Abstract
Downy mildew (DM) is a plant disease that strongly limits maize production. The resistance (R) genes for this disease have been identified and characterized and serve as important tools for plant disease evaluation. In this study, partial genes for PIC15, PO145579, and zmcf5 were identified and analyzed in a panel of 60 public and private maize inbred lines using bioinformatics and statistics tools. These candidate R genes exhibited an average nucleotide diversity value of 0.015. The negative Tajima’s D values for these genes suggested that purifying selection has played a role in the evolution of maize DM resistance genes. In addition, linkage disequilibrium among these genes occurred across an extent of ~ 200 bp. Using association analysis methods, a general linear model, and a mixed linear model, five polymorphisms within the partial PIC15 and PO145579 genes were detected and associated with DM disease. These are new polymorphisms that have not previously been reported in association with DM disease traits. Therefore, correlations between these new polymorphisms and DM disease in these germplasm collections could advance the development of a functional marker for marker-assisted selection of DM disease resistance in maize breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Downy mildew (DM), which is caused by several species in the genera Peronosclerospora, Sclerospora, and Sclerophthora, is one of the most destructive maize diseases in subtropical and tropical regions of the world, including America, Asia, Africa, Europe, and Australia. In Asia, the main causal agent is Peronosclerospora sorghi (Weston and Uppal) C.G. Shaw (Sriwatanapongse et al. 1993; Jeffers et al. 2000; Raymundo 2000; George et al. 2003).
In tropical and subtropical lowland Asia, maize-growing areas report economic losses occasionally as high as 75 % (Exconde and Raymundo 1974) due to DM (Jeffers et al. 2000). While genetic resistance is a cost-effective and environmentally safe method for controlling DM, despite the use of DM resistant cultivars and metalaxyl fungicide as a seed treatment for controlling DM, the incidence of the disease is still severe in localized areas (Dalmacio 2000). New sources of locally adapted DM-resistant lines may alleviate this problem.
Linkage mapping of DM resistance in studies of the genetic basis of DM resistance have been complicated by the polygenic nature of the trait and by the fact that additive effects contribute to resistance (Kaneko and Aday 1980; Singburandom and Renfro 1982; Borges 1987; De Leon et al. 1993). So far, linkage mapping has been the tool of choice for the identification of quantitative trait loci (QTL) that confer resistance to maize DM (Agrama et al. 1999; George et al. 2003; Nair et al. 2005; Sabry et al. 2006; Jampatong et al. 2008). Consequently, QTL from various genomic regions on chromosomes 1, 2, 6, 7, and 10 have been found to confer resistance to DM (George et al. 2003). Despite these discoveries, the QTL approach does have some limitations, including high costs and poor resolution in defining QTL. Furthermore, when using bi-parental crosses between inbred lines, only two alleles at any given locus can be studied at one time by this method.
Recently, sets of cultivars, lines, or landraces have been used to identify marker-trait associations in plants. This method uses linkage disequilibrium (LD) between DNA polymorphisms and genes underlying agronomic traits of interest (Thornsberry et al. 2001; Flint-Garcia et al. 2005; Yu et al. 2006; Buckler and Gore 2007; Zhu et al. 2008) to identify useful markers. The use of association mapping or LD mapping has been broadened to plant studies, and many QTL have recently been identified and confirmed by this approach (Parisseaux and Bernardo 2004; Breseghello and Sorrells 2006; Stich et al. 2006; Agrama et al. 2007; Tommasini et al. 2007; Christopher et al. 2007; Skot et al. 2007; Crossa et al. 2007; Casa et al. 2008; Maccaferri et al. 2011). Gene mapping through association has several advantages in traditional biparental populations because it can precisely pinpoint the genomic region responsible for the expression of the target trait, and has the potential to allow evaluation of a large number of alleles per locus (Buckler and Thornsberry 2002; Flint-Garcia et al. 2003, 2005). Gene mapping through association appears to be a promising approach to overcome some of the previous limitations of conventional linkage mapping in plant breeding (Stich et al. 2005; Yu and Buckler 2006).
R genes confer resistance to pathogens that express matching avirulence genes in a gene-for-gene manner (Flor 1956, 1971). The largest class of known R proteins includes those encoding nucleotide-binding site–leucine-rich repeat (NBS–LRR) proteins and receptor-like kinase enzymes. The major classes of R genes contain a highly conserved NBS domain adjacent to the N terminus and a LRR domain involved in host recognition of pathogen-derived elicitors. Responses to fungal, viral, and bacterial pathogen infections are mediated by genes encoding receptor proteins (Dangl 1995). Association mapping has been used to identify disease resistance genes in several crop species including sugarcane, maize, barley, and potato (Meyers 2003; Flint-Garcia et al. 2005; Yu and Buckler 2006; Wei et al. 2006; Malosetti et al. 2007; Stich et al. 2008; Murray et al. 2009; Inostroza et al. 2009). Although genome scanning is also useful for identifying markers for DM resistance, Phumichai et al. (2012) previously conducted an association mapping study of this trait. In the present study, candidate R genes were used to assess the extent of LD in the target population and to identify single nucleotide polymorphisms (SNPs) that significantly affect DM resistance.
Materials and methods
Plant material and phenotypic evaluation
A panel of this study consisted of 60 maize inbred lines (Table S1) supplied by two public-sector institutions and two private companies in Thailand. The National Corn and Sorghum Research Center (NCSRC-IICRD KU; Suwan Farm) and Nakhon Sawan Field Crop Research Center (NFR) supplied 17 and 15 inbred lines of field corn, respectively. Bangkok Seeds Industry and Sweet Seeds Company provided 15 inbred lines (7 field corn, 4 sweet corn, 2 waxy corn, 2 popcorn), and 13 inbred lines (11 sweet corn, 2 waxy corn), respectively. Although our sample was restricted to 60 inbred lines the average genetic diversity and number of alleles (0.7 and 10.1, respectively) was previously reported by Phumichai et al. 2012.
Artificial inoculation of DM in the field was described in detail in Phumichai et al. (2012) and below (S2). Field experiments were conducted at two locations: Nakhon Sawan Field Crop Research Center (NFR) (15°20′45″N, 100°29′4″E) and the National Corn and Sorghum Research Center, Inseechandrastitya Institute for Crop Research and Development (NCSRC-IICRD KU) (14°24′42″N, 101°25′18″E), Thailand. All maize inbred lines were laid out in a randomized complete block design with three replicates during the 2008 rainy season (May–July). Two-row plots, 5 m in length with 0.75 m row spacing, containing 42 plants per plot were planted using a hand jab planter. Methods for evaluation of disease resistance were described in detail in Phumichai et al. (2012) (S2).
DNA isolation and candidate genes sequencing
Genomic DNA was isolated from fresh young maize leaves using the modified cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle 1990). PCR primers were designed for the three candidate genes based on resistance genes previously identified on maize chromosomes (Table 1). PCR was carried out using 0.5 U/μl Pfu Taq polymerase (Fermentas), 10× PCR buffer (200 mM Tris–HCl pH 8.8, 100 mM (NH4)2SO4, 100 mM KCl, 1 % Triton X-100, 1 mg/ml BSA), 25 mM MgSO4, 1 mM dNTP, 10 μM each of the forward and reverse primers, and 20 ng template DNA. PCR cycles were conducted as follows: 1 cycle of 94 °C for 2 min; then 30 cycles of 94 °C for 30 s, 30 s at annealing temperature specific to each primer pair, and 72 °C for 1 min; followed by 1 cycle of 72 °C for 5 min on a PTC-225 Peltier Thermal Cycler (MJ Research, St. Bruno, Canada). The PCR products were separated by electrophoresis on 1 % (w/v) agarose gels in 1× TAE buffer at 50 V for 30 min, stained with ethidium bromide for 30 min, and visualized under UV light. PCR products were directly purified and sequenced on the ABI 3730XL DNA Analyzers at Macro Gen Company (Seoul, South Korea).
Candidate genes and LD analysis
Candidate R gene loci were chosen as the loci most closely linked to the three SSR loci reported in a previous DM disease association study (Phumichai et al. 2012). Three partial R genes from the chosen candidate genes, PIC15, PCO145579, and zmcf5, located on chromosomes 1, 2, and 9, respectively, were analyzed in this study (Table 1). Sequence analyses were performed using BLAST at the Maize Genetics and Genomics Database (MaizeGDB; http://blast.maizegdb.org/home.php?a=BLAST_UI) and BLASTN (Altschul et al. 1997) at the National Center for Biotechnology Information (NCBI; http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch). The partial sequences used here were assembled using MEGA version 5 software (Tamura et al. 2011). Assembled sequences were then aligned using ClustalW (Chenna et al. 2003). Sequences were manually checked using Sequence Scanner Software v 1.0 (Applied Biosystems 2005). For these three candidate genes, the number of polymorphic sites (S), haplotypes, haplotype diversity, pairwise nucleotide diversity (π), nucleotide polymorphism (θ) and Tajima’s D were computed using DnaSP version 5.1 (Librado and Rozas 2009). LD between pairs of polymorphic sites in each of these three genes was estimated by TASSEL stand-alone software version 3.1 (Bradbury et al. 2007). LD was estimated using squared allele-frequency correlations (r 2). The significance of LD was tested using Fisher’s exact test.
Association analysis and narrow-sense heritability
Analyses of associations between nucleotide polymorphisms and phenotypic values for DM resistance were performed using the general linear model (GLM) and mixed linear model (MLM) functions in TASSEL stand-alone version 3.1 (Bradbury et al. 2007). Principal component analysis (PCA) was used to replace the population structure (Q) (Price et al. 2006; Yu et al. 2006; Zhao et al. 2007; Zhang et al. 2009) and was generated by TASSEL stand-alone version 3.1 (Bradbury et al. 2007). PCA can be used to infer population structure, and the ideal number of principle components can be identified as the component number close to an elbow in the curve of a scree plot between eigenvalues on the y axis and the number of components on the x axis (Linting et al. 2007). An elbow should occur near the last component that contributes significant variation to the trait being analyzed. The kinship matrix for all 60 maize inbred lines was also calculated using polymorphic sites with kinship analysis in TASSEL stand-alone version 3.1 (Bradbury et al. 2007). Type I error was controlled by applying the 1,000 permutation test in TASSEL stand-alone version 3.1 (Bradbury et al. 2007) to control error associated with multiple comparisons and generate strong p values for association analysis of polymorphic sites. The statistical power calculation of the study was calculated using the GWAPower statistical program designed for genome wide association (GWA) studies with quantitative traits where genetic effect was defined as heritability (Feng et al. 2011).
Marker-based narrow-sense heritability (h 2) is defined as the proportion of genetic variance over the total variance, using the restricted maximum likelihood (REML) estimates of V a and V e obtained using TASSEL stand-alone version 3.1 (Bradbury et al. 2007) as: h 2 = V a/V a + V e, where V a is the genetic variance and V e is the residual variance.
Results
Nucleotide diversity
The resulting alignments of PIC15, PO1455579 and zmcf5, from a set of 60 maize accessions, were 587, 712, and 428 bp—in length, respectively (Table 2). Considering all three genes in this study, the average number of polymorphic sites (S) was 43.7, average haplotype diversity was 0.738, and average nucleotide diversity was 0.015. In addition, neutral selection was evaluated using Tajima’s D test with DnaSP version 5.1 software. Tajima’s D test results for these three genes revealed negative values for selection that were significant for the partial zmcf5 gene (Table 2).
Linkage disequilibrium
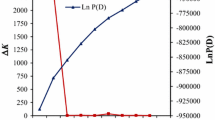
Linkage disequilibrium was estimated between all pairs of polymorphic sites in the partial sequences of the PIC15, PO145579, and zmcf5 genes using TASSEL stand-alone 3.1. Average r 2 values for PIC15, PO145579, and zmcf5 were 0.28, 0.29, and 0.05, respectively. The PIC15 sequence displayed two blocks of polymorphism (from position 238 to 279, and from position 418 to 455) and showed strongly significant LD estimates with r 2 greater than 0.8 (p < 0.0001). The PO14557 sequence displayed three blocks of polymorphism (from position 79 to 148, from position 312 to 369, and from position 383 to 608) with strongly significant r 2 estimates of LD of greater than 0.7 (p < 0.0001). In contrast, only slightly significant LD was observed between a few polymorphic sites in the zmcf5 partial sequence (r 2 ≤ 0.3, p < 0.01) (Fig. 1).
Association analysis with candidate genes and narrow-sense heritability
Analysis of association between these three candidate R genes and phenotypic traits was performed using GLM incorporating PCA, and MLM incorporating both PCA and relative kinship (K) (PCA+K) in TASSEL standalone 3.1. PCA of polymorphic sites from these three candidate genes in 60 maize inbred lines revealed that the top three axes explained 55.6 % of the variation for these three R genes in these populations. PCA was used to remove population effects such as geographic origin or diversifying selection (Price et al. 2006) for analysis of associations between genotypic (PIC15, PO145579, and zmcf5) and phenotypic traits related to DM resistance. Significant association between polymorphisms in the partial PIC15 and PO145579 genes and DM resistance was found, while no such association was identified for zmcf5 (Table 3).
One SNP in the partial PIC15 gene was significantly associated with DM resistance at both the NCSRC-IICRD KU and NFR experimental locations, with r 2 value of 16.4 and 7 %, respectively (Table 3). The SNP site in exon 267 of the PIC15 partial gene sequence encoded a nucleotide transition from A to G. The phenotypic contribution for the A allele was detected in 36 maize inbred lines, of which 21 (58 %) showed resistance to DM. Another SNP detected in the PO145579 partial gene sequence was associated with DM at the NCSRC-IICRD KU experimental location with r 2 value of 7 %, while three more SNPs showed association with DM at the NFR location with r 2 value of 6 % (Table 3). The haplotype (TTGT) characteristic of these SNPs in the partial PO145579 gene contributed 64 % of phenotypic variation for DM resistance. Genetic power was calculated with a GWAPower program (Feng et al. 2011) by using broad sense heritability (Phumichai et al. 2012). The simulation demonstrated that the current sample size (n = 60) has 97.7 % of genetic power to achieve in detecting the studied SNPs. Marker-based narrow-sense heritability (h 2) for these 60 maize inbred lines was calculated using the formula described above for each experimental location by dividing genetic variance by total phenotypic variance obtained from the MLM incorporating PCA and K. Narrow-sense heritabilities were 33 and 29 % at the NCSRC-IICRD KU and NFR experimental locations, respectively (Table 3).
Discussion
The average nucleotide diversity of these genes was 0.015, close to the nucleotide diversity of 0.018 for the non-synonymous maize hm1 disease resistance gene (Zhang et al. 2002). Tajima’s D values were negative and were significant only for zmcf5 (Table 2). The negative value for Tajima’s D in our study, particularly the significant negative value for zmcf5, may be due to purifying selection, which results in a few alleles predominating but most other alleles occurring at low frequencies. Examples of purifying selection have been detected for several plant NBS disease resistance domains (McHale et al. 2006). In the case of lettuce, the Type I RGC2 genes were identified as having undergone diversifying selection, while Type II RGC2 genes were identified as having undergone purifying selection (Kuang et al. 2004). In addition, population studies have shown that balancing selection maintains polymorphism at R gene loci (Meyers et al. 2005). Therefore, balancing, diversifying, and purifying selection may all play roles in the evolution of a particular R gene cluster.
Using mean r 2 = 0.2 as a cutoff point for estimation, the extent of LD for all three of these candidate R genes was ~200 bp in these 60 public and private maize inbred lines (S3). The LD estimator r 2 ranged from 0.05 to 0.29. A low level of LD was observed in the partial sequence of the zmcf5 gene (r 2 = 0.05). Many factors, such as the origin of populations, the choice of populations for analysis, the particular genomic region analyzed, high rates of recombination or mutation, and subdivision of populations, can affect LD. In maize, genome-wide LD decay values have been shown to be in the range of ~200–1500 bp (Remington et al. 2001; Tenaillon et al. 2001). Su et al. (2010) reported that LD of the naked and rab28 drought tolerance genes in maize varied by more than 700 bp. The maize disease resistance gene, glutathione S transferase (GST), exhibited rapid decay of LD in the range of 1 or 2 kb (Wisser et al. 2011) when analyzed in 253 maize inbred lines. Thus, the level of variability in LD in the three candidate R genes in this study could be due to the germplasm origins of these maize inbred lines or to low selection pressure on these genes during the breeding histories of these lines. In addition, the limited partial gene fragments analyzed here might not sufficiently describe the overall patterns of LD in these candidate R genes. Therefore, full-length candidate R genes should be analyzed to better describe patterns of LD among maize DM resistance genes.
Most plant disease resistance genes encode proteins containing NBS and LRR domains, which have been identified in several plant species (McHale et al. 2006). The NBS–LRR domains are involved in the detection of diverse plant pathogens, including bacteria, viruses, fungi, nematodes, insects, and oomycetes (McHale et al. 2006).
Association analysis of partial sequences of three candidate R genes has detected five SNPs associated with DM disease resistance in the PIC15 (maize chromosome 1) and PO145579 (maize chromosome 2) genes. The r 2 values in these SNPs showed ranging from 6 to 16.4 %. The highest r 2 value was found in PIC15 gene with DM resistance at the NCSRC-IICRD KU (16.4 %), while, most revealed minor r 2 was detected in PO145579 gene. The A allele SNP from the PIC15 gene contributes 58 % of the phenotypic variation in DM resistance, while the TTGT haplotype from the PO145579 gene contributes 64 % of the phenotypic variation in DM resistance. Phenotypic contribution information is typically reported in association analysis to estimate the efficiency of detecting associations between traits and particular SNPs (Thornsberry et al. 2001). In soybean, a SNP in the F3H gene appeared in nine of 12 accessions (75 %) that were susceptible to mosaic virus strain SC-7 (Cheng et al. 2010). The lettuce dieback resistance gene, Cntg10192, contains three SNPs, two of which associate perfectly with the resistance allele, while the third SNP explains 40.9 % of the variation in the resistance trait, which was due to variation in association among lines of different origin (Simko et al. 2009).
Broad-sense heritability was previously estimated as the genetic component from ANOVA (H 2 = 97 %) in Phumichai et al. (2012). Marker-based narrow-sense heritability of h 2 = 29–33 % for DM resistance in this study was expected, dominance or epistasis may have caused discrepancies among variance components. In this study, low narrow-sense heritability (29–33 %) may be due to the analysis of few candidate R gene sequences of limited length. Preliminary studies of the inheritance of DM resistance in corn by Gomes et al. (1963) showed that a few partially dominant genes controlled resistance. However, genetic analysis of DM resistance using composite interval mapping (Jampatong et al. 2008) successfully detected QTL with additive and dominant genetic effects, including partial dominance, dominance, and overdominance.
In this study, the significant SNP in the PIC15 partial gene sequence could be detected using either GLM with PCA or MLM with PCA+K. In addition, the A allele of PIC15 and TTGT haplotype of PO145579 also showed strongly significant LD estimates, with r 2 greater than 0.8 (p < 0.0001) and 0.7 (p < 0.0001), respectively. Yu et al. (2006), reported that the Q+K model controls the number of false associations better than other tests using either the K- or Q- models alone. These results indicate that these five SNPs from chromosomes 1 (PIC15) and 2 (PO145579) will be good candidate SNPs to evaluate for use as functional molecular markers for DM resistance in breeding programs.
There were two limitations in the panel used for the GWA. First, the number of inbred lines was small, which 60 could weaken the power of the association and could result in some loci being missed, especially those with small effects. Yang et al. (2010) reported that a panel of 155 inbred lines could obtain 59.2 and 87.6 % of the quantitative genes explaining 5 and 10 % of phenotypic variation, respectively. Secondly, small population size may introduce high frequency of spurious associations when estimating population structure and familial relatedness compared to larger populations and inadequate to identify QTL with low minor allele frequencies (Wang et al. 2012). Bradbury et al. (2011) showed that population sizes of 300 were sufficient to detect QTL for traits with moderate to high heritability (0.75–1.0); however as the number of QTL increased, the larger effect QTL were detected but not the smaller effect QTL. Genetic power was calculated with a GWAPower program (Feng et al. 2011). When the genetic power simulated with the low (0.1) and high (0.97) heritability with the same sample size (n = 60), it is represented with 28.4 and 97.7 % of power to detect the studied SNPs, respectively. Therefore, keeping the same power as well as high heritability (0.97) simulated, the increasing sample size needed is 277 when low heritability simulated. Alternatively, considering combined association analysis with bi-parental linkage analysis to identify the true genetic variants for these traits could help to as the covariance between genotypes and phenotypes can be broken up by generating controlled cross (Yang et al. 2010). In addition, increasing the population size of germplasm panel, consideration of population structure for reducing frequency of spurious associations, may enhance this panel useful in identifying the genetic factors associated with many traits (Yang et al. 2010).
As the small population association analysis in this study could detect only major allele with high heritability traits. In addition, the limitations of association mapping may suffer from a false positives caused by population structure within a germplasm (Stich and Melchinger 2009). Spurious association that found in population could be corrected by using statistical models accounting for population structure (Q matrix) and kinship matrix (K) to reduce the false positive associations (Yu et al. 2006; Price et al. 2006). The results of Zhang et al. (2013) comparison 16 different models for reduction of false-positive associations, the performance of MLM (PCA+K) model was indicated that the best reduction in false-positives frequency in the maize panel.
Therefore, the results of this study provide not only basic genetic information useful for the development of functional molecular markers, but also contribute important information for further research to validate the polymorphic sites by bi-parental mapping and increasing maize populations size to enhance the power of future association analyses.
References
Agrama HA, Moussa ME, Naser ME, Tarek MA, Ibrahim AH (1999) Mapping of QTL for downy mildew resistance in maize. Theor Appl Genet 99:519–523
Agrama HA, Eizenga GC, Yan W (2007) Association mapping of yield and its components in rice cultivars. Mol Breed 19:341–356
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Applied Biosystems (2005) Sequence scanner software version 1.0. Applied Biosystems, Carlsbad
Borges OL (1987) Diallel analysis of maize resistance to sorghum downy mildew. Crop Sci 27:178–181
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Bradbury P, Parker T, Hamblin MT, Jannink JL (2011) Assessment of power and false discovery in genome-wide association studies using the BarleyCAP germplasm. Crop Sci 51:52–59
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177
Buckler ES, Gore M (2007) An Arabidopsis haplotype map takes root. Nat Genet 39:1056–1057
Buckler ES, Thornsberry JM (2002) Plant molecular diversity and applications to genomics. Curr Opin Plant Biol 5:107–111
Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in Sorghum. Crop Sci 48:30–40
Cheng H, Yang H, Zhang D, Gai J, Yu D (2010) Polymorphisms of soybean isoflavone synthase and flavanone 3-hydroxylase genes are associated with soybean mosaic virus resistance. Mol Breed 25:13–24
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res 31:3497–3500
Christopher M, Mace E, Jordan D, Rodgers D, McGowan P, Delacy I, Banks P, Sheppard J, Butler D, Poulsen D (2007) Applications of pedigree-based genome mapping in wheat and barley breeding programs. Euphytica 154:307–316
Crossa J, Burgueno J, Dreisigacker S, Vargas M, Herrera-Foessel SA, Lillemo M, Singh RP, Trethowan R, Warburton M, Franco J, Reynolds M, Crouch JH, Ortiz R (2007) Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177:1889–1913
Dalmacio S (2000) Importance of and growing concern for maize diseases in the Asian region. In: Vasal SK, Gonzalez CF, Fan XM (eds) Proc 7th Asian regional maize workshop. PCARRD, Los Baños, pp 267–276
Dangl JL (1995) Pièce de résistance: novel classes of plant disease resistance genes. Cell 80:363–366
De Leon C, Ahuja VP, Capio ER, Mukherjee BK (1993) Genetics of resistance to Philippine downy mildew in three maize populations. Indian J Genet 4:406–410
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Exconde OR, Raymundo AD (1974) Yield loss caused by Philippine corn downy mildew. Philipp Agric 58:115–120
Feng S, Wang S, Chen CC, Lan L (2011) GWAPower: a statistical power calculation software for genome-wide association studies with quantitative traits. BMC Genet 12:12
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES (2005) Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J 44:1054–1064
Flor HH (1956) The complementary genic systems in flax and flax rust. Adv Genet 8:29–54
Flor HH (1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9:275–296
George ML, Prasanna BM, Rathore RS, Setty TA, Kasim F, Azrai M, Vasal S, Balla O, Hautea D, Canama A, Regalado E, Vargas M, Khairallah M, Jeffers D, Hoisington D (2003) Identification of QTLs conferring resistance to downy mildews of maize in Asia. Theor Appl Genet 107:544–551
Gomez AA, Aquilazan FA, Paison RM, Calub AG (1963) Preliminary studies on the inheritance of the reaction of corn to downy mildew disease. Philipp Agric 47:113–116
Inostroza L, del Pozo A, Matus I, Castillo D, Hayes P, Machado S, Corey A (2009) Association mapping of plant height, yield, and yield stability in recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp. spontaneum as a source of donor alleles in a Hordeum vulgare subsp. vulgare background. Mol Breed 23:365–376
Jampatong C, Jampatong S, Balla C, Grudloyma P, Jompuk C, Prodmatee N (2008) QTL mapping for downy mildew (Peronosclerospora sorghi) resistance in maize. Paper presented at the Maize for Asia: emerging trends and technologies of the 10th Asian regional maize workshop, Makassar, pp 291–298
Jeffers D, Cordova H, Vasal S, Srinivasan G, Beck D, Barandiaran M (2000) Status in breeding for resistance to maize diseases at CIMMYT. In: Vasal SK, Gonzalez CF, Fan XM (eds) Proc 7th Asian regional maize workshop. PCARRD, Los Baños, pp 257–266
Kaneko K, Aday BA (1980) Inheritance of resistance to Philippine downy mildew of maize, Perenosclerospora philippinensis. Crop Sci 20:590–594
Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16:2870–2894
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Linting M, Meulman JJ, Groenen PJF, Van der Kooij AJ (2007) Nonlinear principal components analysis: introduction and application. Psychol Methods 12:336–358
Maccaferri M, Sanguineti MC, Demontis A, El-Ahmed A, Garcia del Moral L, Maalouf F, Nachit M, Nserallah N, Ouabbou H, Rhouma S, Royo C, Villegas D, Tuberosa R (2011) Association mapping in durum wheat grown across a broad range of water regimes. J Exp Bot 62:409–438
Malosetti M, van der Linden CG, Vosman B, van Eeuwijk FA (2007) A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 175:879–889
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS–LRR proteins: adaptable guards. Genome Biol 7:212
Meyers BC (2003) Genome-wide analysis of NBS–LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Meyers BC, Kaushik S, Nandety RS (2005) Evolving disease resistance genes. Curr Opin Plant Biol 8:129–134
Murray SC, Rooney WL, Hamblin MT, Mitchell SE, Kresovich S (2009) Sweet Sorghum genetic diversity and association mapping for brix and height. Plant Genome J 2:48–62
Nair SK, Prasanna BM, Garg A, Rathore RS, Setty TA, Singh NN (2005) Identification and validation of QTLs conferring resistance to sorghum downy mildew (Peronosclerospora sorghi) and Rajasthan downy mildew (P. heteropogoni) in maize. Theor Appl Genet 110:1384–1392
Parisseaux B, Bernardo R (2004) In silico mapping of quantitative trait loci in maize. Theor Appl Genet 109:508–514
Phumichai C, Chunwongse J, Jampatong S, Grudloyma P, Pulam T, Doungchan W, Wongkaew A, Kongsiri N (2012) Detection and integration of gene mapping of downy mildew resistance in maize inbred lines though linkage and association. Euphytica 187:369–379
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Raymundo AD (2000) Downy mildew of maize in Asia: new perspectives in resistance breeding. In: Vasal SK, Gonzalez CF, Fan XM (eds) Proc 7th Asian regional maize workshop. PCARRD, Los Baños, pp 277–284
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98:11479–11484
Sabry A, Jeffers D, Vasal SK, Frederiksen R, Magill C (2006) A region of maize chromosome 2 affects response to downy mildew pathogens. Theor Appl Genet 113:321–330
Simko I, Pechenick DA, McHale LK, Truco MJ, Ochoa OE, Michelmore RW, Scheffler BE (2009) Association mapping and marker-assisted selection of the lettuce dieback resistance gene Tvr1. BMC Plant Biol 9:135
Singburandom N, Renfro BL (1982) Heritability of resistance in maize to sorghum downy mildew (Peronosclerospora sorghi (Weston and Uppal) C.G. Shaw). Crop Prot 1:323–332
Skot L, Humphreys J, Humphreys MO, Thorogood D, Gallagher J, Sanderson R, Armstead IP, Thomas ID (2007) Association of candidate genes with flowering time and water-soluble carbohydrate content in Lolium perenne (L.). Genetics 177:535–547
Sriwatanapongse S, Jinahyon S, Vasal SK (1993) Suwan-1: maize from Thailand to the world. CIMMYT, Mexico
Stich B, Melchinger AE (2009) Comparison of mixed-model approaches for association mapping in rapeseed, potato, sugar beet, maize, and Arabidopsis. BMC Genom 10:94
Stich B, Melchinger AE, Frisch M, Maurer HP, Heckenberger M, Reif JC (2005) Linkage disequilibrium in European elite maize germplasm investigated with SSRs. Theor Appl Genet 111:723–730
Stich B, Melchinger AE, Piepho H-P, Heckenberger M, Maurer HP, Reif JC (2006) A new test for family-based association mapping with inbred lines from plant breeding programs. Theor Appl Genet 113:1121–1130
Stich B, Piepho HP, Schulz B, Melchinger AE (2008) Multi-trait association mapping in sugar beet (Beta vulgaris L.). Theor Appl Genet 117:947–954
Su Z, Li X, Hao Z, Xie C, Li M, Weng J, Zhang D, Liang X, Wang Z, Gao J, Zhang S (2010) Association analysis of the nced and rab28 genes with phenotypic traits under water stress in maize. Plant Mol Biol Rep 29:714–722
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98:9161–9166
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen BD, Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289
Tommasini L, Schnurbusch T, Fossati D, Mascher F, Keller B (2007) Association mapping of Stagonospora nodorum blotch resistance in modern European winter wheat varieties. Theor Appl Genet 115:697–708
Wang H, Smith KP, Combs E, Blake T, Horsley RD, Muehlbauer GJ (2012) Effect of population size and unbalanced data sets on QTL detection using genome-wide association mapping in barley breeding germplasm. Theor Appl Genet 124:111–124
Wei X, Jackson PA, McIntyre CL, Aitken KS, Croft B (2006) Associations between DNA markers and resistance to diseases in sugarcane and effects of population substructure. Theor Appl Genet 114:155–164
Wisser RJ, Kolkman JM, Patzoldt ME, Holland JB, Yu J, Krakowsky M, Nelson RJ, Balint-Kurti PJ (2011) Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis and implicates a GST gene. Proc Natl Acad Sci USA 108:7339–7344
Yang XH, Yan JB, Shah T, Warburton ML, Li Q, Li L, Gao YF, Chai YC, Fu ZY, Zhou Y, Xu ST, Bai GH, Meng YJ, Zheng YP, Li JS (2010) Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor Appl Genet 121:417–431
Yu J, Buckler ES (2006) Genetic association mapping and genome organization of maize. Curr Opin Biotechnol 17:155–160
Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhang L, Peek AS, Dunams D, Gaut BS (2002) Population genetics of duplicated disease-defense genes, hm1 and hm2, in maize (Zea mays ssp. mays L.) and its wild ancestor (Zea mays ssp. parviglumis). Genetics 162:851–860
Zhang Z, Buckler ES, Casstevens TM, Bradbury PJ (2009) Software engineering the mixed model for genome-wide association studies on large samples. Brief Bioinform 10:664–675
Zhang X, Tang B, Yu F, Li L, Wang M, Xue Y, Zhang Z, Yan J, Yue B, Zheng Y, Qiu F (2013) Identification of major QTL for waterlogging tolerance using genome-wide association and linkage mapping of maize seedlings. Plant Mol Biol Rep 31:594–606
Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M (2007) An Arabidopsis example of association mapping in structured samples. PLoS Genet 3:0071–0082
Zhu CS, Gore M, Buckler ES, Yu JM (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Acknowledgments
This study was supported by Generation Challenge Programme (GCP; G4007.04) and the National Centre for Genetic Engineering and Biotechnology (BIOTEC). Three anonymous reviewers are thanked for their valuable suggestions and comments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wongkaew, A., Phumichai, C., Chunwongse, J. et al. Detection of candidate R genes and single nucleotide polymorphisms for downy mildew resistance in maize inbred lines by association analysis. Euphytica 197, 109–118 (2014). https://doi.org/10.1007/s10681-013-1056-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-1056-2