Abstract
Several species of Botryosphaeriaceae and Phaeomoniella chlamydospora are common agents of grapevine decline worldwide. Currently, the use of culture independent PCR based techniques for detection of Botryosphaeriaceae within grapevine tissues has been limited to Botryosphaeria dothidea. In the present study, two Botryosphaeriaceae specific nested PCR assays were developed. One with a narrow target range, to detect Neofusicoccum parvum and the closely related species complex (Neofusicoccum parvum/N. ribis sensu Pavlic et al. Molecular Phylogenetics and Evolution 51:259–268, 2009) and another, with a wider range, to detect all 17 species of Botryosphaeriaceae which have been reported as potential wood pathogens of grapevine. The effectiveness of these assays was validated in vivo on naturally infected wood samples collected from standing vines and dormant grafted rooted cuttings commercialized in Italy by different nurseries in different years. All samples were also screened by means of a previously published nested PCR assay specific for Phaeomoniella chlamydospora. It was found that: 1) propagation material may play an important role as source of primary inoculum, not only of Phaeomoniella chlamydospora, as previously reported, but also for members of the Botryosphaeriaceae, among which Neofusicoccum parvum, Botryosphaeria dothidea and Diplodia seriata are the most common, and 2) multiple infections by different species belonging to Botryosphaeriaceae and/or Phaeomoniella chlamydospora occur frequently both in standing vines and propagation material. This last finding supports the hypothesis that at least some of the non-specific symptoms of grapevine decline may be due to the presence of different pathogens within host tissues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the fungi which are known to cause trunk diseases and grapevine decline (Rolshausen et al. 2010), increasing importance has been given to Phaeomoniella chlamydospora (W. Gams, P. Crous, M. J. Wingf. & L. Mugnai) Crous & W. Gams (Crous and Gams 2000) and to species in Botryosphaeriaceae Theiss. & H. Syd. (Crous et al. 2006). Phaeomoniella chlamydospora alone or together with species of Phaeoacremonium is the causal agent of a complex disease that is characterized by three different syndromes: Brown wood streaking (mostly affecting rooted cuttings), Petri disease and Grapevine leaf stripe disease (previously known as “young esca”) (Surico 2009). The Botryosphaeriaceae comprise cosmopolitan Ascomycetes occurring worldwide as saprophytes, endophytes or pathogens on a wide range of gymnosperms, monocotyledons and dicotyledons, including the grape Vitis vinifera L. (Crous et al. 2006; van Niekerk et al. 2006). To date, 17 potentially phytopathogenic species of this family have been associated with cases of dieback of grapevines, bud mortality, brown streaking or necrosis of the wood, grape rot, delayed bud burst (Luque et al. 2005; van Niekerk et al. 2006; Damm et al. 2007; Martin and Cobos 2007; Carlucci et al. 2009; Aroca et al. 2010) as well as different leaf symptoms (Cristinzio 1978; Larignon et al. 2001). Pathogenicity tests have shown that Neofusicoccum parvum is one of the most virulent Botryosphaeriaceae species on grapevine worldwide (Phillips 2002; van Niekerk et al. 2004; Úrbez-Torres and Gubler 2009).
Together with Neofusicoccum ribis, N. parvum was considered to be part of the Botryosphaeria dothidea complex (Slippers et al. 2004). Currently, these two Neofusicoccum species, together with three cryptic species isolated from Syzygium cordatum in South Africa, are regarded as forming a unique group, named the N. parvum/N. ribis complex (Pavlic et al. 2009). Neofusicoccum parvum has been frequently associated with brown streaking and necrosis of grapevine wood (Phillips 2002; van Niekerk et al. 2006) whereas N. ribis has been isolated only occasionally from grape bunch rots (Pascoe 1998) as well as from discoloured wood (Crous et al. 2000). Nevertheless, the results of pathogenicity tests on grapevine carried out with a strain isolated from Ribes sp. indirectly suggest that this species could also be an important wood pathogen of Vitis sp. (van Niekerk et al. 2004).
The problem of the early diagnosis of P. chlamydospora has been addressed by the development of several PCR based protocols which have been shown to be efficient and reliable to detect the fungus from grapevine tissues, water, soil and grafting tools. Surveys carried out with these tools have shown that vegetative propagation materials are frequently infected by P. chlamydospora (Retief et al. 2006; Edwards et al. 2007). On the other hand, with one exception (Romanazzi et al. 2009), diagnosis of the various species in Botryosphaeriaceae which may infect grapevine tissues, is currently based on their isolation on agar media followed by characterization by PCR fingerprinting (Alves et al. 2007), dot blot hybridization (Martos et al. 2009) or sequence analysis of one or more DNA regions (van Niekerk et al. 2004; Úrbez-Torres et al. 2006). It is well documented that these procedures, especially when carried out using multiple gene sequence data, are extremely reliable for identification. Nevertheless, they are time consuming and their sensitivity is limited by the fact that a minimum density of the target organism within the sample is required for isolation.
In this study, two independent nested PCR assays were developed based on sequences of the ITS1-5,8S-ITS2 rDNA (ITS rDNA) region (White et al. 1990) of species in Botryosphaeriaceae. The first assay (NprcA/NprcB assay) aimed at detecting the fungi belonging to the N. parvum/N. ribis complex sensu Pavlic et al. (2009) whereas the second (BoitsA/BoitsB assay) aimed at specifically amplifying the DNA of all the species in Botryosphaeriaceae, including N. parvum and N. ribis, which have been associated with wood diseases of grapevine. By means of these two molecular tools, as well as one (Pch1/Pch2 assay) specific for P. chlamydospora (Tegli et al. 2000; Edwards et al. 2007), we evaluated the relative incidence of these species within the woody tissues of standing vines and in nursery propagating material currently commercialized in Italy.
Materials and methods
Fungal and bacterial strains
To assess the in vitro specificity of the primer sets developed or employed in this study, 81 reference strains of fungi and bacteria (Table 1) and 43 strains belonging to Botryosphaeriaceae, isolated and characterized during this study (Table 2), were tested. All strains, except those of Dothiorella iberica, Dothiorella sarmentorum, Diplodia corticola and Neofusicoccum ribis, were isolated from grapevine wood in different countries. All fungi were grown in 30 ml 2% potato dextrose broth (Difco laboratories Inc., Detroit, MI, USA) at 25°C in the dark on a rotary shaker (100 rpm) for 7–14 days. Approximately 200 mg of actively growing mycelium were collected aseptically for DNA extraction. Agrobacterium vitis strains were grown for 48 h at 26°C on Nutrient agar (Difco laboratories Inc., Detroit, MI, USA). Single colonies were subjected to DNA extraction.
Plant material
The three molecular assays were tested in vivo on two different types of naturally infected samples: i) chips from the trunk and arms of standing vines or the rootstock trunk of dormant grafted rooted cuttings [chip samples]; ii) pooled samples of the rootstock trunks of dormant grafted rooted cuttings or canes of Vitis vinifera subsp. sylvestris [pooled samples].
Chip samples
Five standing vines (cv. Sauvignon blanc) were uprooted in 2007 from a twelve year old commercial vineyard in the province of Florence (Italy). In the same year five Dormant Grafted Rooted Cuttings (DGRC 2007) were chosen randomly from each of three lots (different combinations of cultivars and/or rootstocks) produced by nursery B (central Italy) and from one lot produced by nursery E (France). Vines and DGRC 2007 were washed with tap water, cross sectioned, surface disinfected as described by Retief et al. (2006), and the bark was removed with a sterile scalpel. The sections of the trunk and arms (standing vines) or rootstock trunk of the DGRC 2007 were then cut longitudinally and, if present, the position and the extent of the wood discolouration noted. Fifty-four samples were taken from discoloured wood of standing vines and 45 from DGRC 2007, of which 24 were from discoloured and 21 were from non-discoloured wood. All samples were processed immediately for DNA extraction and fungal isolation.
Pooled samples
Thirteen lots of dormant grafted rooted cuttings (different combinations of cultivars and/or rootstocks) ready to be commercialized in Italy by four national nurseries [A and B, (central Italy); C and D (northern Italy)] and one French nursery (E), were inspected in 2008 (DGRC 2008). From each lot of material, a minimum of five DGRC 2008 were chosen randomly, the scions and the roots removed with sterile pruning shears and the rootstock trunk was surface disinfected as described above. Each of the five trunks was cut into segments approximately 5–7 cm long and five segments, one from each trunk, were sub-sampled. After bark removal with a sterile scalpel, the resulting woody tissues comprised the pooled sample to be analysed (average weight 13.7 ± 5.8 g). Based on the size of each sampled lot, the sampling procedure was replicated from one to nine times, resulting in a total of 50 pooled samples. Ten canes were taken from two wild Vitis vinifera subsp. sylvestris vines located in the World Wildlife Fund (WWF) Conservation Area of the Burano Lake in Tuscany (Grosseto, Italy) and processed as described above. All samples were immediately processed for DNA extraction.
Isolation and characterization of Botryosphaeriaceae species and P. chlamydospora from plant material
From 10 to 36 wood chips (2–5 mm) per sample of standing vines and DGRC 2007 were placed on 2% potato dextrose agar (PDA, Difco laboratories Inc., Detroit, MI, USA) amended with 0.015% streptomycin sulphate (Sigma-Aldrich Corporation, Saint Louis, MO, USA) and incubated at 25°C in the dark for up to 3 weeks. Isolates were transferred on PDA and, after microscopic examination of fruiting structures and conidia, up to four monoconidial cultures per sample were obtained according to Tuite (1969) from isolates that could be ascribed to Botryosphaeriaceae (Phillips 2002). More than one isolate (up to four) per sample was chosen when different morphometric or colony features were noted. Identification to species level was done by sequence analysis of the ITS rDNA according to Úrbez-Torres et al. (2006) with minor modifications. Isolates that could be ascribed to species belonging to the N. parvum/N. ribis complex, were further characterized by sequencing parts of the β-tubulin (BT) (Úrbez-Torres et al. 2006) and translation elongation factor 1-alpha (EF1-α) (van Niekerk et al. 2004) genes, and subsequent phylogenetic analysis. P. chlamydospora isolates were morphologically identified according to Crous and Gams (2000).
DNA extraction
Fungal mycelia and wood chips (average weight 98 ± 20 mg) from standing vines and DGRC 2007 were ground with sterile inert sand and liquid nitrogen in a sterile ceramic mortar with the addition of 800 μl of the AP1 buffer of the DNeasy Plant Minikit (Qiagen N.V., Venlo, Netherlands). Pooled wood samples (DGRC 2008) were homogenized for 3 min in 100 ml of sterile phosphate buffer (0.05 M, pH 7.2) using a refrigerated Waring blendor (Waring Products, Torrington, CT, USA). One ml of homogenate was centrifuged for 5 min at 1000 × g and the supernatant was transferred to a new sterile tube and centrifuged again for 10 min at 9200 × g and the supernatant discarded. In all cases further steps of DNA extraction were carried out with the DNeasy Plant Minikit according to the manufacturer’s instructions. To ensure that the DNA had been successfully extracted and that no PCR inhibitors were present, the purified DNA was amplified with the universal primer set ITS5/ITS4 (White et al. 1990). DNA was stored at -80°C until further use.
Primer design and set up of two nested PCR assays
The ITS rDNA sequences of 16 Botryosphaeriaceae species reported from grapevine wood were retrieved from the International Nucleotide Sequence Database (INSD; http://www.insdc.org/). Nucleotide sequences were aligned with ClustalX (http://www.clustal.org/) and adjusted manually when necessary. Two primer sets were designed within the regions flanked by the universal primers ITS5 and ITS4 (White et al. 1990): NprcA (5'- AACTCCAGTCAGTGAACT -3') and NprcB (5'- CCGAGGTCAACCTTGAGAAAT -3') to specifically amplify a region of 372 bp in the species belonging to the N. parvum/N. ribis complex (Pavlic et al. 2009); BoitsA (5'- GACCATCAAACTCCAGTCAG -3') and BoitsB (5'- AAAGTTCAGAAGGTTCGTCCGG -3') to specifically amplify a region varying from 359 to 365 bp in the Botryosphaeriaceae species included in this study. Their theoretical specificity was tested in silico using Primer-Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi).
As the quantity of target DNA in the naturally infected samples was expected to be low, nested PCR specific assays were set up. PCR reagents were purchased from Fermentas International Inc., (Burlington, Ontario, Canada) and primers were synthesized by Eurofins MWG Operon (Ebersberg, Germany). All PCR reactions contained the following reagents in a final volume of 25 μl: 0.5 mM MgCl2, 0.2 mM dNTP’s mix, 1x DreamTaq™ Buffer (MgCl2 2 mM + KCl + (NH4)2SO4), 0.2 μM of each primer, 1.25 U of DreamTaq™ DNA Polymerase and 2 μl of DNA template. Reaction mixtures were assembled on ice and performed in a preheated thermal cycler (Bio-Rad laboratories, Inc., Hercules, CA, USA). The first round of the nested PCR, in common to both assays, was carried out with the universal primer set ITS5/ITS4 (White et al. 1990). Thermal cycling conditions for this amplification were 3 min at 94°C followed by 30 cycles, each of which consisted of: 30 s at 94°C, 30 s at 50°C, 30 s at 72°C, and a final extension step of 7 min at 72°C. The resulting PCR products as well as the negative controls of PCR were diluted 1:100 with double distilled sterile water and 2 μl were used as template in the second round of PCR, carried out with the primer set NprcA/NprcB for the N. parvum/N. ribis complex specific assay or with the primer set BoitsA/BoitsB for the Botryosphaeriaceae specific assay. Thermal cycling conditions for the amplification with the primer set NprcA/NprcB were: 5 min at 94°C followed by 40 cycles, each of which consisted of: 45 s at 94°C, 30 s at 61°C, 30 s at 72°C, and a final extension step of 7 min at 72°C. Thermal cycling conditions for the amplification with the primer set BoitsA/BoitsB were the same as for NprcA/NprcB except that 35 cycles instead of 40 were used. First round PCR products were also amplified with the primer set Pch1/Pch2 reported as specific for P. chlamydospora (Tegli et al. 2000), according to the PCR conditions described by Edwards et al. (2007).
All amplified PCR products were resolved by electrophoresis at 100 Vcm-1 in 2% agarose (IBI-Shelton Scientific, Peosta, IA, USA) gels in 1x Tris-borate-EDTA buffer stained with ethidium bromide (0.5 μgml-1), and visualized under a UV transluminator (Bio-Rad laboratories, Inc., Hercules, CA, USA). Fragment size was estimated with Mass Ruler DNA Ladder low range (Fermentas International Inc., Burlington, Ontario, Canada). To reduce the risk of cross-contamination, all manipulations (DNA extraction, PCR reactions set up and electrophoresis) were performed in separate rooms with dedicated tools. Filter pipette tips were always used.
In vitro sensitivity of the nested PCR assays
The genomic DNA concentration of N. parvum PVFi-Np9, Spencermartinsia viticola CBS117009 and P. chlamydospora PVFi-Pch36 (Tables 1 and 2) was measured using a DyNA Quant 200 fluorometer (Hoefer Inc., Holliston, MA, USA) and 1:10 serial dilutions were made with double distilled sterile water. The sensitivity of the nested PCR was estimated at the maximal dilution titre by using the NprcA/NprcB, BoitsA/BoitsB and Pch1/Pch2 primer sets, respectively, both in single step and nested PCR.
Sequencing and phylogenetic analyses
PCR products were purified with the EXOSAP mix (USB Europe GmbH, Staufen, Germany) in accordance with manufacturer’s instructions and sequenced on a ABI prism 310 CE system (Applied Biosystem, Foster City, CA, USA), using the ABI prism Big Dye Terminator Cycle Sequencing Kit v. 1.1 (Applied Biosystem, Foster City, CA, USA). Chromatograms, including sense and antisense sequences, were edited and assembled manually to obtain single consensus sequences. Primer sequences were removed and identity searches were performed on the INSD database.
Bayesian analyses were performed for each of the ITS-rDNA, BT and EF1-α data sets and, after checking their congruence by means of the partition homogeneity test as implemented in PAUP* v.4.0b10 (Swofford 2002), on the concatenated ITS-rDNA and EF1-α data sets, including at least one reference sequence of each of the species in Botryosphaeriaceae that have been reported from grapevine wood. When needed, the reference sequences of Botryosphaeriaceae species isolated from other hosts were also considered. Sequences were aligned with ClustalW using the alignment option in Mega 4.1 (Tamura et al. 2007) and the analyses were performed with MrBayes v 3.1.2 (Huelsenbeck and Ronquist 2001) using B. dothidea CMW8000 (acc. numbers: AY236949, AY236927, AY236898) as the outgroup. The MCMC analysis ran for 1 × 106generations, with a sample frequency of 100 and a burn-in of 2500. Three independent runs were performed for each dataset to check the congruence of the results. The resulting consensus trees were edited with Figtree v 1.2.1 (http://tree.bio.ed.ac.uk/).
The specificity in vivo of the BoitsA/BoitsB and NprcA/NprcB assays and the frequency of samples infected by different species from Botryosphaeriaceae was evaluated using DNA samples that tested positive to both independent assays. To determine the identity of the DNA template, 42 BoitsA/BoitsB amplicons (ten from standing vines, ten from DGRC 2007, 20 from DGRC 2008 and two from V. vinifera subsp. sylvestris) and of 15 NprcA/NprcB amplicons (four from standing vines, four from DGRC 2007, five from DGRC 2008 and two from V. vinifera subsp. sylvestris), were sequenced and included in a phylogenetic analysis as described above using Guignardia bidwelli CBS111145 (acc. number EU683672) as the outgroup.
Statistical analysis
Observed frequencies of positive and negative samples for each of the three independent nested PCR assays or for fungal isolation, were compared as unordered R × C contingency tables using the two-tailed “Fisher-Freeman-Halton” exact test as implemented by StatXact (CYTEL Software Corporation, Cambridge, MA, USA). Differences between compared groups were considered statistically significant at P ≤ 0.05.
Results
Isolation and characterization of the Botryosphaeriaceae and P. chlamydospora
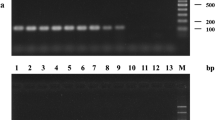
Isolations from discoloured wood from standing vines indicated the presence of Botryosphaeriaceae in 28 and P. chlamydospora in 10 of the 54 samples analyzed (Fig. 1a). Although as a whole Botryosphaeriaceae were present in all and P. chlamydospora in four out of five of the sampled vines, only three samples were co-infected with both taxonomical units (data not shown). With the same analysis on discoloured and non-discoloured wood from DGRC 2007, Botryosphaeriaceae-like morphotypes were isolated from only three out 24 of the discoloured wood samples, while P. chlamydospora-like morphotypes could never be isolated (Fig. 1b). As a whole two out of 20 DGRC 2007 were found to be infected by Botryosphaeriaceae (data not shown). Based on the sequence analysis of the ITS rDNA, 40 Botryosphaeriaceae isolates were obtained from 28 discoloured wood samples of standing vines, of which 27 isolates could be ascribed to N. parvum, ten to D. seriata and three to B. dothidea (Table 2). Different species of Botryosphaeriaceae were isolated from the same plant but never together from the same sample of discoloured wood (data not shown). In the case of DGRC 2007, all three Botryosphaeriaceae isolates analyzed were ascribed to N. parvum (Table 2). BT and EF1-α individual gene sequences, always confirmed the preliminary identification of all the N. parvum isolates. As a whole, two groups of sequences resulted for each one of the ITS (acc. numbers GU187981–GU188010) and EF 1-α (acc. numbers GU188011–GU188040) sequence analyses whereas BT sequences were identical for all the isolates (data not shown). Results of the partition homogeneity test indicated that there was no significant incongruence (P = 0.133) between the EF 1-α and ITS rDNA data sets. Phylogenetic analysis of combined sequences revealed three distinct phylogenetic groups of N. parvum (Fig. 2). Two out of these groups were present both in standing vines and DGRC 2007 (Table 2, Fig. 2). Although at least two of the three phylogenetic groups could be isolated from the same plant, they were never isolated together from the same wood discolouration (data not shown).
Detection of N. parvum/N. ribis, species in Botryosphaeriaceae and P. chlamydospora in naturally infected woody tissue samples (chip samples) by means of three independent nested-PCR specific assays (M) and direct fungal isolation (I). For standing vines a, samples subjected to analysis were taken only from discoloured wood, whereas for DGRC 2007 b both from discoloured and non discoloured wood. An asterisk above a pair of vertical bars indicates a statistically significant difference (P ≤ 0.05) in the frequencies of positive samples between detection methodologies, based on a “Fisher-Freeman-Halton” exact test analysis
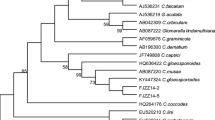
Bayesian consensus phylogram (15002 sampled trees) reconstructed from the concatenated ITS-rDNA and EF1-α data sets of 30 N. parvum strains isolated in this study from standing vines and DGRC 2007. The MCMC analysis ran for 1 × 106generations, with a sample frequency of 100 and a burn-in of 2500. Support values are posterior probabilities (the probability that the clade is correct, given the data and the model parameters). The tree was rooted to B. dothidea strain CMW8000 (acc. numbers: AY236949, AY236927). The bar represents 0.008 changes. The presence of three different phylogenetic groups within the isolated strains is highlighted with grey bars
Evaluation of the PCR assays in vitro
Both the primers NprcA and NprcB showed a theoretical identity equal to 100% toward their respective target regions, whereas the identity values between the primers BoitsA and BoitsB and their respective target regions were variable. All the DNAs extracted from reference strains or fungi isolated during this study were successfully amplified with the universal primer set ITS5/ITS4. The primer sets NprcA/NprcB (N. parvum/N. ribis complex specific), BoitsA/BoitsB (Botryosphaeriaceae specific) and Pch1/Pch2 (P. chlamydospora specific), always discriminated correctly between target and non target DNAs (Tables 1 and 2). The theoretical sensitivity of the nested PCR assay with the primer set BoitsA/BoitsB when applied to the DNA in aqueous solution of N. parvum PVFi-Np9 was 1 pg, three dilution factors below the detection limit that was registered for this primer set when applied in single step PCR reaction (1 ng). When DNA of Spencermartinsia viticola CBS117009, the species of Botryosphaeriaceae that among those considered had shown the lowest theoretical identity with the primers BoitsA and BoitsB, was used, the detection limit was 10 pg and 10 ng in nested and single step PCR, respectively. In the case of the NprcA/NprcB primer set the detection limit was 10 and 100 fg of the DNA of N. parvum PVFi-Np9 in nested and single step PCR, respectively. In the case of the Pch1/Pch2 primer set it was 100 fg and 1 pg of the DNA of the strain PVFi-Pch36 in nested and single step PCR, respectively.
Evaluation of the PCR assays in vivo
With respect to fungal isolation, when the nested PCR assays were applied to the total DNA extracted from the naturally infected samples, a significant increase in the frequency of detection was always observed in the case of standing vines (Fig. 1a) and discoloured and non discoloured tissues of DGRC 2007 (Fig. 1b). The result of the survey carried out on pooled samples of woody tissues from 13 lots of DGRC 2008 revealed the presence of DNA of N. parvum/N. ribis complex, Botryosphaeriaceae and P. chlamydospora in 15, 29 and four out of 50 samples tested, respectively. As a whole ten, 12 and four lots out of 13 could be scored as infected by N. parvum/N. ribis complex, Botryosphaeriaceae and P. chlamydospora, respectively (Figs. 3a, b and c). Moreover, considering the results of each independent PCR assay, the only frequencies of detection that were found to be different were those relative to the N. parvum/N. ribis complex DNA (NprcA/NprcB assay) between different nurseries (P = 0.000) and to the Botryosphaeriaceae DNA (BoitsA/BoitsB assay) between the different lots of DGRC 2008 produced by nursery A (P = 0.004). Nevertheless, when the observed detection frequency of each of the 13 lots were compared in the same analysis (Fig. 3), they were not significant (P = 0.074, P = 0.105 and P = 0.424 for the BoitsA/BoitsB, NprcA/NprcB and Pch1/Pch2 assays, respectively).
Detection of N. parvum/N. ribis a, species in Botryosphaeriaceae b and P. chlamydospora c in naturally infected woody tissue samples (pooled samples) by means of three independent nested-PCR specific assays. From one to nine of pooled samples of woody tissues were taken from each of thirteen lots (1-13) of DGRC produced by five nurseries (A, B, C, D and E) in 2008 and from canes of Vitis vinifera subsp. sylvestris (Vvs). An asterisk above a pair of vertical bars indicates a statistically significant differences (P ≤ 0.05) between lots, based on a “Fisher-Freeman-Halton” exact test analysis
When the results obtained from each sample by each independent PCR assay were compared, evidence of the coexistence of DNA of different species within the same sample, was found (Fig. 4). Considering as a whole 43 (standing vines), 27 (DGRC 2007) and 32 (DGRC 2008) samples whose DNA could be amplified by at least one of the three assays, only the 18.6%, 25.9% and 43.8%, respectively were scored as positive to only one, meanwhile the 81.4% and the 74.1% and 56.2% tested positive to two or to all three independent assays. Both in the case of chip and pooled samples, all the DNA templates that tested positive to the NprcA/NprcB assay, also tested positive to the BoitsA/BoitsB assay or to both the BoitsA/BoitsB and Pch1/Pch2 assays (Fig. 4).
Chromatograms of good quality were obtained from 39 out of 42 BoitsA/BoitsB assay amplicons (three chromatograms could not be used given the presence of more than one sequence) and from all 15 NprcA/NprcB amplicons that were sequenced. The derived phylogenetic analysis showed that in only four cases (one sample from standing vines and three from pooled DGRC 2008) the BoitsA/BoitsB primer set had amplified the DNA of N. parvum/N. ribis complex (Fig. 5a and b). In the remaining 35 cases the DNA of D. seriata (eight out of nine samples from standing vines, five out of nine samples from DGRC 2007, nine out of 19 samples from DGRC 2008), B. dothidea (four samples out of nine from DGRC 2007, six out of 19 samples from DGRC 2008) and Dothiorella sarmentorum (one sample out of 19 from DGRC 2008) were present together with that of N. parvum/N. ribis complex in the sampled tissues (Fig. 5a and b). At last, the presence of Diplodia mutila and Neofusicoccum australe DNA together with that of N. parvum/N. ribis complex could be detected in the pooled samples of canes collected from V. vinifera subsp. sylvestris (Fig. 5a and b). According to sequencing and phylogenetic analysis, no false positive results were produced.
Bayesian consensus phylograms (15002 sampled trees) reconstructed from the portions of the ITS1-5.8-ITS2 nucleotide sequences amplified, from the total DNA extracted from 39 a and 15 b grapevine samples of different origin, by means of nested PCR using the BoitsA/BoitsB a or the NprcA/NprcB b assays, respectively. The MCMC analysis ran for 1 × 106generations, with a sample frequency of 100 and a burn-in of 2500. Support values are posterior probabilities (the probability that the clade is correct, given the data and the model parameters). The tree was rooted to Guignardia bidwelli strain CBS111145 (EU683672). The bars represent 0.9 a and 1.0 b changes. Origin of DNA samples: DSV, discoloured woody tissues from standing vine; NDNSM, non discoloured woody tissues from nursery stock material; DNSM, discoloured woody tissues from nursery stock material; PNSM, pooled woody tissues from nursery stock material; PVvs, pooled woody tissues from V. vinifera subsp. sylvestris canes
Discussion
In this study, two ITS-rDNA based nested PCR assays for the detection of Botryosphaeriaceae species associated with grapevine plants were developed. Thus, one (NprcA/NprcB assay) had a narrow target range to detect DNA of the N. parvum/N. ribis complex, whereas the other (BoitsA/BoitsB assay) had a target range sufficiently wide to amplify the DNA of all the Botryosphaeriaceae species that may colonize grapevine woody tissues. The two protocols were found to be efficient, sensitive and reliable when used with naturally infected grapevine tissues. The NprcA/NprcB assay can cover the variability that exists within N. parvum. The BoitsA/BoitsB assay was able to detect the presence of at least six of the Botryosphaeriaceae species that have been associated with grapevine decline. Moreover, the molecular techniques developed here offer considerable advantages over culture-dependent detection in terms of time, reproducibility and low detection limits and thus may be useful for the early detection of low levels of infection of nursery stock material, or in epidemiological studies. With these tools, together with one specific for P. chlamydospora (Pch1/Pch2 assay, Edwards et al. 2007), the main agent of the Grapevine leaf stripe disease and Petri disease (Surico 2009), we have shown that in Italy nursery stock material may play a significant role as source of primary inoculum in the vineyards of not only P. chlamydospora, as reported previously (Retief et al. 2006; Edwards et al. 2007), but also of N. parvum, D. seriata and B. dothidea.
The results of our investigation allow some other interesting considerations that may be relevant for understanding the epidemiology of grapevine decline. With respect to direct isolation, the results obtained in this study by means of the molecular tools suggest that an intimate association between different agents of trunk diseases occur in nature both in standing vines and nursery propagating material. The concomitant presence of phytopathogenic fungi in the same wood symptoms of standing vines was observed in California by Úrbez-Torres et al. (2006) using isolations on agar media. In that study, 22% of the wood alterations were found to be infected by Botryosphaeriaceae species, but other fungi, such as P. chlamydospora, Phaeoacremonium spp., Eutypa lata, Phomopsis viticola and other species of Ascomycetes were also detected. By means of direct fungal isolation, co-infections of nursery propagating material (grafted grapevines or rooted rootstocks) by several wood pathogens (P. chlamydospora, Phaeoacremonium spp., species in Botryosphaeriaceae, Cylindrocarpon spp. and Phomopsis spp.), were reported by Aroca et al. (2006) in Spain. Of the 97 plants that they found to be infected at the end of their survey, nearly 30% tested positive for the presence of two or more phytopathogenic species. In our study, in the case of standing vines, direct isolation indicated the concomitant presence of different species at the whole plant level, but when single wood discolourations were considered, the different species appeared to be spatially confined in different parts of the plants. Co-infection by Botryosphaeriaceae species and P. chlamydospora occurred in only three out of 54 of the analyzed samples whereas different Botryosphaeriaceae species were never isolated from the same wood discolouration. On the other hand, results of the analysis performed on the same material, using the three molecular protocols, indicated that DNA of two or all three targeted taxonomic groups were present in 81.4% of the samples that tested positive to at least one assay (43 out of 54). A similar situation was also observed in the case of nursery propagating material (DGRC 2007), where 27 of 45 samples tested positive to at least one assay and DNA of two or all three targeted taxonomic groups were present in 74.1% of the samples. Hence, our results support the increasing evidence that a broad range of taxonomically unrelated pathogens are independently contributing not only to a more or less non-specific decline of grapevines (Rolshausen et al. 2010), but perhaps also to more specific syndromes. This could be the case of the Grapevine leaf stripe disease (ex young esca) (Surico 2009), the aetiology of which has been conferred to more than one organism, including Botryosphaeriaceae species (Surico et al. 2006). In this regard, it has recently been shown (Martos et al. 2008; Evidente et al. 2010) that at least some of the Botryosphaeriaceae species isolated from grapevine wood produce the phytotoxic metabolites isosclerone (N. parvum) and exopolysaccharides (N. parvum and N. luteum). Interestingly, such compounds are produced also by P. chlamydospora and are currently considered to be involved in foliar symptom expression in Grapevine leaf stripe disease (Sparapano et al. 2000; Andolfi et al. 2009). Although the cause of the typical foliar chloro-necrotic symptoms of this disease still needs to be fully elucidated (Surico et al. 2006), they could be the result, in toto or in part, of the synergistic action of the phytotoxic matabolites produced by species of Botryosphaeriaceae and P. chlamydospora.
References
Alves, A., Phillips, A. J. L., Henriques, I., & Correia, A. (2007). Rapid differentiation of species of Botryosphaeriaceae by PCR fingerprinting. Research in Microbiology, 158, 112–121. doi:10.1016/j.resmic.2006.10.003.
Andolfi, A., Cimmino, A., Evidente, A., Iannaccone, M., Capparelli, R., Mugnai, L., et al. (2009). A new flow cytometry technique to identify Phaeomoniella chlamydospora exopolysaccharides and study mechanisms of esca grapevine foliar symptoms. Plant Disease, 93, 680–684. doi:10.1094/PDIS-93-7-0685.
Aroca, A., García-Figueres, F., Bracamonte, L., Luque, J., & Raposo, R. (2006). A survey of trunk disease pathogens within rootstocks of grapevines in Spain. European Journal of Plant Pathology, 115, 195–202. doi:10.1007/s10658-006-9008-5.
Aroca, A., Gramaje, D., Armengol, J., García-Jiménez, J., & Raposo, R. (2010). Evaluation of the grapevine nursery propagation process as a source of Phaeoacremonium spp. and Phaeomoniella chlamydospora and occurrence of trunk disease pathogens in rootstock mother vines in Spain. European Journal of Plant Pathology, 126, 165–174. doi:10.1007/s10658-009-9530-3.
Carlucci, A., Lops, F., Raimondo, M. L., Gentile, V., Mucci, M., & Frisullo, S. (2009). The Botryosphaeria species from vineyards of Apulia. Phytopathologia Mediterranea, 48, 180.
Cristinzio, G. (1978). Gravi attacchi di Botryosphaeria obtusa su vite in provincia di Insernia. Informatore Fitopatologico, 6, 21–23.
Crous, P. W., & Gams, W. (2000). Phaeomoniella chlamydospora gen. et. comb. nov., the causal organism of Petri grapevine decline and esca. Phytopathologia Mediterranea, 39, 112–118.
Crous, P. W., Phillips, A. J. L., & Baxter, A. P. (2000). Phytopathogenic fungi from South Africa. Stellenbosch, South Africa: University of Stellenbosch Printers, Department of Plant Pathology Press, p 358.
Crous, P. W., Slippers, B., Wingfield, M. J., Rheeder, J., Marasas, W. F. O., Phillips, A. J. L., et al. (2006). Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology, 55, 235–253.
Damm, U., Crous, P. W., & Fourie, P. H. (2007). Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia, 99, 664–680. doi:10.3852/mycologia.99.5.664.
Edwards, J., Constable, F., Wiechel, T., & Salib, S. (2007). Comparison of the molecular tests-single PCR, nested PCR and quantitative PCR (SYBR®Green and TaqMan®) for detection of Phaeomoniella chlamydospora during grapevine nursery propagation. Phytopathologia Mediterranea, 46, 58–72.
Evidente, A., Punzo, B., Andolfi, A., Cimmino, A., Melck, D., & Luque, J. (2010). Lipophilic phytotoxins produced by Neofusicoccum parvum, a grapevine canker agent. Phytopathologia Mediterranea, 49, 074–079.
Huelsenbeck, J. P., & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755. doi:10.1093/bioinformatics/17.8.754.
Larignon, P., Fulchic, R., Cere, L., & Dubos, B. (2001). Observation on black dead arm in French vineyards. Phytopathologia Mediterranea, 40, S336–S342.
Luque, J., Martos, S., & Phillips, A. J. L. (2005). Botryosphaeria viticola sp. nov. on grapevines: a new species with Dothiorella anamorph. Mycologia, 97, 1111–1121. doi:10.3852/mycologia.97.5.1111.
Martin, M. T., & Cobos, R. (2007). Identification of fungi associated with grapevine decline in Castilla y León (Spain). Phytopatologia Mediterranea, 46, 18–25.
Martos, S., Andolfi, A., Luque, J., Mugnai, L., Surico, G., & Evidente, A. (2008). Production of phytotoxic metabolites by five species of Botryosphaeriaceae causing decline on grapevines, with special interest in the species Neofusicoccum luteum and N. parvum. European Journal of Plant Pathology, 121, 451–461. doi:10.1007/s10658-007-9263-0.
Martos, S., Torres, E., Garcia, F., & Luque, J. (2009). Detection of Botryosphaeriaceae species occurring on grapevines in Spain by cooperational PCR coupled with dot blot hybridization. Phytopathologia Mediterranea, 48, 162.
Pascoe, I. (1998). Trunk diseases of grapevines—perspectives from a tour of California. The Australian Grapegrower & Winemaker, 417, 68–71.
Pavlic, D., Slippers, B., Coutinho, T. A., & Wingfield, M. J. (2009). Multiple gene genealogies and phenotypic data reveal cryptic species of the Botryosphaeriaceae: a case of study on the Neofusicoccum parvum/N. ribis complex. Molecular Phylogenetics and Evolution, 51, 259–268. doi:10.1016/j.ympev.2008.12.017.
Phillips, A. J. L. (2002). Botryosphaeria species associated with diseases of grapevines in Portugal. Phytopathologia Mediterranea, 41, 3–18.
Retief, E., McLeod, A., & Fourie, P. H. (2006). Potential inoculum sources of Phaeomoniella chlamydospora in South African grapevine nursery. European Journal of Plant Pathology, 115, 331–339. doi:10.1007/s10658-006-9025-4.
Rolshausen, P. E., Úrbez-Torres, J. R., Rooney-Latham, S., Eskalen, A., Smith, R. J., & Gubler, W. D. (2010). Evaluation of pruning wound susceptibility and protection against fungi associated with grapevine trunk diseases. American Journal of Enology and Viticulture, 61, 113–119.
Romanazzi, G., Murolo, S., Pizzichini, L., & Nardi, S. (2009). Esca in young and mature vineyards, and molecular diagnosis of the associated fungi. European Journal of Plant Pathology, 125, 277–290. doi:10.1007/s10658-009-9481-8.
Slippers, B., Crous, P. W., Denman, S., Countinho, T. A., Wingfield, B. D., & Wingfield, M. J. (2004). Combined multiple gene genealogies and phenotypic character differentiate several species previously identified as Botryosphaeria dothidea. Mycologia, 96, 83–101.
Sparapano, L., Bruno, G., & Graniti, A. (2000). Effects on plants of metabolites in culture by Phaeocremonium chlamydospora, P.aleophilum and Fomitiporia punctata. Phytopathologia Mediterranea, 39, S169–S177.
Surico, G. (2009). Towards a redefinition of the diseases within the esca complex of grapevine. Phytopathologia Mediterranea, 48, 5.
Surico, G., Mugnai, L., & Marchi, G. (2006). Older and more recent observations on esca: a critical overview. Phytopathologia Mediterranea, 45, S68–S86.
Swofford, D. L. (2002). PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b10 (Alvitec). Sunderland: Sinauer.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599. doi:10.1093/molbev/msm092.
Tegli, S., Bertelli, E., & Surico, G. (2000). Sequence analysis of ITS ribosomal DNA in five Phaeoacremonium species and development of a PCR-based assay for the detection of P. chlamydosporum and P. aleophilum in grapevine tissue. Phytopathologia Mediterranea, 39, 134–149.
Tuite, J. (1969). Plant pathological methods-fungi and bacteria. Minneapolis: Burgess.
Úrbez-Torres, J. R., & Gubler, W. D. (2009). Pathogenicity and epidemiology of Botryosphaeriaceae from grapevines in California. Plant Disease, 93, 584–592. doi:10.1094/PDIS-93-6-0584.
Úrbez-Torres, J. R., Leavitt, G. M., Voegel, T. M., & Gubler, W. D. (2006). Identification and distribution of Botryosphaeria spp. associated with grapevine cankers in California. Plant Disease, 90, 1490–1503. doi:10.1094/PD-90-1490.
van Niekerk, J. M., Crous, P. W., Groenewald, J. Z., Fourie, P. H., & Halleen, F. (2004). DNA phylogeny, morphology and pathogenicity of Botryosphaeria species on grapevines. Mycologia, 96, 781–798.
van Niekerk, J. M., Fourie, P. H., Halleen, F., & Crous, P. W. (2006). Botryosphaeria spp. as grapevine trunk disease pathogens. Phytopathologia Mediterranea, 45, S43–S54.
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR protocols, a guide to methods and applications (pp. 315–322). San Diego: Academic.
Acknowledgements
This research was commissioned from ARSIA-Toscana (Regional Agency for Development and Innovation in Agriculture and Forestry) by fourteen administrative Regions and autonomous provinces and financed with funds provided by the Ministero per le Politiche Agricole e Forestali (Ministry for Agricultural and Forestry Policy) to implement the inter-Regional Project “Grapevine esca: research and experiment in the nursery and in the field for prevention and cure”. Thanks are due to all nurseries and the Stazione Sperimentale per la Viticoltura Sostenibile S.r.l. of Gaiole in Chianti (Siena, Italy) for providing the propagation material that was used in this work, and the WWF of the Burano Lake Conservation Area (Grosseto, Italy) for allowing us to sample V.vinifera subsp. sylvestris plants. A. Phillips was financed by grant number SFRH/BCC/15810/2006 from Fundação para a Ciência e a Tecnologia (FCT) Portugal and project number PPCDT/AGR/56140/2004 from FCT. Prof. Marlene Jaspers, Dr. Jordi Luque, Prof. Sandra Savocchia and Dr. José Ramón Úrbez-Torres provided many of the fungal strains.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Spagnolo, A., Marchi, G., Peduto, F. et al. Detection of Botryosphaeriaceae species within grapevine woody tissues by nested PCR, with particular emphasis on the Neofusicoccum parvum/N. ribis complex. Eur J Plant Pathol 129, 485–500 (2011). https://doi.org/10.1007/s10658-010-9715-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-010-9715-9