Abstract
Escherichia coli can persist in streambed sediments and influence water quality monitoring programs through their resuspension into overlying waters. This study examined the spatial patterns in E. coli concentration and population structure within streambed morphological features during baseflow and following stormflow to inform sampling strategies for representative characterization of E. coli populations within a stream reach. E. coli concentrations in bed sediments were significantly different (p = 0.002) among monitoring sites during baseflow, and significant interactive effects (p = 0.002) occurred among monitoring sites and morphological features following stormflow. Least absolute shrinkage and selection operator (LASSO) regression revealed that water velocity and effective particle size (D 10) explained E. coli concentration during baseflow, whereas sediment organic carbon, water velocity and median particle diameter (D 50) were important explanatory variables following stormflow. Principle Coordinate Analysis illustrated the site-scale differences in sediment E. coli populations between disconnected stream segments. Also, E. coli populations were similar among depositional features within a reach, but differed in relation to high velocity features (e.g., riffles). Canonical correspondence analysis resolved that E. coli population structure was primarily explained by spatial (26.9–31.7 %) over environmental variables (9.2–13.1 %). Spatial autocorrelation existed among monitoring sites and morphological features for both sampling events, and gradients in mean particle diameter and water velocity influenced E. coli population structure for the baseflow and stormflow sampling events, respectively. Representative characterization of streambed E. coli requires sampling of depositional and high velocity environments to accommodate strain selectivity among these features owing to sediment and water velocity heterogeneity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Fecal contamination of water resources introduces enteric pathogens, which can be subsequently transmitted to human populations through recreational exposure and consumption of inadequately processed drinking water or irrigated food crops (Solomon et al. 2002; Reynolds et al. 2008; Yoder et al. 2008). Escherichia coli is the most commonly used fecal indicator bacterium (FIB) for monitoring freshwater resources, where it is assumed to have high fecal specificity and exhibit similar environmental fate and transport to fecal pathogens (Tallon et al. 2005). E. coli remains the primary FIB for water quality monitoring programs and for simulating microbial dynamics in deterministic watershed models, such as SWAT and WATFLOOD (Dorner et al. 2006; Kim et al. 2010). However, the appropriateness of E. coli as a FIB has been questioned given recent evidence that E. coli is capable of long-term persistence in forest soils, beach sands and freshwater sediments (Ishii et al. 2006; Whitman et al. 2006; Ishii et al. 2007; Halliday and Gast 2011). Recognition of persistent sediment-borne E. coli in watershed models through process-based representations of particle settling and resuspension has improved the efficacy of model simulations (Rehmann and Soupir 2009; Pandey et al. 2012).
Uncertainty in predicted stream water E. coli concentrations using these models has been attributed in part to uncertainty in E. coli concentrations within stream sediments, which can vary by several orders of magnitude (Cho et al. 2010; Kim et al. 2010). Although sediment E. coli concentration is an important parameter when modeling waterborne E. coli, and attempts have been made to model temporal variability in sediment E. coli concentrations (Kim et al. 2010), sediment sampling programs for model parameterization often do not consider the potential spatial variability possible within a stream reach. Single or composite grab samples are typically taken at the point of water collection, which is to represent the concentrations and population structure of sediment-borne E. coli along the investigated stream reach (Rehmann and Soupir 2009; Ouattara et al. 2011; Yakirevich et al. 2013). Fluvial sediments are characterized by considerable spatial heterogeneity in sediment properties brought about by differential patterns in water velocity and shear stress (Gordon et al. 2004). Since elevated concentrations of organic matter and percentages of silt and clay affect the environmental persistence of E. coli (Burton et al. 1987; Davies et al. 1995; Craig et al. 2004; Haller et al. 2009; Garzio-Hadzick et al. 2010), spatial variation in sediment E. coli concentrations may exist along a stream reach and lead to uncertainty in monitoring and modeling programs designed to link sediment- and waterborne E. coli. Greater understanding of the spatial variability existing in sediment E. coli concentrations could improve model performance by incorporating representative estimates of E. coli concentration within a stream reach. Model performance has been improved by incorporating spatial heterogeneity in particle properties (Pandey et al. 2012), and better representation of sediment E. coli concentrations may further improve model performance.
Another important consideration in modeling E. coli distribution is the potential for persistent sediment E. coli to dominate the microbial budget of waterborne E. coli in streams (Kim et al. 2010). Previous studies have attempted to associate sediment-borne E. coli populations to waterborne E. coli strains in an effort to link persistent E. coli strains to water quality impairment, often concluding that high variability in sediment and waterborne E. coli populations limited study outcomes (Kinzelman et al. 2004; Lu et al. 2004; Wu et al. 2009). In these studies, sediment sampling occurred at the point of water collection without consideration of possible spatial variability in E. coli population structure within the stream reach. Strain-dependent variability in the response of E. coli to environmental conditions has been demonstrated previously (Topp et al. 2003; Anderson et al. 2005; Pachepsky et al. 2008). Failure to represent the spatial variability inherent in a source population can affect confidence in source assignment of waterborne E. coli (Lu et al. 2004; Johnson et al. 2004). Insight into the spatial patterns of E. coli populations within streambed sediments would aid in guiding sampling design for studies aiming to relate clonal E. coli populations in sediments to waterborne E. coli strains.
The present study evaluated the influence of sediment properties and streambed geomorphology on the concentration and population structure of sediment-borne E. coli in a stream draining a rural, mixed-use watershed. The objectives of the study were to: (1) assess differences in E. coli populations among watershed monitoring sites and streambed morphological features; (2) examine the degree to which spatial and environmental variables explain streambed E. coli population variability; and (3) assess the stability of the observed spatial patterns during baseflow and following stormflow. This information was required to help design future studies examining temporal alterations in streambed strain composition and the relationship between sediment and waterborne E. coli.
Material and methods
Site description and sampling design
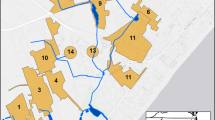
Sampling was conducted in the Thomas Brook Watershed (TBW), which is part of Agriculture and Agri-Food Canada’s Watershed Evaluation of Beneficial Management Practices (WEBs) program, located in the Annapolis Valley of Nova Scotia. The soils of the TBW are predominantly sandy loam textured Humo-Ferric Podzols (CSSC), and the area experiences average annual precipitation and temperatures of 1211 mm and 6.9 °C, respectively (Environment Canada 2013). Previous studies have concluded that high surface water E. coli levels are a critical water quality issue in this watershed (Jamieson et al. 2003; Sinclair et al. 2008). Three stream reaches located downstream from permanent monitoring locations (Sites 2, 3 and 4) were selected for study: Site 2 is downstream from a large dairy operation; Site 3 is influenced by low-density residential development; and Site 4 is in a mixed land-use area downstream from both Sites 2 and 3 (Fig. 1). At each stream reach, five morphological features were identified at stream meanders (point bars and bank scours), pool-riffle sequences and straight segments (i.e., runs). Each stream reach was sampled in the following order: riffle, pool, point bar, bank scour and run (Fig. 2).
At each morphological feature, triplicate sediment samples (200–300 g) were retrieved from the sediment–water interface (0 to 5 cm depth) using a lever-action grab sampler with a 950-cm3 bucket that was rinsed with stream water between each sample. The retrieved samples were stored at 4 °C and cultured for E. coli within 24 h of collection. Geographic coordinates of each sample location were obtained with a HiPerGa GPS system (Topcon Positioning Systems Inc., Livermore, CA, USA) for use in spatial statistics. Flow velocity was measured at each feature using a FlowTracker Acoustic Doppler Velocimeter (SonTek/YSI, San Diego, CA, USA). Channel geometry and water velocity are summarized for each sampling site and event in Table 1.
Duplicate sampling events were conducted to assess the stability of the observed spatial structure. The first sampling event occurred in August 2010 during a period of prolonged baseflow (~2 weeks), and the second event was conducted in September 2010, 3 days following a storm event (40 mm over 24 h) that lead to a 5- to 8-fold increase in discharge in the investigated reaches. Previous storm hydrographs measured on Thomas Brook indicate stormflow recession occurs within 24 to 48 h following a precipitation event. Sampling 3 days following the storm event ensured that no significant sediment redistribution was occurring at the time of sampling.
E. coli enumeration and isolation
Sediment E. coli concentration was measured using membrane filtration. Prior to analysis, sediment samples were sieved (2.38 mm, No. 8; W.S. Tyler, St. Catherines, ON, Canada) to remove coarse particles and homogenize the samples. Twenty grams of the <2.38 mm fraction were resuspended in 180 ml of sterile peptone–saline (0.1 % peptone, 0.85 % NaCl) by handshaking for 60 s. The supernatant was collected after a settling time of 10 min, filtered through 0.45-μm cellulose-nitrate membranes (Whatman Laboratory Division, Maidstone, England), and incubated on mFC basal media supplemented with 3-bromo-4-chloro-5-indolyl-β-glucopyranoside (BCIG; Inverness Medical, Ottawa, ON, Canada) for 2 h at 35 °C, then overnight at 44.5 °C. Distinctly separate, blue E. coli colonies were counted and converted to concentration per dry weight of sediment. Presumptive E. coli were purified on Sorbitol-MacConkey agar (Oxoid, Ltd., Hampshire, England) and confirmed as E. coli through enzymatic (DMACA Indole; Difco Laboratories, Sparks, MD, USA) and molecular procedures described below. Prior to DNA extraction, indole-positive isolates were cultured in tryptic soy broth (TSB; Difco Laboratories) at 37 °C for 24 h. Twelve E. coli isolates were taken from each replicate sample of the morphological features, yielding 36 isolates per morphological feature per site. This generated a total of 540 isolates for each sampling event and a total of 1080 isolates.
DNA extraction and genetic analysis
DNA was extracted from the broth cultures using prepGEM Bacteria DNA kits (ZyGEM Corporation, Ltd., Hamilton, New Zealand) following manufacturer’s protocols. Each isolate was genetically identified as E. coli through the phylogenetic grouping procedure developed by Clermont et al. (2000), which assigns isolates to one of four phylogenetic groups based on the presence of three target genes. Amplification conditions described by Clermont et al. (2000) were used, except the denaturation time was extended to 10 s and the annealing and extension time was extended to 15 s to consistently amplify all three bands in the control strain E. coli ATCC 25922. Strain typing was performed using repetitive element palindromic (rep)-PCR using BOX A1R primers (BOX-PCR), following protocols reported by Rademaker et al. (2004). Polyacrylamide gel electrophoresis was used, with 3.5 % gels run at 7.5 V/cm for 195 min. E. coli strain ATCC 25922 was used as a positive control for intergel comparison, and AmpliSize Molecular Ruler (50–2000 bp; Bio-Rad, Hercules, CA, USA) was used as a size standard. The PAGE gels were stained with ethidium bromide and imaged using an ImageMaster VDS-CL documentation system (Amersham Pharmacia Biotech, Inc., UK).
Computer-assisted image analysis and cluster assignment
BOX-PCR images were analyzed with GeneTools software (Syngene Ltd., Frederick, MD, USA). Band matching was performed using the rolling disk method for background subtraction and band sizes between 50 and 2,000 bp were used in subsequent cluster analysis. Cluster analysis was performed with GeneDirectory software (Syngene Ltd.) through unweighted pair group method with arithmetic mean (UPGMA), using the Dice coefficient of similarity and a 1 % threshold. Isolates exhibiting ≥90 % similarity were classified as clonal strains.
Sediment analyses
The >2.38-mm sieved fraction was combined with the sieved material (<2.38 mm) for determining particle size distribution (PSD). The reconstituted sample was dried at 105 °C for 24 h and mechanically sieved to determine mass distributions for particle classes between 6 and 0.25 mm. For samples where the <0.25-mm fraction was >10 % of the total dry mass of the sediment, a laser in situ scanning and transmissometer (LISST-100X; Sequoia Scientific Inc., Bellevue, WA, USA) was used to determine the PSD between 0.0025 and 0.25 mm. Here, approximately 100 mg of the <0.25-mm sediment was dispersed in 10 ml of 5 % hexametaphosphate solution, diluted with 110 ml deionized water in the LISST-100X mixing chamber attachment, and analyzed for 30 repetitions. Particle size concentrations were converted to proportions by dividing the average for each class by the sum of all classes. The mass of each class was calculated by multiplying the class proportion with the final mass of the <0.25-mm sieve fraction. All mass data were combined and particle size properties were calculated with GRADISTAT software (version 4.0; Blott and Pye 2001). The calculated values included geometric mean diameter, sorting, percent sand (>0.063 mm), percent silt/clay (<0.063 mm), D 75/25 (ratio of interquartile particle diameters), median particle diameter (D 50) and effective particle size (D 10), where 10 % of the particles in that sample (by weight) are of a smaller diameter. Organic carbon concentration was determined by the dichromate redox titration method outlined by Skjemstad and Baldock (2008). Approximately 1 g of dry sediment from the <0.25-mm sample was used in the analysis.
Statistical analysis of E. coli concentrations
The significance of spatial location on sediment E. coli concentration was assessed using two-way analysis of variance (ANOVA) in SigmaPlot (version 11.0; SYSTAT Software Inc., Chicago, IL, USA), where monitoring site and morphological feature served as the main factors. Tukey’s test was used for post-hoc determination of significant differences (p ≤ 0.05) among factors. Prior to analysis, E. coli concentrations were computed per gram of wet weight sediment and log transformed. Least absolute shrinkage and selection operator (LASSO) regression was performed in MATLAB (Version 2012a; The MathWorks Inc., Natick, MA, USA) to determine the environmental variables that explained sediment E. coli concentration. LASSO regression was performed on normalized variables to examine the relative effect of the variables by removing measurement scale, and the model chosen had the lowest mean squared error calculated through 10-fold cross-validation.
Richness and similarity of E. coli populations in stream morphological features
The richness of E. coli strains observed the stream morphological features was estimated through rarefaction procedures described by Lu et al. (2005). Rarefaction curves were generated through the freeware program Analytical Rarefaction 1.3, available at http://www.uga.edu/strata/software/. The rarefaction curves were plotted in SigmaPlot (v11.0; Systat Software Inc.), and the asymptotes were estimated using a one-site saturation ligand model. The asymptote (V max) estimates the strain richness at sampling saturation, and the K d value estimates the number of isolates required to capture 50 % of the estimated richness. All isolates obtained from each morphological feature (n = 36) were used to build the rarefaction curves.
Principle coordinates analysis (PCoA) was conducted in PAST (version 2.11; Hammer et al. 2001) software to visualize similarities in E. coli population structure existing among monitoring sites and fluvial features. The intent of this analysis was to visualize patterns of E. coli population similarity based on the monitoring sites and streambed morphological features. Ordinations were generated using Euclidean distances on Hellinger transformed abundance data (Legendre and Gallagher 2001). All isolates (n = 36) obtained from each fluvial feature were composited to aid in visualization, and a transformation exponent of 4 was chosen.
Variation partitioning between spatial and environmental variables
Variation partitioning was used to inform whether clonal E. coli populations exhibit spatial clustering or random distribution in the streambed, and identify whether these distributions result from responses to environmental and/or spatial gradients. Spatial explanatory variables [S] were produced using topological-based Moran’s eigenvector maps, generated from the MATLAB code produced and distributed by Griffith and Peres-Neto (2006). All of the listed PSD properties, organic carbon and water velocity were used as environmental explanatory variables [E]. Partial canonical correspondence analysis (pCCA) was used to evaluate the influence of environmental variables in the absence of spatial autocorrelation [E|S], and the influence of spatial location without the influence of environmental gradients [S|E]. These analyses were performed using CANOCO Software (version 4.5; Plant Research International, Wageningen, The Netherlands). For all analyses, biplot scaling was used focusing on inter-sample differences, and rare species were downweighted. Automatic selection of variables was conducted using 999 unrestricted Monte Carlo permutations. For pCCA analysis of environmental variables, only significant (p < 0.05) spatial variables were used as covariables to retain high analytical power (Peres-Neto and Legendre 2010).
Results
Influence of sampling site and fluvial morphology on E. coli concentration
For the baseflow sampling event, sediment E. coli concentrations among the fluvial morphological features were not significantly different (p = 0.0697), but varied significantly among monitoring sites (p = 0.002) (Fig. 3a). Following the storm event, E. coli concentration exhibited significant interactions (p = 0.002) indicating that differences among morphological features varied by site. High sediment E. coli concentrations existed in the pools following stormflow, where it was highest at Sites 3 and 4, and second highest at Site 2 (Fig. 3b). Overall, sediment E. coli concentrations were greater at all sites after the September storm event compared to the August baseflow sampling.
Average sediment E. coli concentration (ln CFU/g) collected within fluvial morphological features sampled at each stream reach for the: a baseflow sampling event and b post-stormflow sampling event. From left to right, the grayscale bars indicate point bar, bank scour, pool, riffle and run for all groupings. Error bars represent standard deviation (n = 3)
LASSO regression with normalized variables was used to determine the relative influence of environmental factors on sediment E. coli concentrations in the absence of measurement scale. The resultant beta coefficients demonstrate the magnitude of change in sediment E. coli given one standard deviation change in the predictor variable within the system studied, and should not be interpreted as broadly applicable regression coefficients. During baseflow, sediment E. coli concentrations were observed to be influenced by water velocity and effective particle size (D 10) according to the following equation:
Both variables have negative beta coefficients, suggesting that lower effective particle size (D 10) and velocity are associated with higher sediment E. coli concentrations. Velocity had a greater influence on sediment E. coli concentrations than texture during baseflow, according to the magnitude of the beta coefficients.
Following stormflow, organic carbon, water velocity and median particle diameter (D 50) were included in the regression equation:
Similar to baseflow, sediment texture and velocity are negatively associated with sediment E. coli concentrations, with velocity exhibiting greater influence than texture. Organic carbon is positively associated with E. coli concentrations, suggesting that higher organic carbon is associated with higher E. coli concentrations. Furthermore, organic carbon appears to exhibit a greater relative effect on E. coli concentration than velocity and sediment texture. Average values of the sediment variables and velocity for both baseflow and stormflow are included in the Online Resources.
E. coli strain similarity among sites and morphological features
Repetitive element analysis (BOX-PCR) separated the 1080 E. coli isolates into 274 genotypes, with 82 genotypes uniquely identified during baseflow, 121 genotypes uniquely identified following stormflow, and 71 genotypes present on both sampling occasions. High diversity in E. coli genotypes was observed in the sediments studied. During baseflow, runs and bank scours exhibited the highest estimated E. coli genotype richness, followed by point bars, pools, and then riffles (Table 2). The sampling effort required to characterize 50 % of the total genotypes present ranged from 41 to 100 isolates (Table 2). Following stormflow, all morphological features demonstrated an increase in the number of estimated genotypes present, ranging from 88 to 107 genotypes (Table 2). The sampling effort required to characterize 50 % of the total strains also increased, ranging from 93 to 105 isolates. Within a stream reach, assuming a composite sample of morphological features is obtained, the number of genotypes present was 55 to 77 genotypes during baseflow, increasing to 79 to 117 genotypes following stormflow (Table 2). The sampling effort required to characterize 50 % of the genotypes in a stream reach was 58 to 83 isolates during baseflow, and 77 to 122 isolates following stormflow.
E. coli population similarity among the sites and stream morphological features was visualized with PCoA. For the baseflow event, coordinates 1 and 2 explained 32.9 % and 14.7 % of the variance, respectively (Fig. 4). Samples from Sites 2 and 4 demonstrated clustering indicating higher population similarity, whereas samples from Site 3 were dissimilar on the basis of their distance from Sites 2 and 4 in the PCoA ordination. A common spatial pattern was evident among all sites, where populations associated with point bars, bank scour and pools exhibited similarity, whereas riffles exhibited a lower degree of similarity (i.e., greater multivariate distance) to other morphological features. The likeness of E. coli populations found in runs varied, as these populations were similar to the riffles at Site 2, depositional environments (i.e., pools and point bars) at Site 4, and dissimilar to all features at Site 3.
Principle coordinate analysis ordination diagram to explain variations in the E. coli community composition for the baseflow sampling event. Black circles represent Site 2, crosses denote Site 3 and open boxes indicate Site 4. Morphological features: Pb point bars, Bs bank scours, P pools, R for riffles, S runs
Following stormflow, coordinates 1 and 2 explained 33.0 % and 22.4 % of the E. coli population variance, respectively (Fig. 5). A similar spatial structure to the baseflow event was observed, where Sites 2 and 4 showed greater population similarity compared with Site 3, and the depositional areas exhibited a high degree of similarity. However, streambed features characterized by higher velocities tended to cluster together, particularly at Site 3, where populations at the bank scour, riffle and runs exhibited higher similarity. Bank scours at Sites 2 and 4 showed less similarity to the depositional areas than was observed during baseflow. The riffles of Sites 2 and 4 still clustered together, along with the run of Site 2. The run at Site 4 clustered with the depositional areas, similar to the baseflow event. Overall, other than differences in the bank scour feature, the spatial structure among the sites appeared relatively consistent between the sampling events.
Principle coordinate analysis ordination diagram to explain variations in the E. coli community composition for the post-stormflow sampling event. Black circles represent Site 2, crosses denote Site 3 and open boxes indicate Site 4. Morphological features: Pb point bars, Bs bank scours, P pools, R riffles, S runs
Variation partitioning to determine the influence of spatial and environmental variables on E. coli populations
For both sample events, spatial variables explained a greater proportion of variance in E. coli population structure than environmental variables, with spatial eigenvectors [S] explaining 26.9 % of the population variance during baseflow and 31.7 % following stormflow (Table 3). Comparatively, environmental variables [E] explained 9.2 % of the variance during baseflow and 13.1 % of the variance following stormflow. Variance in E. coli population structure explained jointly by environmental and spatial variables [E ∩ S] was relatively low both for baseflow (1.8 %) and following stormflow (2.9 %), suggesting low spatial structuring of the environmental variables according to the spatial eigenvectors included in the analysis. The residual, or unexplained, variance [R] is 63.9 % during baseflow and 55.2 % following stormflow, indicating that variables other than those modeled greatly influenced E. coli population structure.
A total of 26 Moran's eigenvectors with positive eigenvalues were produced through spatial analysis. For the baseflow event, spatial partial canonical coordinates analysis (pCCA) [S|E] revealed that Moran’s eigenvectors 2 (ME-2) and 10 (ME-10) were significant in explaining strain variation. Comparatively, ME-2 and ME-7 explained E. coli strain variation following stormflow. Plots of these eigenvectors suggest that ME-2 represents spatial autocorrelation, or clustering, based on site location, and ME-7 and ME-10 represent spatial autocorrelation according to morphological features (Online Resources). Environmental pCCA [E|S] revealed that mean particle diameter (p = 0.037) and water velocity (p = 0.025) explained the variance in E. coli population structure during baseflow and following stormflow, respectively.
Discussion
Influence of sampling site, morphological feature and environmental variables on E. coli concentration
The observed differences in sediment E. coli concentrations among sampling sites, and the higher concentration of E. coli following stormflow, are likely associated with variable fecal loading into the stream system affected by upstream land use. Sinclair et al. (2008), in examining the same watershed, found that waterborne bacterial loading was highest in subcatchments containing livestock operations (i.e., Site 2), and increased throughout the growing season and during stormflow events. Likewise, waterborne E. coli concentrations have been reported to be greater during stormflow events and adjacent to livestock operations in other studies (McKergow and Davies-Colley 2010; Pachepsky and Shelton 2011).
During prolonged baseflow, E. coli concentrations were different among sites, but not among morphological features within a stream reach. This result is surprising considering that water velocity and effective particle size (D 10) were identified as important predictors in the LASSO regression equation, and that morphological features are defined by sediment textural differences brought about by differential velocity and shear stress distributions (Charlton 2008). Heterogeneity in sediment texture within the morphological features could explain the non-significant difference among morphological features. For example, Powell (1998) reported sediment fining from the head to tail of depositional bars, indicating textural difference within the same morphological feature. Although sediment properties were different among morphological features (Online Resources), considerable variability was observed within the samples retrieved.
Following stormflow, sediment E. coli concentrations were variable among the morphological features depending on the stream reach, although pools exhibited high E. coli concentrations at each site. Higher deposition in pools is expected since these features are characterized by lower velocity and settling of finer particles, which are in turn associated with greater E. coli attachment (Oliver et al. 2007). Similar to baseflow, water velocity and sediment texture (D 50) explained variance in sediment E. coli concentrations, but organic matter had greater relative influence on E. coli concentrations than both velocity and texture. The importance of organic matter following stormflow, but not during baseflow, supports the postulation of Pachepsky and Shelton (2011) that sediment E. coli concentrations are positively associated with organic matter following runoff events as bacteria enter the stream together with an influx of fecal organic matter, whereas organic production during baseflow is disassociated with aboveground inputs. In this study, sediment organic carbon concentrations were higher following stormflow than during baseflow (Online Resources), reflecting the possibility of runoff inputs.
Sediment textural differences reflect velocity and shear stress distributions within a stream reach. In our study, finer sediments (lower D 10 and D 50) were associated with higher E. coli concentrations. The significance of median particle diameter and effective particle size on E. coli numbers as opposed to explicit percentages of silt and clay is in contrast to other studies that relate silt–clay percentage to sediment E. coli concentration (Haller et al. 2009; Garzio-Hadzick et al., 2010). High E. coli concentrations have been previously observed in fine sand sediments (Cinotto 2005), which may reflect greater nutrient exchange via hyphoreic flow in the pore space of sandy sediments (Grant et al. 2011).
Although sediment texture and organic matter influence E. coli persistence in sediments, there appears to be no significant relationship between E. coli concentrations and fluvial morphological features during baseflow. The lack of statistical relationship could result from sediment heterogeneity and, consequently, statistical homogenization of E. coli persistence within a stream reach. Spatial structuring of E. coli concentration among morphological features within a stream reach only appears to be relevant following stormflow events, where recent inputs and limited die-off have occurred. Adequate characterization of sediment E. coli concentrations for the purposes of monitoring or modeling should capture the variability within a stream reach. There exists a clear relationship of E. coli concentrations to sediment properties and water velocity, but the relationship to particular streambed morphological features is not temporally stable. Sampling programs should focus on the collection of multiple samples of sediments consisting of a variety of textures for representative characterization of a reach, although targeting particular morphological features does not appear to be necessary.
E. coli diversity and strain similarity among sites and morphological features
Site-level differences in E. coli population structure were observed between Sites 2 and 3, presumably due to differences in fecal inputs: large dairy farm versus low-density residential dwellings with on-site wastewater systems. The E. coli population at Site 4 exhibited high similarity to Site 2, illustrating that the higher loading of fecal inputs at Site 2 has a greater influence on the downstream Site 4. Wu et al. (2009) also reported a similar association of downstream waterborne E. coli isolates with isolates from upstream sediments. Walk et al. (2007) observed homogenous sediment E. coli populations across sites within beach sand, suggesting association with a well mixed waterborne population. The association between Sites 2 and 4 could be due to high similarity in waterborne strains owing to high fecal loading at the upstream site, but comparison with the waterborne E. coli population is required to support this assertion.
Within each stream segment, greater similarity in E. coli populations occurred among low water velocity depositional features compared to high velocity features. The population dissimilarity does not appear to be a function of strain richness, as all features demonstrated relatively consistent strain richness, other than the riffle features during baseflow. Thus, it appears that some characteristic of high water velocity features are selecting for different strains within the streambed, possibly due to the capacity for these strains to resist migration either through the production of, or association with, biofilms (Droppo et al. 2009; Hirotani and Yoshino 2010). Conversely, the selection for E. coli strains in depositional environments could be attributed to strain-specific differences in attachment to, and deposition of, suspended particles of various sizes (Oliver et al. 2007; Pachepsky et al. 2008). Strain-dependent survival of E. coli strains in sediments has been reported previously (Anderson et al. 2005). This study shows that sediment heterogeneity along a stream reach affects which strains persist in fluvial morphological features.
The observed spatial pattern appeared to be fairly consistent between the two sampling events, suggesting stability in the observed spatial structuring in terms of certain E. coli genotypes preferentially existing in depositional features while others preferentially exist at high velocity features. However, E. coli populations in bank scours exhibited variability, where the populations were similar to depositional environments during baseflow, but not following stormflow. The similarity in bank scour populations to depositional environments during baseflow could result from comparable texture, as bank scour sediments in this system are fine textured and poorly sorted (Tables S1 and S2), or as a consequence of dispersal from the point bar or pools, which are located in close proximity to the bank scours. Determining the relative importance of spatial and environmental factors in explaining E. coli population structure could provide evidence to the dominant force affecting E. coli populations in these environments.
Variation partitioning to determine the influence of spatial and environmental variables on E. coli populations
Spatial autocorrelation, or clustering, was observed within monitoring sites on both sampling occasions indicating that E. coli populations within each site are more similar than among sites. In this context, spatial autocorrelation is the degree to which E. coli genotypes exhibit higher similarity as a function of sample distance rather than as a response to ecological gradients. However, population similarity was also observed between Sites 2 and 4, suggesting that the site-level autocorrelation results from the disconnected Sites 2 and 3. Indeed, ME-2, which was significant at both sampling events, emphasizes the separation of Site 3 from Sites 2 and 4, further supporting the conjecture that high E. coli loading at Site 2 is contributing to the E. coli found at the downstream Site 4. In the absence of variance explained by environmental gradients, spatial autocorrelation was observed within morphological features, suggesting that E. coli populations within fluvial features are more similar than populations among features within a stream reach, perhaps due to dispersal limitations.
Although spatial variables dominated the explained variance, environmental gradients were also important for structuring E. coli populations. During baseflow, mean particle diameter was found to explain E. coli population variance suggesting that texture selects for different E. coli strains, possibly a result of differences in nutrient acquisition, predation, UV damage or biofilm association (Craig et al., 2004; Haller et al., 2009). Following stormflow, water velocity explained E. coli population variance, likely a result of differences in velocity-dependent particle settling behavior among the morphological features. Since strains vary in their attachment to particles of various sizes (Pachepsky et al. 2008), differential sedimentation could result in a strain sorting effect.
The combination of spatial and environmental influences on E. coli strain composition can be explained through ecological metacommunity theory. According to Cottenie (2005), the combined significance of spatial dependence in the absence of environmental gradients [S|E] and response to environmental gradients after the removal of spatial autocorrelation [E|S] denotes mass-effect, or source-sink, ecological dynamics. For this structuring effect to occur within E. coli populations, strains must exhibit preference for ecological gradients and be subject to sufficient dispersal, such that they emigrate from environments where they are good competitors (source) to environments where they are bad competitors (sink; Leibold et al. 2004). Adaptation, or preferential deposition, of strains to particular sediment environments yields a source effect, whereas dispersal occurring from frequent patterns of sediment resuspension or hyporheic exchange (Grant et al. 2011) disperses the strains to sink environments. The statistical methods used in this study demonstrated the importance of both spatial and ecological gradients in clonal E. coli population structure. However, future research should consider explicit representation of spatial boundaries and downstream dispersal conditions inherent in stream systems by using dendritic ecological networks (Peterson et al. 2013).
The observed source-sink effect is an important consideration when characterizing sediment E. coli populations. Previous studies linking sediment E. coli to waterborne load have taken sediment samples at the point of water collection (Lu et al. 2004; Kinzelman et al., 2004; Wu et al. 2009). Considering the streambed environment affects strain-sorting among low velocity deposition and high velocity features, a targeted sampling campaign is required to capture the diversity of strains found within a reach in order to develop unbiased, representative sediment libraries required to confidently assign waterborne E. coli strains to sources (Johnson et al. 2004). Particular focus should be paid to depositional environments (point bars and pools) and riffles, since these environments demonstrated the greatest dissimilarity of E. coli populations.
Conclusion
Spatial heterogeneity of sediment properties and water velocity exerts a selective effect on E. coli concentration and population structure, although the differences are not necessarily reflected among fluvial morphological features within a stream reach. Sampling programs attempting to characterize sediment E. coli concentrations and population structures should consider heterogeneity in streambed properties, rather than collecting sediments at the point of water collection. At a minimum, representative characterization of E. coli concentrations should be performed at each stream reach of concern, particularly where different land use inputs occur, and should capture differences in sediment properties in depositional areas (pools and point bars) and higher velocity features (riffles and runs). High diversity in stream sediments requires substantial isolate characterization, where up to 120 isolates are required to identify 50 % of the isolates within a stream reach. Strain-sorting based on spatial and environmental variables should be accommodated in sampling programs, by collecting samples from depositional and high velocity features. Better representation of streambed E. coli concentrations and population structure can increase confidence in waterborne E. coli modeling and source assignment.
References
Anderson, K. L., Whitlock, J. E., & Harwood, V. J. (2005). Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Applied and Environmental Microbiology, 71(6), 3041–3048.
Blott, S. J., & Pye, K. (2001). GRADISTAT: a grain size distribution and statistics package for the analysis of unconsolidated sediments. Earth Surface Processes and Landforms, 26, 1237–1248.
Burton, G. A., Jr., Gunnison, D., & Lanza, G. R. (1987). Survival of pathogenic bacteria in various freshwater sediments. Applied and Environmental Microbiology, 53(4), 633–638.
Charlton, R. (2008). Fundamentals of fluvial geomorphology. New York, NY: Routledge. 234 pp.
Cho, K. H., Pachepsky, Y. A., Kim, J. H., Guber, A. K., Shelton, D. R., & Rowland, R. (2010). Release of Escherichia coli from the bottom sediment in a first-order creek: experiment and reach-specific modeling. Journal of Hydrology, 391(3–4), 322–332.
Cinotto, P. J. (2005). Occurrence of fecal-indicator bacteria and protocols for identification of fecal-contaminated sources in selected reaches of the West Branch Brandywine Creek, Chester County, Pennsylvania. U.S. Geological Survey Scientific Investigations Report 2005–5039. 91 p.
Clermont, O., Stephane, B., & Bingen, F. (2000). Rapid and simple determination of Escherichia coli phylogenetic group. Applied and Environmental Microbiology, 66(10), 4555–4558.
Cottenie, K. (2005). Integrating environmental and spatial processes in ecological community dynamics. Ecology Letters, 8, 1175–1182.
Craig, D. L., Fallowfield, H. J., & Cromar, N. J. (2004). Use of microcosms to determine persistence of Escherichia coli in recreational coastal water and sediment and validation with in situ measurements. Journal of Applied Microbiology, 96, 922–930.
Davies, C. M., Long, J. A. H., Donald, M., & Ashbolt, N. J. (1995). Survival of fecal microorganisms in marine and freshwater sediments. Applied and Environmental Microbiology, 61(5), 1888–1896.
Dorner, S. M., Anderson, W. B., Slawson, R. M., Kouwen, N., & Huck, P. M. (2006). Hydrologic modeling of pathogen fate and transport. Environmental Science and Technology, 40(15), 4746–4753.
Droppo, I. G., Liss, S. N., Williams, D., Nelson, T., Jaskot, C., & Trapp, B. (2009). Dynamic existence of waterborne pathogens in river sediment compartments: implications for water quality regulatory affairs. Environmental Science and Technology, 43(6), 1737–1743.
Environment Canada, 2013. Canadian climate normals 1971–2000: Kentville CDA, Nova Scotia. http://climate.weatheroffice.gc.ca/climate_normals/index_e.html
Garzio-Hadzick, A., Shelton, D. R., Hill, R. L., Pachepsky, Y. A., Guber, A. K., & Rowland, R. (2010). Survival of manure-borne E. coli in streambed sediment: effects of temperature and sediment properties. Water Research, 44, 2753–2762.
Gordon, N. D., McMahon, T. A., Finlayson, B. L., Gippel, C. J., & Nathan, R. J. (2004). Stream hydrology: An introduction for ecologists, 2nd Edition. West Sussex, England: John Wiley & Sons.
Grant, S. B., Litton-Mueller, R. M. & Ahn, J. H. (2011). Measuring and modeling the flux of fecal bacteria across the sediment–water interface in a turbulent stream. Water Resources Research, 47(5). doi:10.1029/2010WR009460.
Griffith, D. A., & Peres-Neto, P. R. (2006). Spatial modeling in ecology: the flexibility of eigenfunction spatial analyses. Ecology, 87(10), 2603–2613.
Haller, L., Amedegnato, E., Pote, J., & Wildi, W. (2009). Influence of freshwater sediment characteristics on persistence of fecal indicator bacteria. Water, Air, and Soil Pollution, 203, 217–227.
Halliday, E., & Gast, R. J. (2011). Bacteria in beach sands: an emerging challenge in protecting coastal water quality and bather health. Environmental Science and Technology, 45, 370–379.
Hammer, O., Harper, D. A. T., & Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Paleontologica Electronica, 4(1), 9 pp.
Hirotani, H., & Yoshino, M. (2010). Microbial indicators in natural biofilms developed in the riverbed. Water Science and Technology, 62(5), 1149–1153.
Ishii, S., Ksoll, W. B., Hicks, R. E., & Sadowsky, M. J. (2006). Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Applied and Environmental Microbiology, 72(1), 612–621.
Ishii, S., Hansen, D. L., Hicks, R. E., & Sadowsky, M. J. (2007). Beach sand and sediments are temporal sinks and sources of Escherichia coli in Lake Superior. Environmental Science and Technology, 41, 2203–2209.
Jamieson, R., Gordon, R., Tattrie, S., & Stratton, G. (2003). Sources and persistence of fecal coliform bacteria in a rural watershed. Water Quality Research Journal of Canada, 38, 33–47.
Johnson, L. A. K., Brown, M. B., Carruthers, E. A., Ferguson, J. A., Dombek, P. E., & Sadowsky, M. J. (2004). Sample size, library composition and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Applied and Environmental Microbiology, 70(8), 4478–4485.
Kim, J.-W., Pachepsky, Y. A., Shelton, D. R., & Coppock, C. (2010). Effect of streambed bacteria release on E. coli concentrations: monitoring and modeling with the modified SWAT. Ecological Modelling, 221(12), 1592–1604.
Kinzelman, J., McLellan, S. L., Daniels, A. D., Cashin, S., Singh, A., Gradus, S., & Bagley, R. (2004). Non-point source pollution: determination of replication versus persistence of Escherichia coli in surface water and sediments with correlation of levels to readily measurable environmental parameters. Journal of Water and Health, 2(2), 103–114.
Legendre, P., & Gallagher, E. D. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia, 129, 271–280.
Leibold, M. A., Holyoak, M., Mouquet, N., Amarasekare, P., Chase, J. M., Hoopes, M. F., et al. (2004). The metacommunity concept: a framework for multi-scale community ecology. Ecology Letters, 7, 601–613.
Lu, L., Hume, M. E., Sternes, K. L., & Pillai, S. D. (2004). Genetic diversity of Escherichia coli isolates in irrigation water and associated sediments: implications for source tracking. Water Research, 38(18), 3899–3908.
Lu, Z., Lapen, D., Scott, A., Dang, A., & Topp, E. (2005). Identifying host sources of fecal pollution: diversity of Escherichia coli in confined dairy and swine production systems. Applied and Environmental Microbiology, 71(10), 5992–5998.
McKergow, L. A., & Davies-Colley, R. J. (2010). Stormflow dynamics and loads of Escherichia coli in a large mixed land use catchment. Hydrologic Processes, 24, 276–289.
Oliver, D. M., Clegg, C. D., Heathwaite, A. L., & Haygarth, P. M. (2007). Preferential attachment of Escherichia coli to different particle size fractions of an agricultural grassland soil. Water, Air, and Soil Pollution, 185, 369–375.
Ouattara, N. K., Passerat, J., & Servais, P. (2011). Faecal contamination of water and sediment in the rivers of the Scheldt drainage network. Environmental Monitoring and Assessment, 183, 243–257.
Pachepsky, Y. A., & Shelton, D. R. (2011). Escherichia coli and fecal coliforms in freshwater and estuarine sediments. Critical Reviews in Environmental Science and Technology, 41(12), 1067–1110.
Pachepsky, Y. A., Yu, O., Karns, K. S., Shelton, D. R., Guber, A. K., & van Kessel, J. S. (2008). Strain-dependent variations in attachment of E. coli to soil particles of different sizes. International Agrophysics, 22, 61–66.
Pandey, P. K., Soupir, M. L., & Rehmann, C. R. (2012). A model for predicting resuspension of Escherichia coli from streambed sediments. Water Research, 46(1), 115–126.
Peres-Neto, P. R., & Legendre, P. (2010). Estimating and controlling for spatial structure in the study of ecological communities. Global Ecology and Biogeography, 19, 174–184.
Peterson, E. E., Ver Hoef, J. M., Isaak, D. J., Falke, J. A., Fortin, M.-J., Jordan, C. E., McNyset, K., Monestiez, P., Ruesch, A. S., Sengupta, A., Som, N., Steel, E. A., Theobald, D. M., Torgersen, C. E., & Wenger, S. J. (2013). Modelling dendritic ecological networks in space: an integrated network perspective. Ecology Letters, 16, 707–719.
Powell, D. M. (1998). Patterns and processes of sediment sorting in gravel-bed rivers. Progress in Physical Geography, 22(1), 1–32.
Rademaker, J. L.W., Louws, F. J., Versalovic, J., & de Bruijn, F. J. (2004). Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. Akkermans, & J. D. van Elsas (Eds.), Molecular microbial ecology manual (Vol. 1 and 2, pp. 611–643). Dordrecht, The Netherlands: Kluwer Academic Publishers.
Rehmann, C. R., & Soupir, M. L. (2009). Importance of interactions between the water column and the sediment for microbial concentrations in streams. Water Research, 43(18), 4579–4589.
Reynolds, K. A., Mena, K. D., & Gerba, C. P. (2008). Risk of waterborne illness via drinking water in the United States. Reviews of Environmental Contamination and Toxicology, 192, 117–158.
Skjemstad, J. O., & Baldock, J. A. (2008). Total and organic carbon. In M. R. Carter & E. D. Gregorich (Eds.), Soil sampling and methods of analysis (2nd ed., pp. 225–237). Boca Raton, FL: Canadian Society of Soil Science, Taylor & Francis Group.
Sinclair, A., Hebb, D., Jamieson, R., Gordon, R., Benedict, K., Fuller, K., et al. (2008). Growing season surface water loading of fecal indicator organisms within a rural watershed. Water Research, 43, 1199–1206.
Solomon, E. B., Yaron, S., & Matthews, K. R. (2002). Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Applied and Environmental Microbiology, 68(1), 397–400.
Tallon, P., Magajna, B., Lofranco, C., & Leung, K. T. (2005). Microbial indicators of faecal contamination in water: a current perspective. Water, Air, and Soil Pollution, 166, 139–166.
Topp, E., Welsh, M., Tien, Y.-C., Dang, A., Lazaro, G., Conn, K., & Zhu, H. (2003). Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiology Ecology, 44, 303–308.
Walk, S. T., Alm, E. W., Calhoun, L. M., Mladonicky, J. M., & Whittman, T. S. (2007). Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environmental Microbiology, 9(9), 2274–2288.
Whitman, R. L., Nevers, M. B., & Byappanahalli, M. N. (2006). Examination of watershed-wide distribution of Escherichia coli along southern Lake Michigan: an integrated approach. Applied and Environmental Microbiology, 72(11), 7301–7310.
Wu, J., Rees, P., Storrer, S., Alderisio, K., & Dorner, S. (2009). Fate and transport modeling of potential pathogens: the contribution from sediments. Journal of the American Water Resources Association, 45(1), 35–44.
Yakirevich, A., Pachepsky, Y. A., Guber, A. K., Gish, T. J., Shelton, D. R., & Cho, K. H. (2013). Modeling transport of Escherichia coli in a creek during and after high-flow events: three-year study and analysis. Water Research, 47(8), 2676–2688.
Yoder, J. S., Hlavsa, M. C., Craun, G. F., Hill, V., Roberts, V., Yu, P. A., et al. (2008). Surveillance for waterborne disease and outbreaks associated with recreational water use and other aquatic facility-associated health events – United States. MMWR Surveillance Summaries, 57(9), 1–29.
Acknowledgements
Financial support for this project was provided by the Natural Sciences and Engineering Research Council of Canada (NSERC). The authors would like to acknowledge the assistance of Tristan Goulden and Nick Dourado for assisting with field work and sample processing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Piorkowski, G.S., Jamieson, R.C., Hansen, L.T. et al. Characterizing spatial structure of sediment E. coli populations to inform sampling design. Environ Monit Assess 186, 277–291 (2014). https://doi.org/10.1007/s10661-013-3373-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10661-013-3373-2