Abstract
Enteric viruses, especially human rotaviruses present in aquatic environments, are microbial criteria in quality assessment of water resources. The present research aimed to investigate molecular monitoring of human rotavirus and efficacy evaluation of Isfahan water treatment plant (WTP) in the elimination of viruses. In total, 60 water samples were collected from different units of WTP. Zeta plus electropositive Virosorb cartridge filter and elution buffer was used for concentrating water samples. Enzyme-linked immunosorbent assay (ELISA) was used for detecting rotavirus antigen. Quantitative real-time reverse transcription PCR (qRT-PCR) with SYBR Green I fluorescent dye was performed for molecular detection of rotavirus. Multiplex nested reverse transcription-polymerase chain reaction (RT-PCR) was used for rotavirus G genotyping. Total coliform count varies from 102–103 CFU/mL in the raw water resources. Rotavirus antigen was detected in 17 samples (28.33%) by ELISA, and 13 samples (21.67%) were found positive by RT-PCR. These included 41.18% (7 cases) of raw water influent, 29.41% (5 cases) after sedimentation, 23.52% (4 cases) after ozonation, and 5.88% (1 case) after filtration in ELISA method. The highest number of rotaviruses was detected by qRT-PCR in autumn (46.15% (6 cases)). The commonest circulating G type in the sampling points was the mixed types, which was identified in 6 samples (46.15%), followed by non-typeable (23.07%), G3 (15.38%), G1 (7.69%), and G8 (7.69%), respectively. Despite the presence of rotavirus in raw water, after clarification and ozonation, filtration and treated water did not show the presence of rotavirus. The results of this study showed that multi-stage treatment has a positive effect on virus removal in WTP.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Life is heavily influenced by the presence of intestinal pathogens caused by the sewage entering the water. Despite progress made in water and wastewater treatment in recent decades, diseases caused by contaminated water are still a serious risk to human health worldwide, so about 605 million human beings suffer from unsafe drinking water (Assis et al. 2015; Asami et al. 2016; Hamza et al. 2011). Continuous discharge of sewage into water sources has a direct role in the circulation of gastroenteric viruses into the aquatic environments. A wide range of viruses is found in human excretion, varying from 1010 to 1013 numbers per gram of feces (Fongaro et al. 2012). The World Health Organization (WHO) scientific group estimated that even a virus in a large volume of drinking water can be a life-threatening agent for a human being (WHO 2009). Water quality and the health of people are seriously compromised due to the uncontrolled discharge of effluents from human activities in water resources. Consumption of food and contaminated water is one of the prominent factors in developing some lethal diseases for humans (Delgado-Gardea et al. 2017).
It is possible that enteric viruses (e.g., rotavirus) cause viral gastroenteritis via water-borne routes. Replication of such viruses is done in the intestinal tract of the infected person. Afterwards, these viruses are excreted with feces in great concentrations. Enteric viruses detection was observed in rivers, reservoirs, and groundwater, which are applied as sources of drinking water (Asami et al. 2016). Combining insufficient sanitation, bad hygiene, and insecure drinking water causes the expected annual burden of 0.7 million diarrheal mortalities, which make diarrhea as the 4th important reason for mortality among children under 5 years old (Kiulia et al. 2015).
In general, viruses have low concentrations in water. In addition, they are adequately purified via drinking water treatment plants. Nonetheless, when there is failure in one or more procedures of the water treatment, possibly a high population is subjected to enteric viruses through tap water (Asami et al. 2016). The WHO (2014) reported that vaccine-preventable rotavirus infections cause death by approximately 21,5000 children aged less than 5 years each year, predominantly among those in low-income countries (WHO 2014). Gastroenteritis is an inflammation of the stomach and intestines caused by rotavirus as a contagious virus, whose clinical sings are severe watery diarrhea, vomiting, fever, and abdominal pain. Rotavirus diseases occur among infants and young children as high-risk group, which severely develop dehydration and hospitalization, and even lead to death (CDC 2014).
Rotaviruses are non-enveloped particles (resistant to lipid solvents) with a diameter of 60 to 80 nm and a genome length of 18.5 Kb, whose genome includes double-stranded RNA (dsRNA) with 12 segments that encode 6 nonstructural (NSP1-NSP6) and 6 structural proteins (VP1 to VP4, VP6, and VP7) (Silva et al. 2016; Bosch et al. 2008; Kargar et al. 2013; Kluge et al. 2014). The RNA is surrounded by a three-layered icosahedral capsid. The outer layer of the capsid consists of VP4 and VP7 proteins, the inner layer of VP6 glycoprotein, and the inner core of VP2. VP6 is found abundantly in the protein of virus, and accounts for about 51% of the virion capsid, which is a criterion for classification of viruses into eight groups of A–H. The rotavirus genotypes are divided into VP4 (P-genotype) and VP7 (G-genotype) based on two external proteins of the capsid glycoprotein VP7 and the protease-sensitive VP4 that are targeted by neutralizing antibodies (Silva et al. 2016; Liu et al. 2015).
Asami et al. surveyed drinking water treatment plant for removal efficiency of virus in the coagulation-sedimentation and rapid sand filtration stages in Bangkok, Thailand. Their findings showed that the most abundant viruses in raw waters are pepper mild mottle virus (PMMoV) and JC polyomavirus (JC PyV) at concentrations of 10 2.88 ± 0.35 and 10 3.0 6 ± 0.42 copies/L (geometric mean ± S.D.) (Asami et al. 2016). Ye et al. (2012) detected enteric viruses in the treated drinking water and source water using qRT-PCR technique in Wuhan, China.
Fecal contamination of water sources possibly contributes importantly to the transmission and epidemiology of enteric viruses (Bosch et al. 2008; Adefisoye et al. 2016). Reuse of wastewater for agriculture or industrial purposes may pollute the environment; therefore, environmental monitoring of the rotavirus A (RVA) in the environments is essential. There are a few studies conducted to evaluate the quality of drinking water in developing countries. The studies just focused on the bacterial indicators, including coliforms and heterotrophs, whereas viruses have been neglected. Therefore, it is very important to create a quick and precise detection to protect public health from the risk of enteric viruses (Asami et al. 2016; Ibfelt et al. 2016).
However, the lack of basic information of gastroenteric viruses in water resources used for drinking water and the significance of these viral infections among humans made us to conduct this study aiming at optimization of treatment plant. Until this time, we found only a study by Kargar et al. who used RT-PCR for molecular detection of group A rotavirus in hospital and urban sewage systems in Shiraz, Iran (Kargar et al. 2013). The present research aimed at assessing efficiency of removal group A human rotavirus in water treatment plant (WTP) of Isfahan, Iran.
Materials and methods
Area of study
This study was conducted in WTP of Baba Sheikh Ali located in Zayanderud city of Isfahan, Iran. This treatment plant is the largest water treatment plant in Iran, which provides drinking water for Isfahan, 56 cities, more than 300 villages, and a large number of industries in Isfahan province. The Isfahan WTP provides drinking water for around 4 million people. Therefore, providing safe drinking water for this large population is strategically important (Fig. 1).
Sample collection
From October 2016 to September 2017, sixty water samples were collected from different units of Isfahan water treatment in 5 points (raw water, sedimentation, filtration, disinfection (ozonation), and treated water) using the grab sampling procedure, so that 20 L water samples were obtained from each point, which were poured in sterile plastic bottles and stored at 4 °C (cold chain) (Fout et al. 1996).
Detection of indicator bacteria
Total and fecal coliforms testing was measured from 100 ml raw water and treated water samples by using the standard multiple-tube fermentation method based on the standard methods of water and sewage testing (Rice et al. 2012).
Used filters for virus concentration
Primary concentration was performed for recovery efficiency of the virus by Zeta plus 1MDS cartridge filter (Rames et al. 2016). Twenty-liter water samples were passed through 1MDS filter with air pressure created by a vacuum pump. Zeta plus Virosorb 1 MDS (Cuno, 3M Germany) is an electropositive charged modified glass and cellulose medium. One MDS cartridge filter used in this study was 25.4 cm (10 inches) in length. The elution and flocculation steps were performed after filtration according to Lee et al. (2016). Briefly, elution of viruses loaded on the 1MDS filter was performed by 400 mL of elution buffer (1.5% beef extract and 0.05 M glycine at pH 9.5) using a constant flow at the pressure vessel of 4–8 psi for 5 min. During secondary concentration, polyethylene glycol (PEG 6000, Merck)/sodium chloride (NaCl, Merck) was added and shaken at 4 °C for 1 h and stationery for 1 h at 4 °C to enhance flocculation (Grøndahl-Rosado et al. 2014). Centrifuging the mixture of virus particles was done at 15,000g at 4 °C for 15 min for further concentration, followed by maintenance at − 70 °C (Fig. 2) (Verheyen et al. 2009).
Detection of rotavirus antigen by enzyme-linked immunosorbent assay
ELISA technique through the kit (EIA) (Rotavirus Ag ELISA, DRG/Germany) was used to screen the existence of rotavirus antigens in each concentrated water sample in accordance with the protocol instructed by the manufacturer in the laboratory (Najafi et al. 2012). A sandwich type method was used to coat the microwell plates by monoclonal antibodies against the middle layer antigen of RV (VP6). The specimen with optical density (OD) of more than the cut-off value was considered to be positive, and the OD of the negative control was + 0.15 (Kargar et al. 2013; Yousuf et al. 2017; Maite 2016).
Extraction of nucleic acids and synthesis cDNA
The extraction of viral RNA was done from 140 μL of each antigen-positive water samples. Firstly, the sample suspension was prepared by phosphate-buffered saline (PBS), and centrifugation was conducted at 1500g for 10 min. The RNA was extracted from supernatant using a QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) based on the company instructions, followed by reverse-transcribed to cDNA PrimeScript strand cDNA synthesis kit (Takara, Japan) with the following procedures: 30 °C for 10 min, 42 °C for 10 min, 70 °C for 15 min. Afterwards, it was cooled immediately to 4 °C. The RT product was kept at − 20 °C for subsequent qPCR (Asami et al. 2016; Banerjee et al. 2008; Salvo et al. 2018).
Quantitative real-time reverse transcription PCR
Quantitative real-time reverse transcription PCR (qRT-PCR) was performed for cDNA through SYBR Green I fluorescent dye approach. Each 20 μL RT-PCR mix contained 10 μL 2× SYBR Premix (Takara, Japan), and 2 μL DNA template, and 100 nM each primer. Primers of rotavirus designed for the present research (Accession = LC178569) include RVA-F: 5′-CAAACTGACGAAGCGAATAAATG-3′ and RVA -R: 5′-TGTAGCATCAGTTGTCAAGCATC-3′´[Tm] at 61 °C. The amplification processes were run in Rotor-Gene 6000: 40 cycles involving denaturation at 95 °C for 15 s, annealing at 61 °C for 30 s, and extension at 72 °C for 30 s. Emitting all sample was recorded in thermal cycling, and the raw fluorescence data were included in SDS software (Rotor-Gene 6000) for obtaining the threshold cycle (Ct) values, accompanied by calculation of the standard curve through Ct values of the diluted standards. Next, Ct values were applied for extrapolating absolute amounts for the unspecified specimens (Ye et al. 2012).
Standard curve
For obtaining the representative positive control standards, the former mentioned plasmid pcDNA3.1 (+) was employed for quantifying (Delgado-Gardea et al. 2017). To evaluate the exclusivity of primers used in this study, the primers were firstly BLAST-specific on NCBI website (Accession = LC178569), and it was determined that they are correct for the genome of the rotavirus. Then, a 110 bp PCR amplicon was cloned into the pcDNA3.1 (+) plasmid (pcDNA3.1 vector, Invitrogen, USA.). Afterwards, the primers were transformed into the competent cells of E. coli TOP10F´ by calcium chloride (CaCl2) chemical method. To screen the recombinant vectors, the transformants were selected on LB-ampicillin agar plates.
Nanodrop 2000p equipment (Thermo Fisher Scientific Inc., Boston, MA, USA) was applied for determining DNA concentration (ng/L). After calculating the numbers of RNA copies, a standard curve was plotted from ten-fold serial dilutions of the RNA standard ranged at 102–108. All samples were analyzed in duplicate. Two methods were recruited to obtain the efficiency of PCR process, including the Rotor-Gene 6000 qRT-PCR software and the equation of % Efficiency = [10 (− 1/slope) − 1] × 100 (Delgado-Gardea et al. 2017).
Multiplex nested reverse transcription-polymerase chain reaction for rotavirus G genotyping
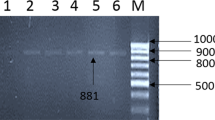
The G typing was administered using cDNA products as templates for amplifying PCR VP7 gene with pair primers. For G genotyping G1 to G4, G8 to G9 and G10 and G12, specific primers were employed in the RT-PCR (Table 1). Briefly, first round PCR was carried out with 5 μL of dsRNA, DMSO, 5× RT buffer, dNTPs, primers Beg9, End9, and distilled water. The cycling variables applied included 30 cycles (94 °C for 1 min, 42 °C for 2 min, & 72 °C for 2 min) and a final extension (72 °C for 5 min). The 2nd round VP7 RT-PCR was performed with 5 μL of the 1st round VP7 amplicons in 40 μL of the 2nd round VP7 reagent mixes. Cycling included 20 cycles of the cycling protocol similar to the 1st reaction. PCR mixes contained MgCl2 (50 mM), deoxy nucleoside triphosphates (10 mM), primers (10 pmole), 10 × PCR buffer, and Taq DNA polymerase (1 U). Analysis of the amplicons was done by 2% agarose gel electrophoresis along with ethidium bromide (10 μg/mL) to increase the products resolution. Primer sequences are shown in Table 1 (Grassi et al. 2010; Kargar et al. 2013).
Statistical analysis
Cultivation or qRT-PCR on 10-fold serially diluted RNA (endpoint dilution) was used to count the virus particles in water. SPSS 16.0 software (SPSS Inc., New York 10504-1722, USA) was applied to analyze the data statistically using chi-square test at the significance level of p value less than 0.05.
Results
Detection of indicator bacteria
Outputs of the physicochemical parameters and bacterial indicator of raw water and the treated water at Isfahan WTP are represented in Table 2. According to bacteriological analyses, mean of total and fecal coliforms was estimated 102–103 MPN/mL in raw water samples of water treatment plant; of course, no samples of treated water were contaminated to coliforms (Fig. 3). Statistical analysis showed a significant association between the concentration of bacterial indicators and rotaviruses (p < 0.01).
The standard curve
The standard curve for each run based on the cycle threshold value (Ct) for each standard was calculated by the SDS software (Fig. 4).
Distribution of rotaviruses
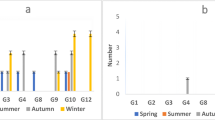
From 60 water samples that were collected from 5 points in Isfahan WTP, rotavirus was detected by ELISA in 17 samples (28.33%) and by qRT-PCR in 13 samples (21.67%). EIA showed that detection of the largest number of rotaviruses is related to the input samples 41.18% (7 case), and the lowest is related to the output samples (0 cases) of the treated water (Fig. 5). Detection of the highest number of rotaviruses by qRT-PCR was observed in autumn (46.15%), followed by winter (38.46%), spring (7.69%), and summer (7.69%), respectively (Table 3). Generally, analyses of the removal efficacy of rotaviruses showed that the highest removal efficiency was in filtration and treated water units. According to the seasonal distribution, the cold seasons showed significantly higher number of rotaviruses.
Distribution of rotavirus genotypes
G typing was determined via the RT-qPCR on 13 positive samples. The result showed the maximum circulating G type in the sampling points for mixed types. It was identified in 6 samples (46.15%), followed by non-typeable (23.07%), G3 (15.38%), G1 (7.69%), and G8 (7.69%), respectively. No individual detection of the G types G2, G4, G9, G10, and G12 was observed during this research. The commonest rotavirus type reported was the mixed types and non-typeable in raw water and clarification in cold seasons. The G types G3 and G8 were detected in ozonation points. None of the rotavirus genotypes was observed after filtration. The rotavirus genotype distribution obtained from various sampling points of the Isfahan water treatment plant is shown in Table 4.
Discussion
The microbiological quality of treated water in water treatment plant is traditionally evaluated via indicator organisms as total and fecal coliforms. Nonetheless, such indicators do not refer to the danger of additional microbial pathogens, including viruses. It is possible that human rotaviruses entered environmental waters via different pathways such as discharging the untreated or insufficiently treated wastewater. Human rotaviruses were suggested as probable viral indices of human fecal pollution in aquatic contexts because of their host specificity, higher prevalence (60 to 70% of viral diarrhea are in children with rotavirus), and stability in the environment (Barril et al. 2016). The detection of viral genomes in treated water of different regions is not a usual event. However, some research reported the existence of rotaviruses in tap water purified by conventional processes (Karim et al. 2009; Verheyen et al. 2009). For proper assessment and management of the risks of infections through enteric viruses in tap water, it is necessary to have a quantitative evaluation for virus removal efficiency for distinct procedures in real drinking water treatment plants (DWTPs). Yet, a limited research was done because of technical problems in quantification of low virus concentrations in water samples (Asami et al. 2016). Because rotaviruses frequently have higher resistance to water treatment processes compared with the bacterial indicators, and the high inefficiency of such would be accompanied by higher risks of transmission in comparison with the bacteria and protozoa, the importance of rotavirus is revealed for evaluating the drinking water quality compared with other viruses.
The viruses recoveries from various water matrixes depends on both the viral concentration techniques employed and the kind of water experimented. Due to the low concentration of viruses in the water matrix compared with the sewage samples, effective concentration methods are essential for large amounts of water. In most studies, the use of electronegative and electropositive filters and buffer for viral elution was suggested to increase the recovery of enteric viruses from smaller amounts of water. In this study, a novel surveillance method was implemented by filtration with cartridge Zeta plus Virosorb 1 MDS filters and glycine buffer elution for the first time in order to enhance the concentration and recovery rotavirus of water samples of Isfahan WTP (Inker et al. 2012; Karim et al. 2009; Verheyen et al. 2009).
The WHO recommends methods for detecting rotavirus, based on viral antigens and molecular methods.
Since outputs obtained from ELISA method may provide improper positive specimens based on the detection of proteins, qRT-PCR process was employed. The antibody-based experiments have a restriction to detect rotaviruses that include the necessity of great concentration of free antigen for generating a positive reaction. The free antigen would be considerably declined during a disease. Hence, such experiments enjoy lower sensitiveness, and are mostly designed for a diagnostic laboratory as compared with qRT-PCR. Quantitative PCR method is a molecular instrument that has high sensitivities and specificity, and is commonly applied on viruses in aquatic matrices (Assis et al. 2015; Atabakhsh et al. 2019; Soltan et al. 2016).
In this study, RVA detected in 17 of the 60 samples (28.33%) examined from water samples through ELISA and 13 samples (21.67%) through qPCR (Table 4, Fig. 5). Group A rotavirus was found in 7 specimens of 12 raw water samples, but no samples were found after filtration. Even though microbiological factor is commonly applied for determining fecal contamination levels of water, various researches published the absence of correlation between the existence of viruses and fecal coliform counts in various water matrices (Assis et al. 2015). The analysis of the bacterial indicators showed that the coliforms are completely removed at ozonation points, but rotaviruses are found in ozonation (15.38%).
Our results showed a significant relationship between seasonal conditions and the presence of rotavirus, indicating maximum prevalence in autumn and winter. Thus, this is in line with previous reports made by Assis et al. (2015) and Asami et al. (2016). Such a finding suggests greater distribution of rotaviruses during the cold seasons of the year (Table 3).
Moreover, the commonest rotavirus genotype published was non-typeable and mixed (each 23.08%) and G3 (15.38%) in raw water and clarification during the cold seasons of the year.
Similar to our study, some international studies found that the rotaviruses pervasiveness was 2–31% in water samples (Ye et al. 2012; Gibson et al. 2011; Barril et al. 2016). The highest circulating genotype was G1 (73.33%), accompanied by G1G4 (20%) and non-typeable (6.67%), that has been reported by Kargar et al. (2013).
Ibrahim et al. (2018) counted human adenoviruses (HAdVs) for genotyping their species and evaluating HAdVs removal efficiency in the samples from hospital pilot wastewater treatment plant (PWWTP) in Tunis City, Tunisia. The sequencing and PCR were used to obtain the genotype of the samples positive for HAdVs. The researchers revealed 64% (65/102) positive wastewater samples (Ibrahim et al. 2018). Thirty-five percent of water samples from drinking water treatment plants in Karachi, Pakistan were contaminated by rotavirus. This might be due to diversity of the methodologies used to detect rotavirus or high pollution of drinking water (Yousuf et al. 2017).
Kargar et al. employed RT-PCR to evaluate the group A rotavirus in samples collected from urban and hospital sewage systems in Shiraz (Iran). They found 25% (15 cases) positive sewage samples. Higher concentration of rotavirus was detected in autumn (Kargar et al. 2013). In their study, temporal distribution showed that rotaviruses isolates were observed in autumn>winter>spring, but there was no rotavirus detected in summer (Rizk and Allayeh 2018). Nevertheless, environmental monitoring of genotyping rotavirus may present worthwhile regional insight into the transmissions of human rotavirus genotypes in Isfahan province for providing accurate vaccine to immunize children. Result showed that filtration unit in Isfahan WTP is an essential section for rotaviruses removal, because all rotaviruses were not removed after clarification and ozonation. Asami et al. applied qRT-PCR to evaluate the indigenous virus removal efficiency by counting viruses during pre and post treatment processes (Asami et al. 2016)
The increased positive samples observed in winter and autumn may be due to favorable environmental conditions such as lower temperature compared with other seasons of the year. Moreover, due to rainfall in these seasons and changes in the parameters of the input water treatment plant, conditions for the survival of rotavirus are available. Despite the presence of rotavirus antigen in raw water, after clarification and ozonation, filtration and treated water revealed absent rotavirus antigens. As observed in the study, the treatment processes did completely remove rotaviruses from the samples collected from river raw water.
Conclusion
For accurate assessment and management of the risks of infections via enteric viruses in tap water, it is necessary to evaluate virus removal efficiency for treatment procedures in DWTPs especially in urban water supply systems with high population. After traditional purification, this reservoir water is employed for supplying a population containing potable water. Yet, its consumption is possibly a potent danger to the public health, because researchers previously showed that the RVA is resistant to the increased concentrations of residual chlorine that is a chemical agent with a widespread application in the disinfection procedure to produce potable water.
This study provided valuable information regarding the prevalence of group A rotavirus in Isfahan WTP; that is, this system has the necessary efficiency for removal of rotaviruses. Moreover, new information of rotaviruses circulating in the environment of Isfahan, Iran, was studied for the first time. Rotaviruses monitoring carried out in our work showed that the existing system in Isfahan water treatment plant, which consists of sedimentation, ozonation, and filtration processes, is an acceptable system.
Therefore, the lack of the detection of rotavirus in the drinking waters suggests that the methodology used in the Isfahan water treatment plant is efficient for the removal of viruses.
Thus, we suggest that this molecular monitoring be done routinely. It is also recommended to monitor other viruses in all water treatment plants. On the other hand, the process in Isfahan WTP is proposed as a pattern. Previous studies suggested comparing the methods for removing other viruses (norovirus, adenovirus, enterovirus, etc.) in order to achieve a precise monitoring protocol. Continuous environmental monitoring of rotaviruses can provide useful information on rotating rotavirus genotypes in order to find out an appropriate pattern for regional vaccination.
Abbreviations
- IWTP:
-
Isfahan water treatment plant
- ELISA:
-
Enzyme-linked immunosorbent assay
- rpm:
-
Revolutions per minute
- MPN:
-
Most probable number
- NT:
-
Non-typeable samples
- CFU:
-
Colony-forming unit
- OD:
-
Optical density
- NSP:
-
Nonstructural proteins
- PMMoV:
-
Pepper mild mottle virus
- Nested RT-PCR:
-
Nested reverse transcription (RT) PCR
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- WHO/CDC:
-
World Health Organization and Centers for Disease Control and Prevention.
- EC:
-
Electrical conductivity
- RVA:
-
Rotavirus A
- PAC:
-
Poly aluminum chloride
- NTU:
-
Nephelometric turbidity unit
- Ct:
-
Cycle threshold
- WTP:
-
Water treatment plant
- DWTP:
-
Drinking water treatment plant
- VP:
-
Viral protein
- PBS:
-
Phosphate-buffered saline
- FC:
-
Fecal coliform
References
Adefisoye, M. A., Nwodo, U. U., Green, E., & Okoh, A. I. (2016). Quantitative PCR detection and characterization of human adenovirus, rotavirus and hepatitis A virus in discharged effluents of two wastewater treatment facilities in the Eastern Cape, South Africa. Food Environmental Virology, 8, 262–274.
Asami, T., Katayama, H., Torrey, J. R., Visvanathan, C., & Furumai, H. (2016). Evaluation of virus removal efficiency of coagulation-sedimentation and rapid sand filtration processes in a drinking water treatment plant in Bangkok, Thailand. Water Research, 101, 84–94. https://doi.org/10.1016/j.watres.2016.05.012.
Assis, A. S. F., Cruz, L. T., Ferreira, A. S., Bessa, M. E., de Oliveira, P. M. A., Vieira, C. B., et al. (2015). Relationship between viral detection and turbidity in a watershed contaminated with group A rotavirus. Environmental Science and Pollution Research, 22, 6886–6897. https://doi.org/10.1007/s11356-014-3874-8.
Atabakhsh, P., Kargar, M., & Doosti, A. (2019). Molecular detection and genotyping of group A rotavirus in two wastewater treatment plants, Iran. Brazilian of microbiology, 1–7. https://doi.org/10.1007/s42770-019-00131-0.
Banerjee, I., Ramani, S., Primrose, B., Iturriza-Gomara, M., Gray, J. J., Brown, D. W., & Kang, G. (2008). Modification of rotavirus multiplex RT-PCR for the detection of G12 strains based on characterization of emerging G12 rotavirus strains from South India. Medical Virology, 79(9), 1413–1421.
Barril, P. A., Prez, V. E., & Nates, S. V. (2016). Rotavirus monitoring in drinking and surface waters of Cordoba, Argentina. Gastrointestinal Diseases, 1–11. https://smjournals.com/ebooks/gastrointestinal-diseases/chapters/GD-16-01.pdf
Bosch, A., Guix, S., Sano, D., & Pinto, M. R. (2008). New tools for the study and direct surveillance of viral pathogens in water. Current Opinion in Biotechnology, 19, 295–301. https://doi.org/10.1016/j.copbio.2008.04.006.
Centers for Disease, C, Prevention. (2014). Rotavirus surveillance-worldwide. https://www.cdc.gov/vaccines/pubs/surv-manual/chpt13-otavirus.html.
Delgado-Gardea, M. C. E., Tamez-Guerra, P., Gomez-Flores, R., Mendieta-Mendoza, A., Zavala-Diaz de la Serna, F. J., et al. (2017). Prevalence of rotavirus genogroup A and norovirus genogroup II in Bassaseachic falls national park surface waters in Chihuahua, Mexico. International Journal of Environmental Research and Public Health, 14, 482.
Fongaro, G., Nascimento, M., Vianccelli, A., Tonetta, D., Petrucio, M., & Barardi, C. (2012). Surveillance of human viral contamination and physicochemical profiles in a surface water lagoon. Water Science and Technology, 66, 2682–2687. https://doi.org/10.2166/wst.2012.504.
Fout, G. S., Schaefer III, F. W., Messer, J. W., Dahling, D. R., & Stetler, R. E. (1996). Information collection rule (ICR) microbial laboratory manual. EPA/600/R-95/178. Washington, DC: U.S. Environmental Protection Agency.
Gibson, K. E., Opryszko, C. M., Schissler, J. T., Guo, Y., & Schwab, K. J. (2011). Evaluation of human enteric viruses in surface water and drinking water resources in Southern Ghana. American Society of Tropical Medicine and Hygiene, 84(1), 20–29.
Grassi, T., Bagordo, F., Idolo, A., Lugoli, F., Gabutti, G., & Donno, A. D. (2010). Rotavirus detection in environmental water samples by tangential flow ultrafiltration and RT-nested PCR. Environmental Monitoring and Assessment, 164, 199–205.
Grøndahl-Rosado, R. C., Yarovitsyna, E., Trettenes, E., Myrmel, M., & Robertson, L. J. (2014). A one year study on the concentrations of norovirus and enteric adenoviruses in wastewater and a surface drinking water source in Norway. Food Environmental Virology, 6, 232–245.
Hamza, I. A., Jurzik, L., Uberla, K., & Wilhelm, M. (2011). Methods to detect infectious human enteric viruses in environmental water samples. International Journal of Hygiene and Environmental Health, 214, 424–436.
Ibfelt, T., Frandsen, T., Permin, A., Andersen, L. P., & Schultz, A. C. (2016). Test and validation of methods to sample and detect human virus from environmental surfaces using norovirus as a model virus. Hospital Infection, 92(4), 378–384. https://doi.org/10.1016/j.jhin.2016.01.003
Ibrahim, C., Hassen, A., Pothier, P., Mejri, S., & Hammami, S. (2018). Molecular detection and genotypic characterization of enteric adenoviruses in a hospital wastewater. Environmental Science and Pollution Research, 25, 10977–10987.
Inker, L. A., Gerba, C. P., & Bright, K. R. (2012). Concentration and recovery of viruses from water: a comprehensive review. Food environmental virology, 4, 41–67.
Kargar, M., Javdani, N., Najafi, A., & Tahamtan, Y. (2013). First molecular detection of group A rotavirus in urban and hospital sewage systems by nested-RT PCR in Shiraz, Iran. Environmental Health Science Engineering, 11, 4. https://doi.org/10.1186/2052-336X-11-4.
Karim, M. R., Rhodes, E. R., Brinkman, N., Wymer, L., & Fout, G. S. (2009). New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Applied and Environmental Microbiology, 75, 2393–2399. https://doi.org/10.1128/AEM.00922-08.
Kiulia, N.M., Hofstra, N., Vermeulen, L.C., Obara, M.A., Medema, G., & Rose, J.B. (2015). Global Occurrence and Emission of Rotaviruses to Surface Waters. Pathogens, 4, 229-255. https://doi.org/10.3390/pathogens4020229.
Kluge, M., Fleck, J. D., Soliman, M. C., Luz, R. B., Fabres, R. B., Comerlato, J., et al. (2014). Human adenovirus (HAdV), human enterovirus (HEV), and genogroup A rotavirus (GARV) in tap water in southern Brazil. Water Health, 12, 26–532. https://doi.org/10.2166/wh.2014.202.
Lee, D. Y., Leung, K. T., Lee, H., & Habas, M. B. (2016). Simultaneous detection of selected enteric viruses in water samples by multiplex quantitative PCR. Water, Air, and Soil Pollution, 227, 107–111. https://doi.org/10.1007/s11270-016-2811-5.
Liu, J., Lurain, K. U., Sobuz, S., Begum, S., & Kumburu, H. (2015). Molecular genotyping and quantitation assay for rotavirus surveillance. Virological Methods, 213, 157–163.
Maite, M. (2016). Human norovirus in artificial and environmental marine water: development of antibody based rapid methods. a dissertation submitted to the graduate faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The School of Nutrition and Food Science.
Najafi, A., Kargar, M., & Jafarpour, T. (2012). Burden and typing of rotavirus group A in children with acute gastroenteritis in Shiraz, Southern Iran. Iranian Red Crescent Medical, 14, 531.
Rames, E., Roiko, A., Stratton, H., & Macdonald, J. (2016). Technical aspects of using human adenovirus as a viral water quality indicator. Water Research, 96, 308–326.
Rice, E. W., Baird, R. B., Eaton, A. D., & Clesceri, L. S. (2012). Standard methods for the examination of water and wastewater Washington. 22nd Edition, American Public Health Association, American Water Works, Water Environment Federation, Washington DC., USA.
Rizk, N. M., & Allayeh, A. K. (2018). Multiplex semi-nested RT-PCR for genotyping of rotaviruses group A in Giza Tap Water, Egypt. Asian Journal of Water, Environment and Pollution, 15, 217–221. https://doi.org/10.3233/AJW-180034.
Salvo, M., Lizasoain, A., Castells, M., Bortagaray, V., Castro, S., Colina, R., Tort, F. L., & Victoria, M. (2018). Human bocavirus: detection, quantification and molecular characterization in sewage and surface waters in Uruguay. Food environmental virology, 10, 193–200. https://doi.org/10.1007/s12560-017-9334-0.
Silva, M., Miagostovich, M., & Victoria, M. (2016). Rotavirus and astrovirus. global water pathogen project.
Soltan, M. A., Tsai, Y-L., Lee, P-YA., Tsai, C-F., Chang, H-FG., Wang, H-TT., et al. (2016). Comparison of electron microscopy, ELISA, real time RT-PCR and insulated isothermal RT-PCR for the detection of Rotavirus group A (RVA) in feces of different animal species. Virological methods, 235, 99–104.
Verheyen, J., Timmen-Wego, M., Laudien, R., Boussaad, I., Sen, S., Koc, A., et al. (2009). Detection of adenoviruses and rotaviruses in drinking water sources used in rural areas of Benin, West Africa. Applied and Environmental Microbiology, 75(9), 2798–2801.
WHO Geneva. (2009). Department of Immunization, Vaccines and Biological manual of rotavirus detection and characterization methods. CH-1211, Switzerland This publication is available on the Internet at: http://whqlibdoc.who.int/hq/2008/WHO_IVB_08.17_eng.pdf.
WHO. (2014). Progress on drinking water and sanitation: update. Geneva: Switzerland.
Ye, Y. X., Ming, X., Zhang, L. Y., Xiao, W. Q., Huang, X. N., Cao, Y. G., & Gu, K. D. (2012). Real-time PCR detection of enteric viruses in source water and treated drinking water in Wuhan, China. Current Microbiology, 65, 244–253. https://doi.org/10.1007/s00284-012-0152-1.
Yousuf, F. A., Siddiqui, R., & Ahmed Khan, N. (2017). Presence of rotavirus and free-living amoebae in the water supplies of Karachi, Pakistan. Sao Paulo Institute of Tropical Medicine, 59, 1–7. https://doi.org/10.1590/s1678-9946201759032.
Acknowledgments
The authors are grateful to the Islamic Azad University of Jahrom and Isfahan water treatment plant for their executive support of this project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not required.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Atabakhsh, P., Kargar, M. & Doosti, A. Molecular surveillance of human rotaviruses in drinking water and investigation of the efficiency of their removal in Isfahan water treatment plant. Environ Monit Assess 191, 759 (2019). https://doi.org/10.1007/s10661-019-7834-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-019-7834-0