Abstract
Small cardamom (Elettaria cardamomum Maton) is extensively cultivated in the Western Ghats of South India either as a monocrop under the forest trees or as an intercrop along with arecanut and coffee plantations. Colletotrichum species responsible for severe outbreaks of anthracnose on small cardamom in South India are reported. Small cardamom anthracnose, popularly known as “Chenthal”, manifests itself on the foliage as yellowish lesions, which later coalesce to form large blighted areas. In advanced stages, the affected leaves dry up giving a burnt appearance to the plant. Twenty-five isolates of Colletotrichum were isolated from leaves of small cardamom in Karnataka, Kerala and Tamil Nadu states of India. The isolates were characterized through morphological studies and multilocus phylogenetic analysis (ITS, ACT, CHS-1, GAPDH, TUB2, CYLH3, GS and ApMat gene regions) to test whether different species are present and identified: C. karstii (2 isolates), C. gloeosporioides (1), C. siamense (7), C. syzygicola (6), Colletotrichum sp (5), and C. guajavae (4), as the cause of anthracnose on small cardamom for the first time. Pathogenicity of the six species was confirmed. To our knowledge, this is the first detailed study of Colletotrichum species which cause anthracnose diseases on small cardamom.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Small cardamom (Elettaria cardamomum Maton), belonging to the family Zingiberaceae, is the world’s third-most-expensive spice crop (Reyes et al. 2006). It is popularly known as the “Queen of spices” and mainly used in culinary and confectionery preparations apart from medicine and perfumes (Reyes et al. 2006). Guatemala, with a production of 23,000 tonnes, is the largest producer of cardamom followed by India and Tanzania (Sasikumar et al. 2012). Other producers include Sri Lanka, Papua New Guinea, El Salvador, Laos, and Vietnam. In India, small cardamom has been commercially cultivated in the hilly tracts of Karnataka, Kerala and Tamil Nadu states for 150 years and it was responsible for establishing the sea route from Europe to the Far East. It is grown in an area of 0.69 million ha either as a mono crop under the shade of forest trees or as an intercrop along with arecanut and coffee with production of 9,470 MT (www.indianspices.com).
Leaf blight, commonly known as “Chenthal”, has emerged as the most destructive foliar disease affecting small cardamom in India (Praveena et al. 2013). The symptoms initially manifest on the leaves as yellow lesions which later elongate to form necrotic streaks that run parallel to the veins. Several such lesions later coalesce to form yellowish-brown to reddish-brown patches, which subsequently wither off. In the advanced stages of disease development, lesions develop on both young as well as older leaves which dry up and give a burnt appearance to the affected plants. The disease, which appears during mid-monsoon, becomes severe during late monsoon periods and declines by March. Intermittent rains and prevalence of misty conditions in the plantations favour the incidence and spread of the disease. Earlier studies reported Colletotrichum gloeosporioides as the casual organism responsible for anthracnose on small cardamom based on morphology (Govindaraju et al. 1998; Praveena and Biju 2012; Saju et al. 2013).
Traditional methods of characterization and identification of Colletotrichum based on morphology, optimal temperature for mycelial growth, host affiliation and benomyl sensitivity (Cai et al. 2009; Freeman et al. 1998; Peres et al. 2005; Sutton 1992) are time- consuming, tedious and are not reliable (Damm et al. 2009; Freeman et al. 1998). Molecular methods involving sequence analysis of the ITS region of the rDNA region (Brown et al. 1996; Mills et al. 1992; Sreenivasaprasad et al. 1994), restriction fragment length polymorphism analysis of rDNA, analysis of A + T-rich mitochondrial DNA, comparison of arbitrarily primed (AP)-PCR or random amplified polymorphic DNA fingerprints (Freeman et al. 1993, 1998, 2000; Talhinhas et al. 2005) have been used to differentiate populations of Colletotrichum species. In view of the recent changes on the species concepts in Colletotrichum (Cai et al. 2009; Hyde et al. 2009; Prihastuti et al. 2009; Phoulivong et al. 2010; Wikee et al. 2011; Cannon et al. 2012; Damm et al. 2012a, b; Weir et al. 2012), the use of a polyphasic approach involving morphology and multiple-locus sequence comparisons of different genes (e.g., partial β-tubulin, actin, calmodulin, glutamine synthase, and glyceraldehyde-3-phosphate-dehydrogenase) have been advocated for resolving the closely-related Colletotrichum species. The objective of the present study was to characterize the species of Colletotrichum responsible for severe outbreaks of anthracnose on small cardamom in India based on the polyphasic taxonomic approach.
Materials and methods
Collection of anthracnose-affected samples
Small cardamom leaves showing typical symptoms of anthracnose (Fig. 1) were collected from small production fields located in Karnataka, Kerala and Tamil Nadu states of South India, where severe disease epidemics occurred, during the southwest monsoon period (June to September) of 2011 and 2012 (Fig. 2). When most of the small cardamom fields in these localities were infected, leaves with single lesions were collected. Each sample consisted of three infected leaves from each of five plants within a disease focus. The details of the isolates used in this study are shown in Table 1.
Isolation of Colletotrichum species
To obtain fungal isolates, leaves were washed in sterile distilled water and dried with sterilized tissue paper. Then, a small piece (5 by 5 mm) of foliage taken from the leading edge of the infected area was washed in sterile distilled water, surface disinfected in 70 % ethanol for 30s and 1 % sodium hypochlorite for 1 min, rinsed three times in sterile distilled water and placed on Potato dextrose agar (PDA) amended with streptomycin (100 μg/ml). Plates were incubated at 25 ± 1 °C with a 12–h photoperiod provided by fluorescent light for 5 days. The growing edges of fungal hyphae developing from the tissues were then transferred aseptically to PDA. Single-spore isolates were derived according to the method described by Goh (1999). Pure cultures were maintained on PDA slants at 5 °C by sub-culturing at 4-week intervals. For long-term storage, three agar plugs (3 mm diam) from actively growing cultures on PDA were suspended in 5 ml of 20 % glycerol: 17 % skimmed milk (1:1) solution and stored at - 80 ° C (Chowdappa et al. 2009).
Morphological assessments
For morphological analyses, 5-mm-diameter mycelial plugs from the actively growing margin of 5 day old cultures were placed in the centre of 90-mm Petri plates containing 15 ml of PDA (Himedia, Mumbai, India) and incubated at 25 ± 1 °C under continuous fluorescent light for 7 days. The colony colour and diameter of the isolates were recorded during 7 days. The colony colour was scored using the mycological colour chart (Rayner 1970). The colony diameter was measured daily and the mycelial growth rate (mm day−1) was calculated. After 7 days, conidia were directly harvested from cultures and mounted in sterile distilled water. Appressoria were produced using the slide culture technique (Johnston and Jones 1997). PDA pieces of 3 cm square were kept on microscope slides and placed in an empty Petri dish containing filter paper moistened with sterile distilled water. Spores were inoculated on the edge of the agar on all four sides and a sterile cover slip was placed over the inoculated agar. The Petri plates were incubated at 25 ± 1 °C on a laboratory bench sealed with parafilm. Appressoria were formed under the cover slip after 4-7 days. For each isolate, length and width of 100 randomly chosen conidia and appressoria were determined at × 400 magnification with a Zeiss bright field microscope using Axio Vision soft ware. For each isolate, three replicates were maintained and each replicate contained six Petri plates. The experiments were repeated thrice.
DNA extraction
Fungal isolates were grown in potato dextrose broth at 25 ± 1 °C for 7 days. Mycelia were harvested from liquid cultures through Whatman no.3 filter paper, dried and subsequently ground into a fine powder in a mortar and pestle using liquid nitrogen. DNA was extracted from mycelial powder according to the method of Raeder and Broda (1985) and slightly modified by Chowdappa et al. (2003) by incubating at 37 °C for 10 min after the phenol: chloroform: isoamyl alcohol (25:24:1) precipitation. This was followed by precipitation with 0.54 volumes of isopropyl alcohol and centrifugation at 10,000 rpm for 2 min. The DNA pellet was washed with 70 % cold ethanol, dried at room temperature overnight (16 h) and then re-suspended in 30 μl of 10 mM TE buffer (pH.8). DNA was stored at -20 °C.
PCR amplification and DNA sequencing
Eight loci including the partial rDNA-ITS (ITS), actin (ACT), chitin synthase 1 (CHS-1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-tubulin (TUB2), Histone (CYLH3), glutamine synthetase (GS) and ApMat were amplified and sequenced using the primer pairs ITS-1 F (Gardes and Bruns 1993) /ITS-4 (White et al. 1990), ACT512F/ACT783R (Carbone and Kohn 1999), CHS-79 F/CHS-354R (Carbone and Kohn 1999), GDF1/GDR1 (Templeton et al. 1992), Bt2a/Bt2b (Glass and Donaldson 1995), CYLH3F/CYLH3R (Crous et al. 2004), GSF1/GSR1 (Stephenson et al. 1997) and AM-F/AM-R (Silva et al. 2012a), respectively.
Each 50-μl PCR mixture included 39.75 μl of PCR-grade water, 5 μl of reaction buffer, 1 μl of 2.5 mM of each dNTP, 1 μl of each primer (10pmol/μl), 1 μl of DNA template and 0.25 μl of Taq DNA polymerase. PCR reactions were carried out in a thermal cycler (Eppendorf Master cycler). The cycling parameters for ITS consisted of a denaturation step at 95 °C for 3 min followed by 35 cycles at 95 °C for 30 s, 55 °C for 45 s, 72 °C for 45 s and a final step at 72 °C for 15 min. The cycling parameters for ACT were initiated at 95 °C for 3 min followed by 35 cycles at 95 °C for 30 s, 54 °C for 45 s, 72 °C for 1 min and a final step at 72 °C for 15 min. The cycling parameters for partial CHS and CYLH3 region consisted of a 4 min denaturing step at 95 °C followed by 35 cycles at 95 °C for 30 s, 58 °C for 45 s, 72 °C for 45 s and a final cycle of 15 min at 72 °C. GAPDH and GS consisted of 94 °C for 4 min, followed by 35 cycles at 94 °C for 45 s, 60 °C for 45 s, 72 °C for 1 min and a final cycle at 72 °C for 15 min. The cycling parameters for TUB2 consisted of 95 °C for 4 min followed by 35 cycles at 95 °C for 30 s, 54 °C for 45 s, 72 °C for 60 s and a final step at 72 °C for 15 min. The cycling parameters for ApMat were initiated at 94 °C for 3 min followed by 30 cycles at 94 °C for 45 s, 62 °C for 45 s, 72 °C for 1 min and a final step at 72 °C for 7 min. PCR amplification products were separated by 1.5 % agarose electrophoresis gels in 1.0 × TBE buffer and were observed under UV light after staining with ethidium bromide (0.5 μg/ ml). Amplified DNA products of ITS, ACT, CHS-1, GAPDH, TUB2, CYLH3, GS and ApMat were purified using a Nucleospin® gel and PCR Clean-up (Macherey-nagel, Germany). Products were sequenced using the sequencing service from Merck Specialities Pvt Ltd, Bangalore. Sequencing of each PCR product was performed in both directions. Sequencing reactions contained the same primers of all genes that were used in the PCR.
Phylogenetic analyses
Reference sequences downloaded from the Fungal Biodiversity Centre (CBS-KNAW) (http://www.cbs.knaw.nl/Colletotrichum/) were included in the analyses along with the combined dataset of ITS, ACT, CHS-1, GAPDH, TUB2, CYLH3, GS and ApMat gene regions of our isolates (Table 1). Sequences were aligned with Clustal W, in Bioedit v.7.0.5.3 (Hall 1999) and manually adjusted where necessary, the alignment gaps were treated as missing data for each gene, then nucleotide alignments of all genes were concatenated in MEGA v.5 (Tamura et al. 2011). Nucleotide substitution models were determined using Mr-Model-test v.2.3 (Nylander 2004) for each gene region under the Akaike Information Criterion (AIC) and included in the analyses. Phylogenetic reconstructions of concatenated and individual gene trees were performed using Bayesian (BI) Markov Chain Monte Carlo (MCMC) and Maximum Likelihood (ML) criteria. Bayesian analyses (BI) were performed using Mr-Bayes v.3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). MCMC were run for 20 million generations and for every 1000 generations sampling was done with the first 50 % of samples discarded as burn-in. Maximum likelihood (ML) analyses were performed in RAxML v7.0.4 (Stamatakis 2006), all free modal parameters were estimated by RAxML for the concatenated dataset with ML estimate of 25 per site rate categories. In the RAxML platform, the concatenated dataset was partitioned by locus. The RAxML software accommodated the GTR model of nucleotide substitution with the additional options of modeling rate heterogeneity (Γ) and proportion invariable sites (I) and the thorough bootstrap algorithm of RAxML with 1000 replications was implemented with nodal support. Phylogenetic trees and data files were viewed in MEGA v. 5 (Tamura et al. 2011) and Fig-Tree v1.2.2 (Rambaut and Drummond 2008). The sequences of all small cardamom isolates used in multi-gene analyses were deposited in GenBank (Table 1).
Pathogenicity testing
The pathogenicity of the isolates was determined by inoculating intact small cardamom leaves of variety Appangala1 according to the method of Silva et al. (2012b) with slight modifications. Fully matured second leaves from the spindle leaf of one-year-old plants were inoculated. The leaves were surface sterilized by smearing with 1 % sodium hypochlorite solution before inoculation, then washed three times in sterile distilled water and subsequently air dried. The middle portions of the leaves were then pinpricked and placed on wire mesh platforms. Inoculation was carried out on the wounded leaves by placing 20 μl of a conidial suspension (106/ml) prepared from 20-day-old PDA cultures. The fungal inoculum was produced in Petri dishes containing PDA, which were incubated for 20 days at 28 °C under a 12-h photoperiod. Spore suspensions were prepared by adding 20 ml of sterile distilled water to the surface of the cultures, brushing with a soft bristle brush, and filtering through a double layer of cheesecloth. Spore concentration was determined using a haemocytometer and adjusted to 106 conidia ml−1 with sterile water. Leaves used as control were inoculated with 20 μl of sterile distilled water. After inoculation, the leaves were placed in moist chambers and maintained at 25 °C ± 1 °C, RH 90 % under a 14-h photoperiod for 7 days. The lesion size was recorded on each leaf seven days after the inoculation.
Results
Collection of isolates
Twenty-five isolates of Colletotrichum were obtained from small cardamom showing typical symptoms of anthracnose in Karnataka, Kerala and Tamil Nadu states of India (Figs. 1 & 2). The detailed morphological and multilocus phylogenetic analyses (ITS, ACT, CHS-1, GAPDH, TUB2, CYLH3, GS and ApMat genes) revealed six species: C. karstii (2 isolates), C. gloeosporioides (1), C. siamense (7), C. syzygicola (6), Colletotrichum sp (5), and C. guajavae (4).
Morphological characters
The Colletotrichum isolates clustered into six morphological groups based on colony characteristics, growth rate and conidial morphology (Fig. 3 and Table 2).
In the morphological group 1 (C. karstii) isolates, the upper surface color of the colony is pale olivaceous gray with a growth rate ranging from 8.7 to 11.1 mm with an average of 10.0 ± 0.7 mm/day. The conidia length and width varied from 14.2 to 16.1 × 5.1–7.0 μm (15.3 ± 0.8 × 6.0 ± 0.6 μm) respectively and were cylindrical, hyaline, straight and obtuse at both apexes. Appressoria were circular or clavate measuring 6.0–10.0 × 5.0–7.0 μm (7.8 ± 1.7 × 6.1 ± 0.9 μm).
The isolates of morphological group 2 (C. gloeosporioides) produced pale olivaceous grey colonies on PDA with an average growth rate of 11.1 ± 0.2 mm/day ranging from 7.8 to 11.1 mm. The conidia were all sub cylindrical with blunt round ends. The average length and width of the conidia were 12.5–2 × 4.5–6.1 μm (14.7 ± 1.6 × 5.3 ± 0.6 μm), respectively. Appressoria were clavate, regular or irregular in outline and weakly lobed measuring 8.2–11.0 × 5.2–7.0 μm (9.4 ± 0.8 × 6.0 ± 0.6 μm).
The morphological group 3 isolates (C. siamense) on PDA exhibited dense grayish-white aerial mycelium, pale yellowish to pinkish colony with visible orange conidial masses at the inoculum point with a growth rate ranging from 7.5 to 11.0 mm with an average of 11.0 ± 0.9 mm/day. Setae were not found. Conidia were single-celled, smooth-walled, guttulate, hyaline, fusiform with obtuse to slightly rounded ends, sometimes cylindrical with 9.00–17.1 × 3.0–6.0 μm (13.8 ± 3.1 × 4.7 ± 0.9 μm) in size. Appressoria were brown and ovoid measuring 4.8–12.3 × 4.9–7.1 μm (8.5 ± 2.7 × 6.0 ± 0.9 μm).
Colonies of the morphological group 4 (C. syzygicola) isolates exhibited white to grey with orange conidial mass on the upper surface and grey on the reverse side with an average growth rate of 10.8 ± 1.3 mm/day ranging from 7.2 to 11.0 mm. Conidia were hyaline, ovoid to cylindrical with rounded apices 6.0–12.5 × 5.2–6.1 μm (13.8 ± 1.2 × 5.6 ± 0.3 μm). Appressoria were brown to dark brown, circular to clavate and lobed measuring 17.7–19.7 × 7.1–7.9 μm (18.6 ± 0.7 × 7.4 ± 0.3 μm).
The morphological group 5 isolates (Colletotrichum sp) produced light grey to dark grey colonies on PDA with a growth rate ranging from 7.0 to 11.0 mm with an average of 10.7 ± 2.4 mm/day. The conidia were cylindrical, straight, with each apex rounded, often tapering slightly towards the base, measuring 8.5–16.7 × 4.3–7.2 μm (12.1 ± 3.3 × 5.9 ± 1.0 μm). Appressoria were variable in shape, clavate to irregular and lobed 5.1–10.2 × 4.8–7.1 μm (8.0 ± 1.9 × 6.0 ± 0.8 μm).
The morphological group 6 (C. guajavae) isolates had flat colonies with white or pale olivaceous grey to rosy buff color on the upper side while on the reverse side, the color was grey olivaceous to olivaceous black; growth rate ranged from 7.2 to 11.0 mm with an average of 10.7 ± 1.8 mm/day. Conidia were hyaline, smooth-walled, aseptate, straight, cylindrical to fusiform with both ends slightly acute, with or without guttules 6.0–12.5 × 2.6–4.7 μm (8.8 ± 2.2 × 3.7 ± 0.7 μm). Appressoria formed singly, were medium brown, smooth-walled, subglobose or elliptical to clavate, measuring 8.1–9.5 × 4.5–6.1 μm (8.8 ± 0.5 × 5.2 ± 0.6 μm).
Phylogenetic analyses
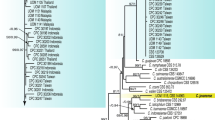
The trimmed sequences of the ITS region ranged from 597 to 618 bp, the ACT gene ranged from 259 to 281 bp, CHS1 gene ranged from 289 to 299 bp, GAPDH ranged between 262 and 277 bp, TUB2 gene ranged from 447 to 701 bp, CYLH3 varied from 392 to 411 bp, GS ranged between 789 and 865 bp and the ApMat gene ranged from 863 to 906 bp. The dataset of combined eight genes used for phylogenetic analyses included 4454 characters including the alignment gaps, the gene boundaries are ITS: 1–621 bp, ACT: 622–906 bp, CHS1: 907–1203 bp, GAPDH: 1204–1511 bp, TUB2: 1512–2230 bp, CYLH3: 2231–2653 bp, GS: 2654–3523 bp and ApMat: 3524–4454 bp, of which 2564 characters were conserved, 1866 characters variable and 1254 characters were parsimony informative. For Bayesian analysis, a GTR + G + I model was selected for combined multi-gene data analysis and incorporated in the analysis. The consensus tree obtained from Bayesian analyses confirmed the tree topology of rapid bootstrapping estimations of RAxML (Fig. 4). The combined analyses resulted in the detection of six well separated clades: two isolates included in group 1 corresponded to C. karstii with a bootstrap support of 98 % and a Bayesian posterior probability value of 1; group 2 had one isolate, representing C. gloeosporioides with a bootstrap support/Bayesian posterior probability value of 100/1; group 3 had seven isolates representing C. siamense with 91/1; group 4 had six isolates representing C. syzygicola with 100/1; group five had five isolates of Colletotrichum sp. with 97/1; and the group six had four isolates representing C. guajavae with 90/0.97.
Phylogenetic tree of Colletotrichum species, isolated from small cardamom in India, constructed with concatenated sequences of the ACT, TUB2, CHS-1, CYLH3, GAPDH, GS and ITS genes. Bayesian posterior probability values above 0.50 are shown at the nodes and bootstrap support values (500 replicates) above 50 % below BPP values. The colour shadings indicate the detection of six well separated Colletotrichum species from small cardamom. Red letters indicate the isolate numbers of Colletotrichum species obtained from small cardamom
Pathogenicity testing
Typical symptoms of sunken and oval streaks with reddish margin surrounded by yellow halo were observed on the leaves of the small cardamom variety Appangala1 inoculated with isolates of: C. karstii (2 isolates), C. gloeosporioides (1), C. siamense (7), C. syzygicola (6), Colletotrichum sp (5), and C. guajavae (4), after seven days. All of the isolates were re-isolated from the leaves and showed the same morphological characteristics that were observed upon the initial isolation.
Discussion
This study represents the first attempt to characterize Colletotrichum species associated with anthracnose of small cardamom in India using a polyphasic approach. Colletotrichum gloeosporioides was previously considered as the only causal organism of the disease based on morphological characters (Govindaraju et al. 1998; Praveena and Biju 2012; Saju et al. 2013). No Colletotrichum species reported on small cardamom have been analyzed previously using molecular approaches. Detailed morphological and molecular phylogenetic analyses of twenty-five isolates revealed six species as the causal agent of small cardamom anthracnose, i.e., C. karstii (C. boninnense species complex), C. gloeosporioides, C. siamense, C. syzygicola, Colletotrichum sp. (C. gloeosporioides species complex), and C. guajavae (C. acutatum species complex).
Colletotrichum karstii a species in the C. boninense species complex was reported on several host plants in the world, including orchid plants (Yang et al. 2011; Damm et al. 2012a; Jadrane et al. 2012), Citrus (Peng et al. 2012), Mangifera indica , Carica papaya, Eugenia uniflora, Bombax aquaticum (Damm et al. 2012a; Weir et al. 2012; Lima et al. 2013) and apple (Velho and Stadnik 2014). Some isolates from Passiflora edulis in Brazil that were initially identified as C. boninense (Tozze et al. 2010) were subsequently revealed to be C. karstii based on GAPDH phylogeny (Damm et al. 2012a). Colletotrichum karstii isolates identified in this study are fitted to the description of Damm et al. (2012a).
Colletotrichum gloeosporioides was epitypified by Cannon et al. (2008) and it has been possible to compare strains of Colletotrichum with that of the epitype, using multi-locus analysis (Cannon et al. 2012; Damm et al. 2012a, b; Weir et al. 2012; Doyle et al. 2013; Sharma et al. 2013; Hyde et al. 2014; Yan et al. 2014). Our C. gloeosporioides isolate fit the description of C. gloeosporioides based on morphology and phylogeny (Cannon et al. 2008; Weir et al. 2012). Previously, there were a few reports of occurrence of C. gloeosporioides on large cardamom based on morphology (Saju et al. 2013) but those isolates are not available for comparison to confirm whether they belong to C. gloeosporioides.
Colletotrichum siamense was originally described as a causal agent of anthracnose on coffee berries in northern Thailand (Prihastuti et al. 2009). C. siamense is thought to be geographically diverse with a wide host range (Weir et al. 2012) and is considered to be a species complex on several hosts (Doyle et al. 2013; Sharma et al. 2013; Udayanga et al. 2013; Lima et al. 2013). The ApMat marker has been reported to be highly useful in resolving the C. siamense species complex (Silva et al. 2012a; Sharma et al. 2013). Our isolates of C. siamense fit within the morphological description of C. siamense (Prihastuti et al. 2009) and this was further confirmed by multi-gene analysis including of the ApMat gene (Sharma et al. 2013, 2014). There are no previous reports of the association of C. siamense as a pathogen of small cardamom.
Colletotrichum syzygicola was first reported from northern Thailand on Citrus aurantifolia and Syzygium samarangense (Udayanga et al. 2013). This C. syzygicola was earlier closely related to C. cordylinicola originally described from Cordyline fruticosa, an ornamental plant in the Asparagaceae in Thailand ((Phoulivong et al. 2010). Our isolates fit the description of C. syzygicola (Udayanga et al. 2013) based on morphology and multi-locus phylogeny .
The unknown Colletotrichum sp., forming a separate clade which is closely related to the C. kahawae clade within a group with high bootstrap support (Fig. 4), needs to be examined in detail.
Colletotrichum guajavae was described within the C. acutatum species complex. Peres et al. (2002) identified C. acutatum (s. lat.) from a guava fruit in Brazil based on ITS sequences which was identical to that of C. guajavae in the C. acutatum complex identified by Damm et al. (2012b). Later, Guerber et al. (2003) identified C. acutatum (s. lat.) based on a phylogeny from combined GAPDH and GS sequences. But it belongs to the clade of C. guajavae where the GAPDH sequence generated by that study differs at 5 bp from that of the C. guajavae ex-holotype strain IMI 350839 of Damm et al. (2012b). Our C. guajavae isolates corresponded to the description of Damm et al. (2012b).
The results of the pathogenecity studies clearly indicate that six species of Colletotrichum produced typical symptoms of anthracnose on leaves of small cardamom and the fungal isolates were re-isolated. Thus, this study revealed that six species of Colletotrichum are responsible for small cardamom anthracnose in India. The present study provides basic information for developing management strategies which, in the absence of resistant or tolerant small cardamom cultivars, should include looking for effective bio-control agents and green fungicides with acceptable levels of residue contamination for use in commercial small cardamom production in India as it is an export-oriented crop.
References
Brown, A. E., Sreenivasaprasad, S., & Timmer, L. W. (1996). Molecular characterization of slow-growing orange and key lime anthracnose strains of Colletotrichum from citrus as C. acutatum. Phytopathology, 86, 523–527.
Cai, L., Hyde, K. D., Taylor, P. W. J., Weir, B., Waller, J., Abang, M. M., Zhang, J. Z., Yang, Y. L., Phoulivong, S., Liu, Z. Y., Prihastuti, H., Shivas, R. G., McKenzie, E. H. C., & Johnston, P. R. (2009). A polyphasic approach for studying Colletotrichum. Fungal Diversity, 39, 183–204.
Cannon, P. F., Buddie, A. G., & Bridge, P. D. (2008). The typification of Colletotrichum gloeosporioides. Mycotaxon, 104, 189–204.
Cannon, P. F., Damm, U., Johnston, P. R., & Weir, B. S. (2012). Colletotrichum—current status and future directions. Studies in Mycology, 73, 181–213.
Carbone, I., & Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia, 91, 553–556.
Chowdappa, P., Brayford, D., Smith, J., & Flood, J. (2003). Molecular discrimination of Phytophthora isolates on cocoa and their relationship with coconut, black pepper and bell pepper isolates based on rDNA repeat and AFLP fingerprints. Current Science, 84, 1235–1238.
Chowdappa, P., Reddy, G. S., Kumar, A., Rao, B. M., & Rawal, R. D. (2009). Morphological and molecular characterisation of Colletotrichum spp. causing anthracnose disease of grapes. The Asian and Australian Journal of Plant Science Biotechnology, 3, 71–77.
Crous, P. W., Gams, W., Stalpers, J. A., Robert, V., & Stegehuis, G. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology, 50, 19–22.
Damm, U., Woudenberg, J. H. C., Cannon, P. F., & Crous, P. W. (2009). Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity, 39, 45–87.
Damm, U., Cannon, P. F., Woudenberg, J. H. C., Johnston, P. R., Weir, B., et al. (2012a). The Colletotrichum boninense species complex. Studies in Mycology, 73, 1–36.
Damm, U., Cannon, P. F., Woudenberg, J. H. C., & Crous, P. W. (2012b). The Colletotrichum acutatum species complex. Studies in Mycology, 73, 37–113.
Doyle, V. P., Oudemans, P. V., Rehner, S. A., & Litt, A. (2013). Habitat and host indicate lineage identity in Colletotrichum gloeosporioides s. l. from wild and agricultural landscapes in North America. PLoS One, 8, e62394.
Freeman, S., Pham, M., & Rodriguez, R. J. (1993). Molecular genotyping of Colletotrichum species based on arbitrarily primed PCR, A-T-rich DNA, and nuclear DNA analyses. Experimental Mycology, 17, 309–322.
Freeman, S., Katan, T., & Shabi, E. (1998). Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Disease, 82, 596–605.
Freeman, S., Minz, D., Jurkevitch, E., Maymon, M., & Shabi, E. (2000). Molecular analyses of Colletotrichum species from almond and other fruits. Phytopathology, 90, 608–614.
Gardes, M., & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Molecular Ecology, 2, 113–118.
Glass, N. L., & Donaldson, G. (1995). Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Applied Environmental Microbiology, 61, 1323–1330.
Goh, T. K. (1999). Single-spore isolation using a handmade glass needle. Fungal Diversity, 2, 47–63.
Govindaraju, C., Thomas, J., & Sudarshan, M. R. (1998). ‘Chenthal’ disease of cardamom caused by Colletotrichum gloeosporioides Penz. and its management. Plantation crops; Development in Plantation Crops Research, 255–259.
Guerber, J. C., Liu, B., Correll, J. C., & Johnston, P. R. (2003). Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia, 95, 872–895.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Huelsenbeck, J. P., & Ronquist, F. (2001). MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Hyde, K. D., Cai, L., Cannon, P. F., Crouch, J. A., Crous, P. W., Damm, U., Goodwin, P. H., Chen, H., Johnston, P. R., Jones, E. B. G., Liu, Z. Y., McKenzie, E. H. C., Moriwaki, J., Noireung, P., Pennycook, S. R., Pfenning, L. H., Prihastuti, H., Sato, T., Shivas, R. G., Tan, Y. P., Taylor, P. W. J., Weir, B. S., Yang, Y. L., & Zhang, J. Z. (2009). Colletotrichum—names in current use. Fungal Diversity, 39, 147–182.
Hyde, K. D., Nilsson, R. H., Alias, S. A., Ariyawansa, H. A., Blair, J. E., Cai, L., de Cock, A. W. A. M., Dissanayake, A. J., Glockling, S. L., Goonasekara, I. D., Gorczak, M., Hahn, M., Jayawardena, R. S., van Kan, J. A. L., Laurence, M. H., Lévesque, C. A., Li, X. H., Liu, J. K., Maharachchikumbura, S. S. N., Manamgoda, D. S., Martin, F. N., McKenzie, E. H. C., McTaggart, A. R., Mortimer, P. E., Nair, P. V. R., Pawłowska, J., Rintoul, T. L., Shivas, R. G., Spies, C. F. J., Summerell, B. A., Taylor, P. W. J., Terhem, R. B., Udayanga, D., Vaghefi, N., Walther, G., Wilk, M., Wrzosek, M., Xu, J. C., Yan, J. Y., & Zhou, N. (2014). One stop shop: backbones trees for important pytopathogenic genera: I. Fungal Diversity, 67, 21–125.
Jadrane, I., Kornievsky, M., Desjardin, D. E., He, Z. H., Cai, L., & Hyde, K. (2012). First Report of flower anthracnose caused by Colletotrichum karstii in white Phalaenopsis orchids in the United States. Plant Disease, 96, 1227.
Johnston, P. R., & Jones, D. (1997). Relationships among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia, 89, 420–430.
Lima, N. B., De Batista, M. V., DeMorais, M. A., Barbosa, M. A. G., Michereff, S. J., Hyde, K. D., & Câmara, M. P. S. (2013). Five Colletotrichum species are responsible for mango anthracnose in northeastern Brazil. Fungal Diversity, 61, 75–88.
Mills, R. R., Sreenivasaprasad, S., & Brown, A. E. (1992). Detection and differentiation of Colletotrichum gloeosporioides isolates using PCR RAPD bind patterns of Colletotrichum isolates from banana from Australia. FEMS Microbiology Letters, 98, 137–144.
Nylander, J. A. A. (2004). MrModeltest v2. Program distributed by the author. Sweden: Evolutionary Biology Centre, Uppsala University.
Peng, L., Yang, Y., Hyde, K. D., Bahkali, A. H., & Liu, Z. (2012). Colletotrichum species on Citrus leaves in Guizhou and Yunnan provinces, China. Cryptogamie Mycologie, 33, 267–283.
Peres, N. A. R., Kuramae, E. E., Dias, M. S. C., & de Souza, N. L. (2002). Identification and characterization of Colletotrichum spp. affecting fruit after harvest in Brazil. Journal of Phytopathology, 150, 128–134.
Peres, N. A. R., Timmer, L. W., Adaskaveg, J. E., & Correll, J. C. (2005). Lifestyles of Colletotrichum acutatum. Plant Disease, 89, 784–796.
Phoulivong, S., Cai, L., Chen, H., McKenzie, E. H. C., Abdelsalam, K., et al. (2010). Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Diversity, 44, 33–43.
Praveena, R., & Biju C. N. (2012) Leaf blight of cardamom. Technical Bulletin, 1–9.
Praveena, R., Biju, C. N., Senthilkumar, R., Darshana, C. N., & Jashmi, K. C. (2013). Preliminary evaluation of cardamom accessions against leaf blight/chenthal disease. Indian Phytopathology, 66, 112–113.
Prihastuti, H., Cai, L., Chen, H., Mckenzie, E. H. C., & Hyde, K. D. (2009). Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Diversity, 39, 89–109.
Raeder, U., & Broda, P. (1985). Rapid preparation of DNA from filamentous fungi. Applied Microbiology, 1, 17–20.
Rambaut, A., & Drummond, A. (2008). FigTree: Tree figure drawing tool, version 1.2. 2. Scotland: Institute of Evolutionary Biology, University of Edinburgh.
Rayner, R. W. (1970). A mycological colour chart. Kew: Commonwealth Mycological Institute.
Reyes, T., Luukkanen, O., & Quiroz, R. (2006). Small cardamom—precious for people, harmful for mountain forests. possibilities for sustainable cultivation in the East Usambaras, Tanzania. Mountain Research and Development, 26, 131–137.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574.
Saju, K. A., Deka, T. N., Gupta, U., Biswas, A. K., Sudharshan, M. R., Vijayan, A. K., & Thomas, J. (2013). Identity of Colletotrichum infections in large cardamom (Amomum subulatum Roxb.). Journal of Spices and Aromatic Crop, 22, 101–103.
Sasikumar, B., Dinesh, R., & Anandaraj, M. (Eds.). (2012). The Capsule-Golden Jubilee Souvenir, Cardamom Research Centre, Appangala. Kozhikode: Indian Institute of Spices Research.
Sharma, G., Kumar, N., Weir, B. S., Hyde, K. D., & Shenoy, B. D. (2013). The ApMat marker can resolve Colletotrichum species: a case study with Mangifera indica. Fungal Diversity, 61, 117–138.
Sharma, G., Pinnaka, A. K., & Shenoy, B. D. (2014). Resolving the Colletotrichum siamense species complex using ApMat marker. Fungal Diversity. doi:10.1007/s13225-014-0312-7.
Silva, D. N., Talhinas, P., Várzea, V., Cai, L., Paulo, O. S., & Batista, D. (2012a). Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Mycologia, 104, 396–409.
Silva, M. R., Martinelli, J. A., Federizzi, L. C., Chaves, M. S., & Pachec, M. T. (2012b). Lesion size as a criterion for screening oat genotypes for resistance to leaf spot. European Journal of Plant Pathology, 134, 315–327.
Sreenivasaprasad, S., Mills, P. R., & Brown, A. E. (1994). Nucleotide sequence of the rDNA spacer 1 enables identification of isolates of Colletotrichum as C. acutatum. Mycological Research, 98, 186–188.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688–2690.
Stephenson, S. A., Green, J. R., Manners, J. M., & Maclean, D. J. (1997). Cloning and characterisation of glutamine synthetase from Colletotrichum gloeosporioides and demonstration of elevated expression during pathogenesis on Stylosanthes guianensis. Current Genetics, 31, 447–454.
Sutton, B. C. (1992). The genus Glomerella and its anamorph Colletotrichum. In J. A. Bailey & J. J. Jeger (Eds.), Colletotrichum: Biology, pathology and control (pp. 1–26). Wallingford: CAB International.
Talhinhas, P., Sreenivasaprasad, S., Neves-Martins, J., & Oliveira, H. (2005). Molecular and phenotypic analyses reveal association of diverse Colletotrichum acutatum groups and a low level of C. gloeosporioides with olive anthracnose. Applied and Environmental Microbiology, 71, 2987–2998.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., & Nei, M. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology Evolution, 28, 2731–2739.
Templeton, M. D., Rikkerink, E. H. A., Solon, S. L., & Crowhurst, R. N. (1992). Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene, 122, 225–230.
Tozze, H. J., Fischer, I. H., Camara, M. P. S., & Massola, N. S., Jr. (2010). First report of Colletotrichum boninense infecting yellow passion fruit (Passiflora edulis f.flavicarpa) in Brazil. Australasian Plant Disease Notes, 5, 70–72.
Udayanga, D., Manamgoda, D. S., Liu, X. Z., Chukeatirote, E., & Hyde, K. D. (2013). What are the common anthracnose pathogens of tropical fruits? Fungal Diversity, 61, 165–179.
Velho, A.C., & Stadnik, M.J. (2014). First report of Colletotrichum karstii causing Glomerella leaf spot on apple in Santa Catarina State, Brazil. Plant Disease Notes, 98.
Weir, B., Johnston, P. R., & Damm, U. (2012). The Colletotrichum gloeosporioides species complex. Studies in Mycology, 73, 115–180.
White, T. J., Bruns, T. D., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In T. J. White, J. J. Sninsky, D. H. Gelfand, & M. A. Innin (Eds.), PCR protocols: A guide to methods and applications (pp. 315–322). San Diego: Academic.
Wikee, S., Cai, L., Pairin, N., McKenzie, E. H. C., Su, Y. Y., et al. (2011). Colletotrichum species from Jasmine (Jasminum sambac). Fungal Diversity, 46, 171–182.
Yan, J. Y., Jayawardena, M. M. R. S., Goonasekara, I. D., Wang, Y., Zhang, W., Liu, M., Huang, J. B., Wang, Z. Y., Shang, J. J., Peng, Y. L., Bahkali, A., Hyde, K. D., & Li, X. H. (2014). Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal Diversity. doi:10.1007/s13225-014-0310-9.
Yang, Y. L., Cai, L., Yu, Z. N., Liu, Z. Y., & Hyde, K. D. (2011). Colletotrichum species on Orchidaceae in southwest China. Cryptogamie Mycologie, 32, 229–253.
Acknowledgments
We are highly thankful to the Indian Council of Agricultural Research, New Delhi for financial support in the form of ALCOCERA, an outreach programme on Alternaria, Colletotrichum and Cercospora diseases of field and horticultural crops.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We hereby state that this manuscript has not been submitted to any other journal other than European Journal of Plant Pathology. The Co- authors have no conflicts of interest to declare; and no human participants or animals were used in the current research. All the authors provided the informed consent for the submission of the manuscript.
Rights and permissions
About this article
Cite this article
Chethana, C.S., Chowdappa, P., Biju, C.N. et al. Molecular and phenotypic characterization revealed six Colletotrichum species responsible for anthracnose disease of small cardamom in South India. Eur J Plant Pathol 146, 465–481 (2016). https://doi.org/10.1007/s10658-016-0931-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-016-0931-9