the susceptible phenotype. In the 66 resistant plants, only 7 plants showed higher resistance (both NoU and SL) than the resistant parent Xiao Jing Huang but all resistant plants produced uredinia and spores. A Chi square (χ2) test revealed that the observed segregation fitted the expected segregation ratio of single dominant resistance gene: 3:1 (resistant: susceptible) in the F2 generation (Table 2). The degree of dominance for NoU and SL were 1.09 and 1.12, indicating that the putative ASR resistant allele primary controlling resistance in Xiao Jing Huang is completely dominant with a little heterosis (Table 3).
The POP-B plants segregated into two clearly separated ranges, 90 with resistant and 30 with susceptible phenotype (Table 2). The observed segregation perfectly fitted the 3:1 (resistant: susceptible) ratio (χ2 = 0.00, P = 1.00) in the F2 generation, suggesting that a single dominant gene controls the resistance in Himeshirazu mainly. However, 7 of 30 susceptible F2 plants showed higher NoU and SL than susceptible parent BRS184 (Fig. 1c, d). The degree of dominance for both NoU and SL was 1.00, indicating that the resistant loci is completely dominant in Himeshirazu (Table 3).
The F2 progeny of POP-C showed a wider variation in values of both NoU and SL than that of POP-A and POP-B. The NoU in the F2 population ranged from 0.00 to 2.53, while that in parental Iyodaizu B and BRS184 were 0.00 and 1.62, respectively (Fig. 1e). Similarly, the value of SL in F2 population ranged from 0.00 to 2.90, while the resistant and susceptible parents had mean SL of 0.00 and 1.97, respectively (Fig. 1f). As a result, the frequency distributions of NoU and SL in the F2 population were bimodal, however, some F2 plants couldn’t be classified into two differentiated classes, i.e. resistant and susceptible phenotype because no clear boundary between resistant and susceptible phenotype was not observed in these plants. Thus, we decided to perform a QTL analysis for mapping of ASR resistance in POP-C.
Mapping of resistance loci to ASR in three populations
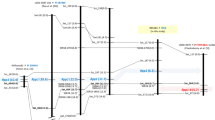
Initial genotyping of a small sub set of POP-A (24 F2 plants) with one marker linked to each of the Rpp1 to Rpp6 and subsequent test by ANOVA showed that the F2-inferred NoU data were significantly associated with Sat_064 linked to the Rpp1 gene (P < 0.0001, data not shown). Genotype data of full set of POP-A with Sat_064 also revealed significant (P < 0.0001) association with variation in NoU and SL (Table 3). The coefficient of determination (R 2) of each phenotypic variation by this marker was 0.48 for NoU and 0.45 for SL, respectively (Table 3). Additional SSR markers Sat_372, Satt191, Sat_117, and Satt199 around Rpp1 were found polymorphic between the parents of POP-A. Genotyping of the full set of POP-A with Sat_064, Sat_372, Satt191, Sat_117, and Satt199, followed by a χ2 test revealed that all SSR markers mapped in the POP-A satisfactorily fitted the expected ratio for co-dominant inheritance (1:2:1). The major resistance locus of Xiao Jing Huang controlling the phenotypic variation of NoU and SL was mapped between Satt191 and Sat_064 where Rpp1 was previously mapped by Ray et al. (2009) (Fig. 2).
Genetic linkage map around Rpp1, the resistance locus against Phakopsora pachyrhizi on linkage group G (chromosome 18) constructed from POP-A (Xiao Jing Huang × BRS184) and POP-B (BRS184 × Himeshirazu). The values in parenthesis next to marker name are cumulative distances (cM) from the top of linkage group in each mapping population. The linkage maps of Hyten et al. (2007), Chakraborty et al. (2009), Ray et al. (2009), and Hossain et al. (2014) are included for comparison reference. The maps of PI 594767A and PI 587905 by Hossain et al. (2014) were re-constructed in this study
Similar to POP-A, primary genotyping of a small subset of POP-B (24 F2 plants) with SSR markers linked to six Rpp loci followed by ANOVA test showed that the genotype at SSR marker Sat_064 was significantly associated with variation in NoU (P < 0.0001, data not shown). The linkage was further confirmed by testing the full mapping population with this marker and six additional SSR markers, Sct_199, Satt191, Sat_117, Sct_187, SSR66, and Sat_372 near Rpp1. A χ2 test revealed that all SSR markers used to map the resistance in the POP-B fitted the expected ratio for co-dominant inheritance and followed a 1 resistant: 2 segregating: 1 susceptible ratio. The major ASR resistance locus of Himeshirazu was mapped between Sct_187 and Sat_064 where Rpp1 was previously mapped in PI200492 by Hyten et al. (2007) and in PI 587886 by Ray et al. (2009) (Fig. 2). Genotype data of POP-B with Sct_187, the nearest linked marker to Rpp1 showed significant (P < 0.0001) association with variations in NoU and SL (Table 3). The R 2 of each phenotypic variation by this marker was 0.65 for NoU and 0.66 for SL, respectively (Table 3).
On the contrary in POP-C, initial marker-trait analysis of 24 F2 plants by ANOVA indicated a significant linkage between NoU and SSR marker Satt380 for Rpp2 (P = 0.0114, data not shown). The linkage was confirmed by subsequent screening and analysis of full set of POP-C with Satt380 and additional markers from the same region. SSR markers Satt380, Sct_001, Sat_255, Satt620, and Sat_366 around Rpp2 were polymorphic and used for mapping. A χ2 test revealed that all the tested SSR markers satisfactorily fitted the expected ratio for co-dominant inheritance (1:2:1). A linkage map was constructed with the five SSR markers that spanned a total length of 15.9 cM (Fig. 3). An interval mapping was performed and the loci for the resistant characters, NoU and SL were identified. As shown in Fig. 3, a peak of LOD score for NoU was detected in the interval between the SSR markers Satt620 and Sat_366 at 9.6 cM from the top of this linkage group. Similarly, a peak of LOD score for SL was detected in the similar position of NoU between Satt620 and Sat_366 at 10.1 cM from the top of this linkage group (Fig. 3). The peak LOD scores for the loci for NoU and SL were 57.05 and 58.23, respectively (Fig. 3). Composite interval mapping also generated a single locus for each trait with same peak position of interval mapping (data not shown). These results indicated that a single locus located close to Satt620 where Rpp2 was previously mapped (Garcia et al. 2008; Silva et al. 2008) contributes to reduce each of NoU and SL mainly in Iyodaizu B. The additive effects in this locus were estimated to reduce NoU and SL by 0.81 and 0.96, respectively (Table 3). The degree of dominance for NoU was 0.63 and that for SL was 0.55; indicating that resistance allele of this locus was dominant but incomplete. The R 2 of each phenotypic variation was 0.70 for NoU and 0.72 for SL, respectively (Table 3).
Genetic linkage map around Rpp2, the resistance locus against Phakopsora pachyrhizi on linkage group J (chromosome 16) constructed from POP-C (Iyodaizu B × BRS184). Resistance locus of Iyodaizu B is estimated by peak positions (closed triangles) of logarithm of the odds (LOD) curves obtained by quantitative trait loci (QTLs) analysis for number of uredinia (NoU) and sporulation level (SL). The values in parenthesis next to marker name are cumulative distances (cM) from the top of linkage group in each mapping population. The linkage maps of Silva et al. (2008) and Garcia et al. (2008) are included for comparison reference
Infection types of ten candidates carrying Rpp1 or Rpp2
ASR reaction profile of the ten candidates carrying Rpp1 or Rpp2 to four Brazilian ASR isolates was obtained by detached leaf method as shown in Table 4. Soybean lines PI 587905, PI 594767A, and PI 587880A which were previously mapped as Rpp1 in separate studies by Hossain et al. (2014) and Ray et al. (2009) showed the same “Highly resistant” phenotypes against two of four tested ASR isolates: BRP-2.49 and BRP-2.1 (Table 4). They also showed resistant phenotype to BRP-2.6 but degree of resistance was different for each. The infection by BRP-2.5 produced the only susceptible reaction in these three lines. On the other hand, the resistant parental lines, Xiao Jing Huang and Himeshirazu of POP-A and POP-B, and the two other known Rpp1 lines, PI 200492 and PI 587886 showed susceptible reaction to all four Brazilian ASR isolates. Therefore, these seven lines clearly revealed two different reaction patterns against infection by four tested ASR isolates.
Iyodaizu B, the resistant parent of POP-C, and the known Rpp2 carrying lines, PI 230970 and Hougyoku showed different ASR reaction profile to the tested ASR isolates (Table 4). Hougyoku was resistant to all ASR isolates, except for BRP-2.6 against which “Slightly resistant” phenotype was observed. PI 230970 was clearly susceptible to all tested ASR isolates. Iyodaizu B showed “Resistant” against BRP-2.49 and “Slightly resistant” against BRP-2.1, while susceptible phenotype was observed against two other ASR isolates, BRP-2.6 and BRP-2.5.
Discussion
Since only six kinds of resistant genes against ASR have been identified so far and South American ASR pathogens are highly virulent and diverse (Yamanaka et al. 2010, 2011; Akamatsu et al. 2013), new ASR resistance resources are needed for soybean breeding in South America. On the other hand, pyramiding available ASR resistance genes in a single soybean cultivar is known to provide highly resistance against the ASR by candidate interactions among Rpp genes (Lemos et al. 2011; Yamanaka et al. 2013b). Therefore, new resistance genes may be useful for gene-pyramiding even though they don’t show strong resistance individually. Soybean lines, Xiao Jing Huang, Himeshirazu, and Iyodaizu B were identified as having ASR resistance in the previous studies (Benitez et al. 2008; Yamanaka et al. 2008, 2010, 2011). However, the genetic basis of the ASR resistance in these lines was not determined in previous studies. Our results suggested that single dominant gene, candidate Rpp1 primarily control the ASR resistance in Xiao Jing Huang and Himeshirazu However, ‘Iyodaizu B’ had a single gene at or near the Rpp2 locus which was incompletely dominant. The level of observed dominance might depend on the P. pachyrhizi isolate used to obtain rust reaction data. It seems quite possible that the degree of dominance against a more aggressive isolate may be less than towards a less aggressive isolate.
Chakraborty et al. (2009) suggested that PI 594538A carries ASR resistance gene Rpp1-b that is likely a different resistant allele or closely linked different gene from Rpp1 of PI 200492 based on their slightly different map positions and different reactions to Zimbabwe ASR isolate. Hossain et al. (2014) also reported clearly different reactions by two kinds of Rpp1 carrying lines against 64 South American ASR populations. Rpp1-carrying lines with weak resistance included PI 200492, PI 368039, and PI 587886, while those with strong resistance were PI 594767A, PI 587905, and PI 587880A. These lines were predicted to share different resistant alleles of Rpp1 or tightly linked loci on the same genomic region. In this study, resistance gene of Xiao Jing Huang was mapped between Satt191 and Sat_064, and resistance gene of Himeshirazu was mapped between Sct_187 and Sat_064 onto the soybean chromosome 18 (LG-G), where Rpp1 of PI 200492 and PI 587886 were previously mapped (Hyten et al. 2007; Ray et al. 2009) (Fig. 2). These Rpp1 loci are slightly different from Rpp1 (Rpp1-b[?]) loci of PI 594538A, PI 594767A, and PI 587905 which were mapped between two SSR markers Sat_064 and Sat_372 (Chakraborty et al. 2009; Hossain et al. 2014). According to those previously reported studies and the present study, soybean lines PI 200492, PI 368039, PI 587886, Xiao Jing Huang, and Himeshirazu may carry the original Rpp1 as identified by Hartwing & Bromfield (1983) that shows weak resistance to South American ASR pathogens. In contrast, PI 594538A, PI 594767A, PI 587905, and PI 587880A may carry the Rpp1-b as identified by Chakraborty et al. (2009) that shows strong resistance to South American ASR populations. ASR reaction profile of seven Rpp1 carrying candidates in this study also supported this assumption by showing clear differences between Rpp1 and Rpp1-b carrying lines in their reaction to three Brazilian ASR isolates (Table 4). However, Kim et al. (2012) suggested that ASR resistance genes from PI 587880A and PI 587886 were located in the same region between Satt191 and Sat_064. In addition, the order of molecular markers between genetic maps sometimes differs among the maps constructed by different parental combinations in soybean. As the Rpp1 linkage maps of Hyten et al. (2007) and Ray et al. (2009) were previously reported to be different from the soybean consensus map of Song et al. (2004) by showing an inversion at Sat_372 and Sat_064. Furthermore, disease-resistance genes are known to cluster in some chromosomal regions in soybean (Jeong et al. 2001). Rpp1/Rpp1-b carrying lines may share different genes in such gene cluster. Thus, allelism test, fine mapping of resistant loci with DNA markers, or map-based cloning of genes are needed to be carried out to conclude if these two groups share different resistance alleles of same Rpp gene or tightly linked different ASR resistance genes.
The NoU and SL data in POP-C produced imperfect bimodal distributions, but interval mapping indicated that most of the variation is likely to be due to the incomplete dominance of a single gene that mapped to the vicinity of the Rpp2 locus on chromosome 16. This incomplete dominance may be brought by the virulence of E1-4-12, since the kinds of ASR isolates are known to influence the level of dominance (Garcia et al. 2011; Lemos et al. 2011). It is also possible that some portion of the phenotypic variation was due to the different genetic backgrounds of the progeny and/or environmental factors. The resistant locus Rpp2 was previously reported to confer resistance in PI 230970 and PI 224270 (Hougyoku), which was placed on chromosome 16 between SSR marker Sat_255 and Satt620 (Silva et al. 2008) and between SSR marker Satt215 and Sat_361 respectively Garcia et al. 2008). A direct comparison between our map and those of Silva et al. (2008) and Garcia et al. (2008) revealed that the order of markers that were common among the maps and resistance locus of these three maps were very similar except that Rpp2 locus of Silva et al. (2008) seems to be far from Sat_366 compared with other two maps (Fig. 3). However, significant difference in reactions to ASR isolates was observed among three Rpp2 carrying varieties. Resistance of PI 230970 was weakest to show susceptible against all four Brazilian ASR isolates, while Hougyoku was resistant against all tested isolate even though degree of resistance was different among ASR isolates used for inoculation (Table 4). Reaction profile of Iyodaizu B was different from those of PI 230970 and Hougyoku. Since resistance genes of them were mapped in the same region (Fig. 3), these three Rpp2 lines may carry different Rpp2 alleles, different tightly linked resistance genes or some other unknown genes except for Rpp2.
In conclusion, our study successfully mapped genes conditioning the resistance against ASR in three soybean lines, Xiao Jing Huang, Himeshirazu, and Iyodaizu B mainly. Since resistant alleles of these lines do not possess very high level of resistance against highly virulent Brazilian ASR isolates, these resistance genes may not be solely useful for ASR resistance in South America. However, pyramiding these genes with other available ASR resistance gene(s) in a commercial soybean cultivar may be proved useful in providing durable and enhanced resistance to strong and diverse P. pachyrhizi population in South America.
References
Akamatsu H, Yamanaka N, Yamaoka Y, Soares RM, Morel W, Ivancovich AJG, Bogado AN, Kato M, Yorinori JT, Suenaga K (2013) Pathogenic diversity of soybean rust in Argentina, Brazil, and Paraguay. J Gen Plant Pathol 79:28–40. doi:10.1007/s10327-012-0421-7
Benitez ER, Yamaoka Y, Takahashi R, Takahashi K, Paiva WM, U SP, Dow MTH (2008) Evaluation of selected Japanese varieties for resistance to Phakopsora pachyrhizi. Breed Res 10 (Supplement 1):70
Brogin RL (2005) Mapeamento de genes de resistência à ferrugem e de QTLs envolvidos na resistência à septoriose em soja. Dissertation, Universidade de São Paulo
Chakraborty N, Curley J, Frederick R, Hyten D, Nelson R, Hartman G, Diers B (2009) Mapping and confirmation of a new allele at Rpp1 from soybean PI 594538A conferring RB lesion-type resistance to soybean rust. Crop Sci 49:783–790. doi:10.2135/cropsci2008.06.0335
Garcia A, Calvo ES, Kiihl RAS, Harada A, Hiromoto DM, Vieira LGE (2008) Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistance genes: discovery of a novel locus and alleles. Theor Appl Genet 117:545–553. doi:10.1007/s00122-008-0798-z
Garcia A, Calvo ES, Kiihl RAS, Souto ER (2011) Evidence of a susceptible allele inverting the dominance of rust resistance in soybean. Crop Sci 51:32–40. doi:10.2135/cropsci2010.01.0037
Goellner K, Loehrer M, Langenbach C, Conrath U, Koch E, Schaffrath U (2010) Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol Plant Pathol 11:169–177. doi:10.1111/J.1364-3703.2009.00589.X
Grant D, Nelson RT, Cannon SB, Shoemaker RC (2010) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 38:D843–D846. doi:10.1093/nar/gkp798
Hartwig EE, Bromfield KR (1983) Relationships among three genes conferring specific resistance to rust in soybeans. Crop Sci 23:237–239. doi:10.2135/cropsci1983.0011183X002300020012x
Hossain MM, Akamatsu H, Morishita M, Mori T, Yamaoka Y, Suenaga K, Soares RM, Bogado AN, Ivancovich AJG, Yamanaka N (2014) Molecular mapping of Asian soybean rust resistance in soybean landraces PI 594767A, PI 587905 and PI 416764. Plant Pathol. doi:10.1111/ppa.12226
Hyten DL, Hartman GL, Nelson RL, Frederick RD, Concibido VC, Narvel JM, Gregan PB (2007) Map location of the Rpp1 locus that confers resistance to soybean rust in soybean. Crop Sci 47:835–838. doi:10.2135/cropsci2006.07.0484
Hyten DL, Smith JR, Frederick RD, Tucker ML, Song Q, Cregan PB (2009) Bulked segregant analysis using the GoldenGate assay to locate the Rpp3 locus that confers resistance to soybean rust in soybean. Crop Sci 49:265–271. doi:10.2135/cropsci2008.08.0511
Jeong SC, Hayes AJ, Biyashev RM, Maroof MS (2001) Diversity and evolution of a non-TIR-NBS sequence family that clusters to a chromosomal “hotspot” for disease resistance genes in soybean. Theor Appl Genet 103:406–414. doi:10.1007/s001220100567
Kendrick MD, Harris DK, Ha BK, Hyten DL, Cregan PB, Frederick RD, Boerma HR, Pedley KF (2011) Identification of a second Asian soybean rust resistance gene in Hyuuga soybean. Phytopathology 101:535–543. doi:10.1094/PHYTO-09-10-0257
Kim KS, Unfried JR, Hyten DL, Frederick RD, Hartman GL, Nelson RL, Song Q, Diers BW (2012) Molecular mapping of soybean rust resistance in soybean accession PI 561356 and SNP haplotype analysis of the Rpp1 region in diverse germplasm. Theor Appl Genet 125:1339–1352. doi:10.1007/s00122-012-1932-5
Lander E, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 11:174–181
Laperuta LDC, Arias CAA, Ribeiro AS, Rachid BF, Pierozzi PHB, Toledo JFFD, Pípolo AE, Carneiro GEDS (2008) New genes conferring resistance to Asian soybean rust: allelic testing for the Rpp2 and Rpp4 loci. Pesqui Agropecu Bras 43:1741–1747
Lemos NG, Braccini AL, Abdelnoor RV, Oliveira MCN, Suenaga K, Yamanaka N (2011) Characterization of genes Rpp2, Rpp4, and Rpp5 for resistance to soybean rust. Euphytica 182:53–64. doi:10.1007/s10681-011-0465-3
Levy C (2005) Epidemiology and chemical control of soybean rust in southern Africa. Plant Dis 89:669–674. doi:10.1094/PD-89-0669
Li S, Smith JR, Ray JD, Frederick RD (2012) Identification of a new soybean rust resistance gene in PI 567102B. Theor Appl Genet 125:133–142. doi:10.1007/s00122-012-1821-y
Miles MR, Frederick RD, Hartman GL (2006) Evaluation of soybean germplasm for resistance to Phakopsora pachyrhizi. Plant Health Prog. doi:10.1094/PHP-2006-0104-01-RS
Monteros M, Missaoui A, Phillips D, Walker D, Boerma H (2007) Mapping and confirmation of the ‘Hyuuga’ red-brown lesion resistance gene for Asian soybean rust. Crop Sci 47:829–836. doi:10.2135/cropsci06.07.0462
Oliveira ACB, Godoy CV, Martins MC (2005) Avaliação da tolerância de cultivares de soja à ferrugem asiática no Oeste da Bahia. Fitopatol Bras 30:658–662
Ray J, Morel W, Smith J, Frederik R, Miles M (2009) Genetics and mapping of adult plant rust resistance in soybean PI 587886 and PI 587880A. Theor Appl Genet 119:271–280. doi:10.1007/s00122-009-1036-z
Ray JD, Smith JR, Morel W, Bogado N, Walker DR (2011) Genetic resistance to soybean rust in PI 567099A in at or near the Rpp3 locus. J Crop Improv 25:219–231. doi:10.1080/15427528.2011.555833
Ribeiro AS, Moreira JUV, Pierozzi PHB, Rachid BF, De Toledo JFF, Arias CAA, Soares RM, Godoy CV (2007) Genetic control of Asian rust in soybean. Euphytica 15:15–25. doi:10.1007/s10681-007-9404-8
Schneider R, Sikora E, Padgett B, Sciumbato G (2008) Managing late-season soybean diseases and soybean rust: a southern perspective. In: Dorrance AE, Draper MA, Hershman DE (eds) Using of foliar fungicides to manage soybean rust. Ohio State University, Columbus, pp 72–75
Silva DCG, Yamanaka N, Brogin RL, Arias CAA, Nepomuceno AL, Di Mauro AO, Pereira SS, Nogueira LM, Passianotto ALL, Abdelnoor RV (2008) Molecular mapping of two loci that confer resistance to Asian rust in soybean. Theor Appl Genet 117:57–63. doi:10.1007/s00122-008-0752-0
Song Q, Marek L, Shoemaker R, Lark K, Concibido V, Delannay X, Specht J, Cregan P et al (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128. doi:10.1007/s00122-004-1602-3
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Accessed 15 March 2013
Yamanaka N, Silva DCG, Passianotto ALL, Nogueira LM, Polizel AM, Pereira SS, Santos JVM, Brogin RL, Arias CAA, Hoffmann-Campo CB, Nepomuceno AL, Abdelnoor RV (2008) Identification of DNA markers and characterization of the genes for resistance against Asian soybean rust. In: Kudo H, Suenaga K, Soares RM, Toledo A (eds) JIRCAS Working Report No. 58: Facing the challenge of soybean rust in South America. JIRCAS, Tsukuba, pp 99–107
Yamanaka N, Yamaoka Y, Kato M, Lemos NG, Passianotto ALL, Santos JVM, Benitez ER, Abdelnoor RV, Soares RM, Suenaga K (2010) Development of classification criteria for resistance to soybean rust and differences in virulence among Japanese and Brazilian rust populations. Trop Plant Pathol 35:153–162
Yamanaka N, Lemos NG, Akamatsu H, Yamaoka Y, Silva DCG, Passianotto ALL, Abdelnoor RV, Soares RM, Suenaga K (2011) Soybean breeding materials useful for resistance to soybean rust in Brazil. JARQ 45:385–395
Yamanaka N, Akamatsu H, Yamaoka (2013a) Laboratory manual for studies on soybean rust resistance. JIRCAS website. http://www.jircas.affrc.go.jp/english/manual/soybean_rust/soybean_rust.html. Accessed 1 Dec 2013
Yamanaka N, Lemos NG, Uno M, Akamatsu H, Yamaoka Y, Abdelnoor RV, Braccini AL, Suenaga K (2013b) Resistance to Asian soybean rust in soybean lines with the pyramided three Rpp genes. Crop Breed Appl Biotechnol 13:75–82
Yamaoka Y, Yamanaka N, Akamatsu H, Suenaga K (2014) Pathogenic races of soybean rust Phakopsora pachyrhizi collected in Tsukuba and vicinity in Ibaraki, Japan. J Gen Plant Pathol 80:184–188. doi:10.1007/s10327-014-0507-5
Yorinori JT (2008) Soybean germplasms with resistance and tolerance to Asian rust and screening methods. In: Kudo H, Suenaga K, Soares RM, Toledo A (eds) JIRCAS Working Report No. 58: Facing the challenge of soybean rust in South America. JIRCAS, Tsukuba, pp 70–87
Acknowledgments
We thank M. Morishita, T. Mori, and M. Hasegawa (JIRCAS) for their technical assistance, advice, and encouragement. We are grateful to the Brazilian Agricultural Research Corporation (Embrapa) in Brazil, the Jilin Academy of Agricultural Sciences (JAAS) in China, the National Institute of Agrobiological Science (NIAS) and the National Institute of Crop Science (NICS) in Japan for providing seeds of soybean varieties. This study was financially supported and conducted by the JIRCAS research Project “Development of Breeding Technologies toward Improved Production and Stable Supply of Upland Crops.” The author, M. M. H. was financially supported by JIRCAS Visiting Research Fellowship Program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yamanaka, N., Hossain, M.M. & Yamaoka, Y. Molecular mapping of Asian soybean rust resistance in Chinese and Japanese soybean lines, Xiao Jing Huang, Himeshirazu, and Iyodaizu B. Euphytica 205, 311–324 (2015). https://doi.org/10.1007/s10681-015-1377-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1377-4