Abstract
Flour color is an important trait in the assessment of flour quality for the production of many end products. In this study, quantitative trait loci (QTLs) with additive effects, epistatic effects, and QTL × environment (QE) interactions for flour color in bread wheat (Triticum aestivum L.) were studied, using a set of 168 doubled haploid (DH) lines derived from a Huapei 3 × Yumai 57 cross. A genetic map was constructed using 283 simple sequence repeats (SSR) and 22 expressed sequence tags (EST)-SSR markers. The DH and parents were evaluated for flour color in three environments. QTL analyses were performed using QTLNetwork 2.0 software based on a mixed linear model approach. A total of 18 additive QTLs and 24 pairs of epistatic QTLs were detected for flour color, which were distributed on 19 of the 21 chromosomes. One major QTL, qa1B, closely linked to barc372 0.1 cM, could account for 25.64% of the phenotypic variation of a* without any influence from the environments. So qa1B could be used in the molecular marker-assisted selection (MAS) in wheat breeding programs. The results showed that both additive and epistatic effects were important genetic basis for flour color, and were also sometimes subject to environmental modifications. The information obtained in this study should be useful for manipulating the QTLs for flour color by MAS in wheat breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) is one of the most important staple crops worldwide. Flour color is an important trait in the assessment of flour quality for the production of many end products (Parker and Langridge 2000). Flour with high levels of yellow pigmentation is preferred for Chinese and Japanese alkaline noodle production. In many Asian countries, noodles are prepared from flour specifically selected to enhance the color of the final product (Kruger et al. 1994). Miskelly (1984) detected a significant positive correlation between flour color and noodle sheet yellowness in both Chinese and Japanese noodles. However, other end products such as bread, steamed bread, dry white noodles, and dumplings require white flour with extremely low levels of yellow pigmentation. Color readings are usually expressed as L* (lightness), a* (red-green chromaticity), and b* (yellow-blue chromaticity) of the flour sample (CIE 1976). Theoretically, a pure white flour should have zero values for a* and b*, and one hundred for L* (Sun et al. 2002). Low values of L* with high positive values of a* result in a gray and dull color. Significant variations in flour color exist among genotypes of wheat. Moreover, environment and management practices may have impacts on flour color. Grain protein content, hardness, vitreousness, seed coat color, and kernel size and shape may all contribute to variations in flour color (Zhang and Tian 2008). The flour color from low-protein-content wheat is more white than the flour from high-protein-content wheat at the same milling extraction ratio. Flour from wheat with a red seed coat has higher a* values than flour from the wheat with a white seed coat (Zhang and Tian 2008). Some recently released varieties show good agronomic characteristics and are high yielding but contain high levels of flour carotenoids, limiting the end product uses (Parker and Langridge 2000). It is of great value to understand the wheat cultivar’s molecular genetic regulation and to select them effectively for genetic and plant breeding purposes. Flour color behaves as a typical quantitative trait. With conventional breeding methods, the selection efficiency for flour color is low.
Recently, the development of molecular markers such as restriction fragment length polymorphisms (RFLPs) and simple sequence repeats (SSRs), and their maps, have provided powerful tools for elucidating the genetic basis of quantitatively inherited traits, including most of the agriculturally important and quality traits, which makes them feasible to detect and map QTLs in a wheat population, even from a cross between two wheat cultivars (Huang et al. 2003). QTLs associated with important traits have been identified in wheat, including agronomic traits, grain yield, disease characters, and quality traits (Börner et al. 2002; Liu et al. 2002; Ma et al. 2005; Huang et al. 2006). Furthermore, DNA MAS has made it possible to develop near-isogenic lines and chromosome segment substitution lines for QTL regions affecting agronomic and quality traits (Lin et al. 2000; Ma et al. 2002). There are several reports for flour color associated with markers. Parker et al. (1998) identified ten RFLP marker loci that showed significant associations with QTLs for flour color, located on chromosomes 3A and 7A, by using 150 single seed descent (SSD) lines. The estimated heritability of flour color was calculated as 0.68, indicating that a large part of the expression of this trait is genetically controlled, making it easier to manipulate at the genetic level in a breeding program. Mares and Campbell (2001) reported that xanthophyll content was very strongly associated with QTLs located on chromosomes 3B and 7A in 163 DH lines derived from Sunco × Tasman. Zhang et al. (2006) identified eight QTLs for b* located on chromosomes 1DS, 2DL, 3A, 4D, 5D, 6AL, 6D, and 7AL. Knott (1984) reported that a gene for b* was linked to Lr19 in ‘Agatha’ wheat. One SSR and sequence related amplified polymorphism (SRAP) marker closely linked to a Lr19 resistance trait was obtained, and were named wms44 and M73 with genetic distances 0.9 and 2.6 cM, respectively (Li et al. 2005; Liu et al. 2007a). A co-dominant amplified fragment length polymorphism (AFLP) marker linked to a major locus controlling flour color in wheat has been converted to an sequence tagged sites (STS) marker for wider applicability in MAS (Parker and Langridge 2000). However, most previous studies have focused on b*; very few studies have reported on a* and L* values, which are also very important components of flour color in wheat.
Epistatic effects have been considered to be important for complex traits by many researchers (Cao et al. 2001; Fan et al. 2005; Ma et al. 2005, 2007). Determining the contribution of epistasis to complex traits is important for understanding the genetic basis of complex traits. Hence, genetic models for QTL mapping assuming no epistasis could lead to a biased estimation of QTL parameters. Large numbers of epistatic effects have recently been detected in rice and maize by using polymorphism markers in the whole genome (Cao et al. 2001; Fan et al. 2005; Ma et al. 2007). Ma et al. (2005) provided strong evidence for the presence of epistatic interactions on dough rheological properties in a wheat DH population. So far, no information has been found available on the role of epistasis in controlling flour color in wheat.
Genotype-by-environment interactions have received much attention in crop-breeding programs (Dudley et al. 1996), which are important for MAS in crop improvement (Wang et al. 1999). A mixed-model-based composite interval mapping (Wang et al. 1999) methodology was proposed for mapping QTLs with additive effects and epistatic effects as well as their QE interactions, and the software QTLNetwork 2.0 (Yang and Zhu 2005) has been developed for analyzing the experimental data. This method has been successfully applied in recent QTL mapping studies (Liu et al. 2007b; Ma et al. 2007; Rebetzke et al. 2007).
In the present study, we investigated flour color for a DH population by using the software of QTLNetwork 2.0 (Yang and Zhu 2005), based on the method of mixed linear models (Wang et al. 1999). The primary goal of the study described here was to detect QTLs with additive effects, epistatic effects, and QE interactions for flour color (a*, b*, and L*), with the aim of identifying markers that could be used for MAS. The results should be useful in relevant wheat breeding programs.
Materials and methods
Plant materials
A population of 168 DH lines was produced by hybridization with maize pollen of wheat F1 hybrid plants from the cross between two Chinese bread wheats Huapei 3 × Yumai 57 and used for the construction of a genetic linkage map. Huapei 3 and Yumai 57 were registered by Henan in 2006 (Hai and Kang 2007) and by the State (China) in 2003 (Guo et al. 2004), respectively. Huapei 3 is a hard wheat with a higher grain protein content and wet gluten content. Yumai 57 is a soft wheat with higher gluten index and sodium dodecyl sulfate (SDS) sedimentation volume and is more productive under different ecological conditions (Hai and Kang 2007; Guo et al. 2004). The parents both have white seed coats.
Field trials and flour color evaluation
Field trials were conducted under three environments during 2005 and 2006 in Taian, Shandong Province, and in 2006 in Suzhou, Anhui Province. The experimental design followed a completely randomized block design with three replications at each location. In autumn 2005, all lines and parental lines were grown in 2-m-long three-row plots (25 cm apart), whereas in autumn 2006, they were grown in 2-m-long four-row plots (25 cm apart). Suzhou and Taian show large differences in climate, soil conditions, and day length. At Taian, there were remarkable differences in temperature and soil conditions between the years 2005 and 2006. Management was in accordance with local practice. The lines were harvested individually at maturity to prevent yield loss from overripening. Harvested grain samples per line from the three replicates at each environment were mixed and cleaned prior to conditioning and milling in order to maintain a manageable number of samples for quality testing.
One thousand grams of grain samples from each line at each environment were milled by using a Buhler experimental mill (model-MLU 300 M/s Buhler, Uzwil Switzerland). Color measurements of flour samples were carried out using a Minolta Color Meter 310 (Minolta Camera Co, Ltd, Japan) on the basis of L*, a*, and b* values (CIE 1976). The L* value indicates the lightness, with 0–100 representing darkness to lightness. The a* value gives the degree of the red–green color, with a higher positive a* value indicating more red. The b* value indicates the degree of the yellow–blue color, with a higher positive b* value indicating more yellow (Hutchings 1994). Colorimeter scores were taken five times per sample. The mean values of flour color in each environment were used for statistical analyses.
DNA marker assay and construction of genetic map
DNA was extracted from freeze-dried leaves as described by Ellis et al. (2005). A total of 2,002 polymorphism markers were determined using parental DNA. Primer sequences for 260 Xgwm (wms) and 82 Xgdm microsatellite markers were available from Röder et al. (1998) and Pestsova et al. (2000). Markers, 621 Xwmc and 480 Xbarc, were available from the Graingenes website (http://www.wheat.pw.usda.gov/ggpages/SSR/WMC/) and the US wheat and barley scab initiative website (http://www.scabusa.org/pdfs/BARC_SSRs_011101.html), respectively. Thirty BE markers were available from Chen et al. (2005). Fifty Xcfa, 130 Xcfd, 48 Xcwem, and 301 Xcfe markers were kindly provided by Dr. Xianchun Xia, Chinese Academy of Agricultural Sciences, Beijing, China.
Amplification reactions were carried out in a 20 μl reaction mixture, containing 3.6 μl genomic DNA (20 ng/μl), 1.5 μl MgCl2 (25 mmol/l), 1.2 μl dNTP mixtures (2.5 mmol/l), 2.0 μl 10× PCR buffer, 0.5 μl primer (10 μmol/l), 0.2 μl Taq polymerase enzyme (5 units/μl), and 11.0 μl ddH2O. The polymerase chain reactions (PCR) were performed in 96-well microtiter plates by using an Eppendorf AG 22331 Hamburg thermal cycler (Eppenf, Hamburg, Germany). DNA amplification was programmed at 95°C for 5 min, followed by 36 cycles of 95°C for 1 min, 50–65°C (depending on the primer combinations) for 50 s, 72°C for 1 min, and a final extension of 10 min at 72°C before cooling to 4°C. After amplification, the PCR products were mixed with 5.6 μl loading buffer (2.5 mg/ml bromophenol blue, 2.5 mg/ml diphenylamine blue, 10 mmol/l EDTA, 95% (v/v) formamide), denatured for 5 min at 95°C, and chilled on ice for 5 min. The PCR products were separated using 6% (W/V) denatured polyacrylamide gel and were detected by silver staining (Karakousis et al. 2003).
Statistical analysis
Analysis of variance (ANOVA) was carried out using the SPSS version 13.0 (SPSS, Chicago, USA) program. QTL analyses were performed using QTLNetwork software version 2.0 (Yang and Zhu 2005) based on a mixed linear model (Wang et al. 1999). Composite interval analysis was undertaken using forward–backward stepwise, multiple linear regression with a probability into and out of the model of 0.05, and window size set at 10 cM. QTL was declared if the phenotype was associated with a marker locus at P < 0.005. The final genetic model incorporated significant additive effects and epistatic effects as well as their environment interactions.
Results
Statistical analysis of the phenotypic assessments
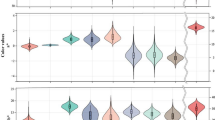
Mean values of flour color for the parents Huapei 3 and Yumai 57, as well as the 168 DH lines under three environments in 2005 and 2006 cropping seasons, are shown in Table 1 and Fig. 1. Huapei 3 had a higher value for b*, while Yumai 57 had higher values for both a* and L*. Transgressive segregants were observed for the three traits among the DH lines in the three environments. Three traits of the DH population segregated continuously and followed a normal distribution (Fig. 1), and both absolute values of skewness and kurtosis were less than 1.0 (Table 1), indicating its polygenic inheritance and suitability of the data for QTL analysis (Cao et al. 2001).
The correlations among the a*, b*, and L* values are shown in Table 2. Significantly negative correlations were detected between L* and b* (r = −0.559), and between a* and b* (r = −0.494).
Construction of the genetic map
A genetic map of the DH population with 305 SSR markers, including 283 SSR and 22 ESTs loci, was constructed, covering a total length of 2141.7 cM, with an average distance of 7.02 cM between adjacent markers in the genome. Ten SSR markers remained unlinked. These markers formed 24 linkage groups at logarithm of the odds ratio (LOD) ≥3.0. The chromosomal locations and marker orders in the map were in accordance with the microsatellite map for Triticum aestivum L. (Somers et al. 2004). The recommended map distance for genome-wide QTL scanning is an interval length less than 10 cM (Doerge 2002). Therefore the map is suitable for genome-wide QTL scanning.
QTLs for flour color
Eighteen QTLs with additive effects and/or additive × environment (AE) effects were detected for flour color in three environments (Table 3, Fig. 2), ranging from four to eight QTLs for each trait, and were distributed on 12 of the 21 chromosomes.
Twenty-four pairs of QTLs with epistatic effects and/or epistasis × environment (AAE) effects were detected for flour color in the three environments (Table 4, Fig. 2), ranging from four to ten QTLs for each trait. The highest numbers of QTLs with epistatic effects were found in b* and L* with ten pairs of QTLs. In contrast, only four pairs of QTLs were detected for a*.
QTLs for a*
Six additive QTLs were detected for a*, on chromosomes 1B, 3B, 5D, 6A, 6D, and 7D. They increased a* from 0.03 to 0.10, explaining phenotypic variance from 2.24% to 25.64%. qa1B had the most significant effect, accounting for 25.64% of the phenotypic variance. Three QTLs (qa1B, qa5D, and qa6D) had negative effects on a* and were contributed by Yumai 57 alleles while the other loci had positive effects on a* and were transmitted by Huapei 3 alleles. This suggested that alleles which increased a* were dispersed within the two parents, resulting in small differences of phenotypic values between the parents and the transgressive segregants among the DH population. The total contribution of all the six QTLs to a* was 41.29%. qa5D was also involved in AE interactions in two environments. The AE interactions explained 2.90% of the phenotypic variance of a*.
Four pairs of epistatic effects were identified for a* and located on chromosomes 1A–7B, 2A–4B, 4D–6A, and 5B–7B. They explained the phenotypic variance ranging from 0.60% to 2.35%. qa4D/qa6A had a positive effect of 0.03. However, the other three pairs showed negative effects on a*. The general contribution of four pairs of epistatic QTLs was 7.10%, while no main-effect QTL was detected in epistatic effects. No AAE interactions were identified for a*.
For a*, the total QE interactions could explain 2.90% of the phenotypic variance.
QTLs for b*
Four QTLs with additive effects significantly influenced b* were located on chromosomes 2B, 3D, 4D, and 5A. The qb5A made the highest contribution and explained 4.30% of the phenotypic variance. All four QTLs could account for 11.21% of the phenotypic variance and were derived from Huapei 3 alleles, which was in accordance with Huapei 3 having much larger b*. No AE interactions were identified for b*.
Ten pairs of epistatic effects were resolved for b* and were located on chromosomes 3D–5B, 1A–1B, 2A–3D, 2B–7A, 3A–5B, 3D–4D, and 6B–7D. Among them, qb3A/qb5B had the highest contribution and accounted for 6.15% of the phenotypic variance. Three pairs of epistatic QTLs (qb3D-2/qb5B, qb3A/qb5B, and qb6B/qb7D) had negative effects on b*. However, the other seven pairs showed positive effects on b*. The total contribution of ten pairs of epistatic QTLs was 31.88%, so epistasis played a strong effect on b*. One main-effect QTL, qb3D-2, was involved in three pairs of epistatic effects. No AAE interactions were identified for b*.
For b*, no QE interactions were detected in this study.
QTLs for L*
Eight main-effect QTLs were identified for L*. These QTLs increased the L* from 0.06 to 0.16, and accounted for the phenotypic variance ranging from 0.82% to 6.14%. The total contribution of the main-effect QTLs could explain 22.28% of the phenotypic variance of L*. Among them, three alleles (ql1B-1, ql4B, and ql7B) came from the parent Huapei 3 and the rest derived from the parent Yumai 57 alleles. This suggested that alleles for increased L* were dispersed within the two parents. This result was also in accordance with the presence of a wide range of variation and transgressive segregations of L* in the DH population. Two QTLs were involved in AE interactions, which could explain 3.01% of the phenotypic variance of L*.
Ten pairs of epistatic QTLs for L* were resolved, explaining the phenotypic variance ranging from 0.80% to 6.24%. ql4D-1/ql5A had the largest effect, with a negative effect of −0.17, accounting for 6.34% of the phenotypic variance. The total contribution of epistatic QTLs was 21.36%. Three main-effect QTLs (ql3D, ql7B, and ql7D) were detected in epistatic effects. Two pairs of epistatic QTLs were also involved in AAE interactions, accounting for 3.88% of the phenotypic variance.
For L*, the total QE interactions could explain 6.89% of the phenotypic variance.
Discussion
Epistasis is an important genetic basis of flour color in wheat
Epistasis as an important genetic basis of complex traits has been well demonstrated in recent QTL mapping studies (Cao et al. 2001; Fan et al. 2005; Ma et al. 2005, 2007). Ma et al. (2005) provided strong evidence for the presence of epistatic effects on dough rheological properties in a wheat DH population. In the present study, 24 pairs of QTLs with epistatic effects and/or AAE effects were detected for flour color in three environments (Table 4, Fig. 2), ranging from four to ten QTLs for each trait. Epistatic effects could explain 7.10%, 31.88%, and 21.36% of the phenotypic variance of a*, b*, and L*, respectively. Furthermore, epistatic effects had a significant effect on b*, which could account for 31.88% of the phenotypic variance. So epistasis is an important genetic basis of flour color in wheat. The results indicate the complexity of the genetics of wheat flour color.
Relationship between additive effects and epistatic effects of flour color in wheat
It would be interesting to study the relationships between additive QTLs and epistatic QTLs identified. The majority (92.59%) of loci involved in the epistatic interactions did not appear to have significant additive effects on flour color in wheat. For example, ten pairs of epistatic effects were identified for L*; 15 single loci involved in epistatic effects had no additive effects. Similarly, Ma et al. (2007) observed that 37% of the main-effect QTLs were involved in the epistatic interactions in maize grain yield and its components. This indicated that many loci in epistatic effects might not have significant effects for flour color alone but might affect its expression by epistatic effects with the other loci. The results also suggest that some of the additive QTLs might be detected with effects confounded by epistatic effects, if the epistatic effects were ignored in QTL mapping. Thus, breeders must take into account such complexity and test for the effects of individual loci in the targeted genetic background in order to obtain the expected phenotypes of the interested genes.
Comparison of the present study with previous researches
In previous studies (Knott 1984; Parker et al. 1998; Mares and Campbell 2001; Zhang et al. 2006), QTL studies for wheat flour color were conducted based on the additive-dominant model and usually assumed absence of epistasis among QTLs. However, epistasis between nonallelic QTLs for some complicated quantitative traits may exist. In the present study, both additive and epistatic effects of QTLs were obtained based on the method of mixed linear models. Furthermore, epistasis was an important component for b*, accounting for 31.88% of the phenotype variation (Table 4).
Knott (1984) reported a gene for b* linked to Lr19 in ‘Agatha’ wheat. One SSR and SRAP marker closely linked to Lr19 resistance trait were obtained, and were named wms44 and M73 on chromosome 7D with genetic distances 0.9 and 2.6 cM, respectively (Li et al. 2005; Liu et al. 2007a). At a similar location, additive QTLs associated with a* and L* were also detected in this study. Near this location, one epistatic QTL was detected for b*. Parker et al. (1998) identified ten RFLP marker loci that showed significant associations with QTLs for flour color, located on chromosomes 3A and 7A, by using a SSD population of 150 lines. Zhang et al. (2006) identified eight QTLs for b*, located on chromosomes 1DS, 2DL, 3A, 4D, 5D, 6AL, 6D, and 7AL. Mares and Campbell (2001) reported that xanthophyll content was very strongly associated with QTLs located on chromosomes 3B and 7A in 163 DH lines derived from Sunco × Tasman. In this study, three epistatic QTLs were also detected for b* on chromosomes 3A, 5B, and 7A. Four QTLs with additive effects were resolved for b* on chromosomes 2B, 3D, 4D, and 5A, and ten pairs of epistatic effects were detected for b* on chromosomes 3D–5B, 1A–1B, 2A–3D, 2B–7A, 3A–5B, 3D–4D, and 6B–7D in the present study.
These differences might be attributed to following possibilities. First, differences in mapping populations and experimental environments were studied in various QTLs mapping. Second, different QTL mapping approaches were used in various studies. Third, some of the genes involved in the wheat flour color pathway did not show any allelic variation between the two parents (Huapei 3 and Yumai 57). Consequently, several loci could not be identified on some chromosomes. Fourth, the coverage of the genome was not complete in the present map; some QTLs associated with b* could not be detected, which might possibly affect the QTLs in the DH population. For complete analysis, a detailed map with total genome coverage is required.
To date, very few studies reported the QTLs which are associated with a* and L*. A significantly negative correlation was detected between L* and b* (r = −0.559) and a* and b* (r = −0.494). So it was very important to study the genetic basis of a* and L* so as to achieve ideal flour color in wheat. In this study, six additive QTLs and four pairs of epistatic QTLs were detected for a*. One major QTL, qa1B, was closely linked to barc372 0.1 cM and could account for 25.64% of the phenotypic variance of a* without any environmental influence. So, qa1B can be used for wheat MAS. This is a new and promising locus to improve our knowledge of wheat flour genetic mechanism for a*. To better understand it, a marker-assisted backcrossing procedure had been initiated in our group. Near-isogenic line development and fine mapping of qa1B are now ongoing following a strategy described by Saito et al. (2004). Closely linked flanking markers associated with major QTL could be further developed by converting them to STS markers for application in MAS. Eight additive QTLs and ten pairs of epistatic QTLs were identified for L*.
The present investigation might be the first report of epistatic QTLs associated with flour color using molecular markers. The results indicated that epistasis was an important genetic basis of flour color in wheat. Therefore, genetic models for QTL mapping assuming no epistasis could lead to a biased estimation of QTL parameters. The results showed that both additive and epistatic effects were important genetic bases of flour color. In addition, additive effects and epistatic interactions were also sometimes subject to environmental modifications.
In summary, a total of 18 additive QTLs and 24 pairs of epistatic QTLs were detected for flour color in 168 DH lines derived from a Huapei 3 × Yumai 57 cross, which were distributed on 19 of the 21 chromosomes. One major QTL, qa1B, was closely linked to barc372 0.1 cM and could account for 25.64% of the phenotypic variation of a* without any influence from the environment. Therefore, qa1B could be used in MAS in wheat breeding programs. The results showed that both additive and epistatic effects were important in the genetic basis of flour color, and were also sometimes subject to environmental modifications.
References
Börner A, Schumann E, Furste A, Coster H, Leithold B, Röder MS, Weber WE (2002) Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor Appl Genet 105:921–936
Cao G, Zhu J, He C, Gao Y, Yan J, Wu P (2001) Impact of epistasis and QTL×environment interaction on the developmental behavior of plant height in rice (Oryza sativa L.). Theor Appl Genet 103:153–160
Chen JF, Ren ZL, Gao LF, Jia JZ (2005) Developing new SSR markers from EST of wheat. Acta Agro Sini 31:154–158
CIE (1976) CIE, Committee TC-1.3 CIE, Technical note. J Opt Soc Am 64:896–897
Doerge RW (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev 3:43–52
Dudley JW, Lamkey KR, Geadelmann JL (1996) Evaluation of populations for their potential to improve three maize hybrids. Crop Sci 36:1553–1559
Ellis MH, Rebetzke GJ, Azanza F, Richards RA, Spielmeyer W (2005) Molecular mapping of gibberellin responsive dwarfing genes in bread wheat. Theor Appl Genet 111:423–430
Fan CC, Yu XQ, Xing YZ, Xu CG, Luo LJ, Zhang QF (2005) The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population. Theor Appl Genet 110:1445–1452
Guo CQ, Bai ZA, Liao PA, Jin WK (2004) New high quality and yield wheat variety Yumai 57. China Seed Ind 4:54
Hai Y, Kang MH (2007) Breeding of Hupei 3 new wheat variety with high yield and early maturing. Henan Agric Sci 5:36–37
Huang XQ, Coster H, Ganal MW, Röder MS (2003) Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor Appl Genet 106:1379–1389
Huang XQ, Cloutier S, Lycar L, Radovanovic N, Humphreys DG, Noll JS, Somers DJ, Brown PD (2006) Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor Appl Genet 113:753–766
Hutchings JB (1994) Food color and appearance. Chapman and Hall, Great Britain; Blackie Academic and Professional, London
Karakousis A, Gustafson JP, Chalmers KJ, Barr AR, Langridge P (2003) A consensus map of barley integrating SSR, RFLP, and AFLP markers. Aust J Agric Res 54:1173–1185
Knott DR (1984) The genetic nature of mutations of a gene for yellow pigment linked to Lr19 in ‘Agatha’ wheat. Can J Genet Cytol 26:392–393
Kruger JE, Anderson MH, Dexter JE (1994) Effect of flour refinement on raw cantonese noodle color and texture. Cereal Chem 71:177–182
Li X, Yang W, Li Y, Liu D, Yan H, Meng Q, Zhang T (2005) A SSR marker for leaf rust resistance gene Lr19 in wheat. Sci Agric Sini 38:1156–1159
Lin HX, Yamamoto T, Sasaki T, Yano M (2000) Characterization and detection of epistatic interactions of three QTLs, Hd1, Hd2 and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet 101:1021–1028
Liu DC, Gao MQ, Guan RX, Li RZ, Cao SH, Guo XL, Zhang AM (2002) Mapping Quantitative trait loci for plant height in wheat (Triticum aestivum L.) using a F2:3 population. Acta Genet Sini 9:706–711
Liu GF, Yang J, Xu HM, Zhu J (2007a) Influence of epistasis and QTL × environment interaction on heading date of rice (Oryza sativa L.). J Genet Genomics 34:608–615
Liu Y, Yan H, Yang W, Meng Q, Zhang T, Liu D (2007b) Identification of a SRAP markers linked to leaf rust resistance gene Lr19 in wheat. Acta Agric Boreali Sini 22:193–196
Ma JF, Shen RF, Zhao ZQ, Wissuwa M, Takeuchi Y, Ebitani T, Yano M (2002) Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol 43:652–659
Ma W, Appels R, Bekes F, Larroque O, Morell MK, Gale KR (2005) Genetic characterisation of dough rheological properties in a wheat doubled haploid population: additive genetic effects and epistatic interactions. Theor Appl Genet 111:410–422
Ma XQ, Tang JH, Teng WT, Yan JB, Meng YJ, Li JS (2007) Epistatic interaction is an important genetic basis of grain yield and its components in maize. Mol Breed 20:41–51
Mares DJ, Campbell AW (2001) Mapping components of flour and noodle color in Australian wheat. Aust J Agric Res 52:1297–1309
Miskelly DM (1984) Flour components affecting paste and noodle color. J Sci Food Agric 35:463–471
Parker GD, Langridge P (2000) Development of a STS marker linked to a major locus controlling flour color in wheat (Triticum aestivum L.). Mol Breed 6:169–174
Parker GD, Chalmers KJ, Rathjen AJ, Langridge P (1998) Mapping loci associated with flour color in wheat (Triticum aestivum L.). Theor Appl Genet 97:238–245
Pestsova E, Ganal MW, Röder MS (2000) Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 43:689–697
Rebetzke GJ, Ellis MH, Bonnett DG, Richards RA (2007) Molecular mapping of genes for Coleoptile growth in bread wheat (Triticum aestivum L.). Theor Appl Genet 114:1173–1183
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Saito K, Hayano-Saito Y, Maruyama-Funatsuki W, Sato Y, Kato A (2004) Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor Appl Genet 109:515–522
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Sun XD, Wang LK, Ren HB, Lan J (2002) The application of tristimulus colorimeter in the determination of flour color. Technol Oil Food 10:31–33
Wang DL, Zhu J, Li ZK, Paterson AH (1999) Mapping QTLs with epistatic effects and QTL×environment interactions by mixed linear model approaches. Theor Appl Genet 99:1255–1264
Yang J, Zhu J (2005) Predicting superior genotypes in multiple environments based on QTL effects. Theor Appl Genet 110:1268–1274
Zhang X, Tian JC (2008) The color advantage of Chinese wheat with high whiteness and analysis of factors affecting color formation. Sci Agri Sini 41:347–353
Zhang LP, Yan J, Xia XC, He ZH, Sutherland MW (2006) QTL mapping for kernel yellow pigment content in common wheat Mapping QTLs for polyphenol oxidase activity in a DH population from common wheat. Acta Agro Sini 32:41–45
Acknowledgements
The authors gratefully thank Professor Yan Hai (Henan Academy of Agricultural Sciences, Zhengzhou, China) for kindly providing the research materials and Dr. Xianchun Xia (Chinese Academy of Agricultural Sciences, Beijing, China) for donating some primers. The present research was supported by the National Natural Science Foundation of China (30671270), Hi-Tech Research and Development (863) Program of China (2006AA10Z1E9 and 2006AA100101), and Improved Variety Project of Shandong Province (LN2006-6).
Author information
Authors and Affiliations
Corresponding author
Additional information
Kun-Pu Zhang and Guang-Feng Chen contributed equally to this study.
Rights and permissions
About this article
Cite this article
Zhang, KP., Chen, GF., Zhao, L. et al. Molecular genetic analysis of flour color using a doubled haploid population in bread wheat (Triticum aestivum L.). Euphytica 165, 471–484 (2009). https://doi.org/10.1007/s10681-008-9756-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-008-9756-8