Abstract
Papaya ringspot virus (PRSV) HA 5-1, a nitrous acid-induced mild mutant of severe strain HA, widely applied for control of PRSV by cross-protection, was used to study the genetic basis of attenuation. Using infectious clones, a series of recombinants was generated between HA 5-1 and HA and their infectivity was analyzed on the systemic host papaya and the local lesion host Chenopodium quinoa. The recombinants that contained mutations in P1 and HC-Pro genes caused attenuated infection on papaya without conspicuous symptoms, similar to HA 5-1. The recombination and sequence analyses strongly implicated two amino acid changes in the C-terminal region of P1 and two in HC-Pro of HA 5-1 involved in the attenuated infection on papaya. The recombinants that infected C. quinoa plants without local lesions contained the same mutations in the C-terminal region of HC-Pro for attenuated infection on papaya. We conclude that both P1 and HC-Pro bear important pathogenicity determinants for the infection on the systemic host papaya and that the mutations in HC-Pro affecting pathogenicity on papaya are also responsible for the inability to induce hypersensitive reaction on C. quinoa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Papaya ringspot virus (PRSV) is a member of the genus Potyvirus in the family Potyviridae. Virion particles are 780 × 12 nm and have a genome of a ssRNA(+) of 10326 nucleotides (Purcifull et al. 1984; Yeh et al. 1992). PRSV encodes a large polyprotein that is proteolytically processed into eight to nine final products by three virus-coded proteinases, P1, HC-Pro and NIa. The genetic organization of PRSV (protein sizes in parentheses) is VPg-5′ leader-P1 (63 K)-HC Pro (52 K)-P3 (46 K)-CI (72 K)-6 K-NIa (48 K)-NIb (59 K)-CP (35 K)-3′ non-coding region-poly(A) tract (Yeh et al. 1992).
PRSV is a major limiting factor for papaya production in tropical and subtropical areas, causing mosaic and distortion on papaya leaves, ringspots on fruits, and water-soaked streaks on stems and petioles. The virus stunts papaya plants and drastically reduces the size of fruits. Many control measures, including cross-protection (Yeh and Gonsalves 1984), have been used to protect papaya plants from PRSV infection. HA is a severe strain of PRSV collected from Hawaii (Gonsalves and Ishii 1980), causing the described symptoms and severe yield loss. A mild strain PRSV HA 5-1, derived from the severe HA strain by nitrous acid-induced mutagenesis, infects papaya without conspicuous symptoms (Yeh and Gonsalves 1984). This mild mutant has been widely used to protect papaya against virulent strains in Taiwan and Hawaii (Wang et al. 1978; Yeh and Gonsalves 1984; Yeh et al. 1988). Sequence comparison of the 3′-terminal 2235 nucleotides of PRSV HA 5-1 with that of HA revealed 99.4% nucleotide identity (Wang and Yeh 1992).
Pathogenicity of a plant virus and symptom severity on infected plants are largely dependent upon the ability of the virus to replicate, assemble and translocate in the host body. Previous studies have shown that several regions of the potyviral genome are critical in affecting symptom induction. These include the 5′ NTR of Plum pox virus (PPV) (Simon-Buela et al. 1997); the HC-Pro genes of Tobacco vein mottling virus (TVMV) (Atreya et al. 1992; Atreya and Pirone 1993) and PPV (Saenz et al. 2001); the P3 genes of PPV and Turnip mosaic virus (Jenner et al. 2002, 2003; Saenz et al. 2000); the P3 and CIP/6K2/NIa genes of Tobacco etch virus (Chu et al. 1997); the VPg/NIa and NIb genes of Potato virus Y (Fellers et al. 2002; Masuta et al. 1999); the CP gene of ZYMV (Ullah and Grumet 2002); and the 3′ NTR of TVMV (Rodriguez-Cerezo et al. 1991).
Recombinant DNA techniques have made it possible to study the molecular basis of host-pathogen interaction and identify viral genes or genetic regions that are involved in pathogenicity. In ZYMV, a point mutation in HC-Pro gene alters symptom expression in cucurbits (Gal-On 2000). In Cucumber mosaic virus (CMV), one amino acid change in RNA polymerase affects virulent/avirulent phenotypes in cowpea (Akira et al. 1999). In Lettuce mosaic virus (LMV), analysis of recombinants constructed by two different virus isolates indicates that HC-Pro is the pathogenicity determinant in susceptible lettuce cultivars (Redondo et al. 2001). Recent studies with LMV recombinants implied interaction of P1 and C1 in causing lethal wilting in lettuce cv. Ithaca (Krause-Sakate et al. 2005). Studies with recombinant viruses between two symptom-type isolates of Turnip mosaic virus showed the dual role of P3 as a symptom and avirulence determinant in Brassica spp. (Jenner et al. 2003). Genetic analyses of chimeric virus strains constructed by exchanging genomic regions between two infectious isolates of Potato virus A suggested the distribution of multiple virulence/avirulence determinants in the genome and also their coordinate functioning (Paalme et al. 2004). Correlative comparison of sequence data of chimeric potato virus genomes and the symptoms they induced identified involvement of two amino acid residues from the C-terminal part of HC-Pro (Tribodet et al. 2005). Analysis of recombinant viruses between two infectious isolates of Turnip mosaic virus (TuMV) suggested involvement of P3 components in virus accumulation and long-distance movement (Suehiro et al. 2004).
In this study, different genomic regions of the severe strain PRSV HA were replaced with the corresponding fragments of the mild mutant HA 5-1 to create a series of recombinants throughout the viral genome. The infectivity of chimeric transcripts generated in vitro was analyzed by mechanical inoculation on the systemic host Carica papaya and on the local lesion host Chenopodium quinoa. Our results suggested that the mutations in the C-terminal regions of both P1 and HC-Pro of the mild mutant HA 5-1 underlie the PRSV symptom attenuation in papaya and that the same mutations in HC-Pro influencing the pathogenicity on papaya also underlie the failure of PRSV mutants to elicit hypersensitive reaction in the local lesion host C. quinoa.
Materials and methods
Construction of in vitro infectious clones of the mild mutant HA 5-1
PRSV HA is a severe type P strain originating from Hawaii (Gonsalves and Ishii 1980). A full-length cDNA of HA was previously constructed in pT3-HAG from which in vitro infectious transcript driven by a T3 promoter was generated (Chiang and Yeh 1997). The mild strain PRSV HA 5-1, a nitrous acid-induced mutant derived from HA, infects papaya without conspicuous symptoms (Yeh and Gonsalves 1984). In this investigation, the full-length cDNA, pHA51, corresponding to the mild mutant HA 5-1 was constructed downstream from a T3 promoter. The strategy for the construction of pHA51 containing the full-length cDNA to HA 5-1 RNA is summarized in Fig. 1.
Three cDNA clones corresponding to HA 5-1 RNA, pM31, pW21, and pW29, were selected from a λZAP II cDNA library constructed following the method described previously (Yeh et al. 1992). These three clones covered the entire HA 5-1 genome except the first six nucleotides at the 5′ end (Fig. 1). Plasmid pNru20 was constructed by ligating the NruI digested fragment (3511 bp), generated from pW21, with the NruI-digested pM31. The construct pL50 was obtained by inserting the SpeI fragment (4020 bp) from pW21 into SpeI-digested pW29. Previous analysis by RNA direct sequencing indicated that HA and HA 5-1 contained identical sequences in the 5′-terminal 391 nucleotides (Yeh et al. 1992). Therefore, the NcoI-AvrII fragment generated from pM31 was ligated with NcoI-SpeI-digested pT3-HAG to obtain the plasmid pM3530 that contained a cDNA fragment of HA 5-1 from nt 1–3530 and a fragment (1144 bp) from pT3-HAG at the 3′ end.
Plasmid pHA-Nhe/Bsiw was constructed by ligating the NheI-BsiwI fragment from pNru20 with pT3-HAG at the same restriction sites. Plasmid pHA-Nhe/Pml was constructed by insertion of the BsiwI-PmlI fragment of pL50 into the corresponding position of pHA-Nhe/Bsiw. Since the 3′ end 315 nt sequence of HA and HA 5-1 are identical (Wang and Yeh 1992), clone pHA-Nhe/Pml contained a sequence corresponding to HA 5-1 RNA from nt 950–10326. Finally, the full-length cDNA to HA 5-1 RNA was completed in pHA51 by ligation of the NheI-PmlI fragment from pHA-Nhe/Pml with pM3530 digested with the same enzymes.
Construction of recombinant cDNA clones between HA and HA 5-1
The previously constructed pT3-HAG containing the full-length cDNA of the severe strain HA (Chiang and Yeh 1997) and the newly constructed pHA51 containing the full-length cDNA of the mild mutant HA 5-1 were used for genetic recombination at the cDNA level. Restriction enzymes with one to three cutting sites in both HA and HA 5-1 full-length cDNAs were used to generate recombinants by DNA replacement. These restriction enzymes included NheI, AatII, MscI, MluI, AvrII, BsiwI, and PmlI (Fig. 2). Standard molecular biology techniques were adopted for cloning, DNA sequencing, restriction enzyme digestion, ligation, agarose gel electrophoresis, and plasmid preparation (Sambrook et al. 1989). Putative full-length cDNA recombinants were obtained by cloning into pBluescript II KS (−) (Stratagene, La Jolla, CA 92037) and were amplified in Escherichia coli XL-1 Blue.
Schematic strategy for the construction of various chimeric hybrids between the severe PRSV HA strain and its mild mutant HA 5-1. Arrows represent the specific primers used for nucleotide sequencing at different regions of PRSV HA 5-1. The black boxes represent the cDNA regions of HA from clone pT3-HAG. The white boxes represent the cDNA of HA 5-1 fragments from clone pHA51. An EcoRI unique to pHA51 and located at residues 2940–2945 is indicated. (A) Recombinants constructed by exchanging the homologous ‘a + b + c’ regions along the cDNA to PRSV genome. (B) Recombinants constructed by replacement of the single or double regions ‘a’, ‘b’, and ‘c’ from HA 5-1. The character ‘a’ represents the region for nt 950–1653, ‘b’ for 1653–2860, and ‘c’ for nt 2860–3261. Symptoms on papaya induced by the recombinants were graded as type I (mosaic and distortion), II (mild mosaic and less distortion), III (mild mottle and yellowing along leaf veins) and IV (inconspicuous symptom of near-normal appearance). DAS-ELISA values recorded at 405 nm, 1 h after substrate hydrolysis, were graded as + = 0.4–0.6; ++ = 0.6–0.8; +++ = A > 0.8. The readings of mock-inoculated control plants had a range from 0.1 to 0.2. Plants were considered infected when A > 2.5 times the mean A of the mock-inoculated controls. Symptoms on C. quinoa induced by the viruses are summarized. ‘+’ indicates the recombinant viruses were able to induce local lesions and ‘−’ represents no symptoms on C. quinoa
The recombinant pHA-Mlu/Not and pHA-Nhe/Mlu were constructed by exchanging the MluI-NotI and NheI-MluI fragments from pT3-HAG with the corresponding fragments of pHA51, respectively (Fig. 2A). The recombinants p51-Nhe/Not, p51-Mlu/Not, and p51-Nhe/Mlu were constructed by replacing the NheI-NotI, MluI-NotI, and NheI-MluI fragment of pHA51 with the corresponding segments from pT3-HAG, respectively (Fig. 2A). The recombinants pHA-Nhe/Bsiw was constructed as described in Fig. 1.
Verification of the recombinants
The EcoRI site at nt 2940–2945 was found unique to pHA51 and it was used to identify the recombinants between HA and HA 5-1 with the exchanges covering this site (Fig. 2A). Oligonucleotide primers (20-mer) in sense orientation: p5′ HA950 (nt 931–950), p5′ HA1653 (nt 1634–1653), and p5′ HA6576 (nt 6557–6576); and in antisense orientation p3′ HA3261 (nt 3261–3242) were designed for sequencing the encompassed corresponding regions of cDNA versions of HA (i.e., pT3-HAG) and HA 5-1 (i..e., pHA51) (Fig. 2). When a recombinant is sequenced, the nucleotide variations in HA 5-1, in relation to HA (Table 1), were used as molecular markers for the confirmation of the intended genomic replacements. In restriction analyses of recombinants, HindIII restrictions were opted, since the frequency and distribution of HindIII sites in HA and HA 5-1 were convenient for mapping. EcoRI digestion of pHA51 and pT3-HAG revealed the presence of the EcoRI site unique to pHA51 at nt 2940–2945 (as well as the absence of the same in pT3-HAG) (Fig. 2). This distinction was employed as a unique marker to confirm the exchange of the segment of genetic material covering this region between the recombinants (Fig. 2A, B). Whenever required, RT-PCR of leaf RNA from infected plants with the aforementioned primers and subsequent EcoRI restriction of RT-PCR products were carried out to confirm the presence of recombinant viruses in the host plants (Table 1).
Recombination within the region of nt 950–3261
In order to analyze the pathogenicity-related mutations in the region 950–3261 of PRSV HA 5-1, seven recombinants were further constructed. The recombinants pHA-Aat/Mlu and pHA-Nhe/Aat were obtained by exchanging the AatII-MluI and NheI-AatII fragments of pT3-HAG with the corresponding segments from pHA51 (Fig. 2B). Recombinant construction using MscI site in this region was complicated because of the presence of three MscI sites in both cDNA genomes of HA and HA 5-1 (Fig. 2B). Therefore, plasmid p51–56 was obtained by cloning the NheI-AvrII segment from the cDNA of HA 5-1 into pBluescript II KS (−). Plasmid pHA76 was a deletion cDNA clone derived from pT3-HAG by the removal of the AvrII-SpeI (3530–9182) segment. Plasmids p51–56mm and pHA76mm were constructed by exchanging the MscI-MluI segments of p51–56 and pHA76 with the corresponding segments of HA and HA 5-1, respectively. Clones pHA-Nhe/Msc and pHA-NA/MM were constructed by replacing the AatII-MluI segment of pHA-Nhe/Mlu with the corresponding segments from clones p51–56mm and pHA76mm, respectively (Fig. 2B). Clones pHA-Aat/Msc and pHA-Msc/Mlu were constructed by replacing the AatII-MluI segment of pT3-HAG with the corresponding segments from clones p51–56mm and pHA76mm, respectively (Fig. 2B). Clone p51-Msc/Mlu was constructed by replacing the AatII-MluI of pHA51 with the corresponding segment from clone p51–56mm (Fig. 2B). As described in the previous section, all the above constructs were verified by restriction enzyme analyses or by sequencing with the designed primers to confirm the replacement.
Infectivity assay with recombinant transcripts
Infectivity of individual PRSV constructs was assayed using in vitro transcripts, as described by Chiang and Yeh (1997). All the PRSV constructs were purified by CsCl gradient centrifugation (Sambrook et al. 1989). In vitro transcription of NotI-linearized PRSV constructs were carried out with mCapmRNA capping system (Strategene, La Jolla, California, USA). The integrity and translationally functional nature of the products were ascertained, respectively, by agarose gel electrophoresis and in vitro translation in rabbit reticulocyte lysate system (Stratagene). An aliquot of 20 μl of each transcription product with 2–3 μg of RNA was mechanically applied onto two carborundum-dusted leaves of a C. papaya plant at the stage of 2–3 true leaves. Since papaya latex interferes with the induction of local lesions on plants of C. quinoa, recombinant viruses were transferred from infected papaya leaves, three weeks post-inoculation, to the propagation host plants of Cucumis metuliferus (Naud.) Mey. (Acc. 2459). Extracts from infected C. metuliferus were then mechanically introduced to C. quinoa three weeks post-inoculation. All inoculated plants were kept in a temperature-controlled (25–28°C) greenhouse for observation of symptom development.
The accumulation of the PRSV and recombinant viruses in papaya plants were monitored by double-antibody sandwich (DAS) enzyme-linked immunosorbent assay (ELISA) with the antiserum to PRSV three weeks post-inoculation (Gonsalves and Ishii 1980). Individual samples containing a total of nine leaf disks, 0.5 cm diam, were collected from three fully expanded leaves (three disks per leaf) of each papaya plant inoculated with HA, HA 5-1, or the recombinants. The ELISA-positive leaf tissues of the infected papaya plants, which displayed very mild symptoms of near-normal appearance, were analyzed by western blotting and examined by serologically specific electron microscopy (SSEM) with PRSV antiserum (Yeh et al. 1984) to verify the presence of the recombinants.
DNA sequencing of the genomic region responsible for attenuation
Following recombination analyses, the genomic region from nt 950–3261 was subcloned into pBluescript SK (−). DNA sequencing was performed from both orientations. Nucleotide sequences and predicted amino acid residues from PRSV HA 5-1 were assembled and analyzed by the PC/GENE 6.85 software (IntelliGenetics, Inc., University of Geneva, Switzerland).
Results
Mild symptoms of near-normal appearance on papaya induced by in vitro transcript of HA 5-1
The full-length cDNA corresponding to HA 5-1 RNA was constructed in pHA51, downstream from a T3 promoter (Fig. 1). This construct contains one extra guanine residue upstream to the 5’ end of the viral sequence and a stretch of poly(A)36 followed by 12 nonviral nucleotides with a NotI site downstream to the 3′ end of viral sequence. In a batch of 20 papaya plants inoculated with the in vitro transcript derived from pHA51, none of the individuals showed conspicuous symptoms, though twelve of them proved ELISA positive three weeks post-inoculation. The inconspicuous symptoms of near-normal appearance developed on the papaya plants infected with the transcript from pHA51 was similar to that induced by HA 5-1 virion.
Confirmation of recombinant construction
Thirteen full-length chimeric constructs were generated, as depicted schematically in Fig. 2A, B. All clones were confirmed by the above described HindIII and EcoRI mapping and by DNA sequencing with specific primers (Table 1; Fig. 2). The recombinant clones that carried the EcoRI restriction site unique to pHA51 (nt 2940–2945) could be differentiated by their EcoRI restriction banding patterns (which differed from that of pT3-HAG). However, the recombinants pHA-Nhe/Bsiw, p51-Mlu/Not, pHA-Nhe/Mlu, pHA-Aat/Mlu, pHA-NA/MM, and pHA-Msc/Mlu, which possessed the EcoRI site unique to pHA5-1, were not distinguishable from pT3-HAG by their HindIII restriction banding patterns. The other seven clones displayed HindIII and EcoRI restriction banding patterns identical to those of pT3-HAG (data not shown).
Symptom types induced by recombinants on papaya plants
The in vitro pT3-HAG transcript induced typical severe mosaic symptoms and leaf distortion on papaya plants 9–12 days post-inoculation; whereas the in vitro transcripts from different recombinant constructs induced different levels of symptoms, ranging from severe mosaic and leaf distortion to mildest symptoms of near-normal appearance, graded Type I–IV, as depicted in Fig. 2. The recombinant viruses derived from pHA-Mlu/Not, p51-Nhe/Not, and p51-Nhe/Mlu induced Type I severe symptoms of mosaic and distortion on leaves, similar to those induced by the HA strain (Fig. 3A, I). These chimeric viruses shared a common region of nt 950–3261 from HA which included the C-terminal half of P1, the complete HC-Pro, and N-terminal part of P3 (Fig. 2A).
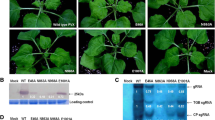
(A) Symptoms on papaya plants inoculated with different recombinant transcripts were graded as Type I–IV (as mentioned in Fig. 2). (B) Western blot analysis of papaya plants inoculated with different recombinant transcripts, using the antiserum to PRSV HA. Type I symptoms are represented by the transcript from pT3-HAG and the recombinant derived from pHA-Mlu/Not; Type II symptoms are represented by the virus derived from pHA-Nhe/Aat; Type III symptoms are represented by the viruses from pHA-Aat/Mlu and pHA-Aat/Msc (III), and Type IV infections are represented by the recombinants from pHA-Nhe/Mlu and pHA-NA/MM (IV). H represents a mock-inoculated plant. Molecular weight markers, in kDa (M)
For comprehensive interpretations of the observations of recombinant analyses, the region of nt 950–3261 was distinguished into three contiguous segments: ‘a, b, and c’ (Fig. 2B). The preliminary results indicated that the recombinant viruses that caused Type I symptoms on papaya contained the genomic segments, ‘a + b + c’, from HA. On the contrary, the recombinant viruses which bore one or two of the ‘a, b, and c’ regions from the parental strain HA 5-1 showed reduced pathogenicity on papaya plants. These recombinant viruses were analyzed for their infectivity on papaya. The recombinant viruses derived from pHA-Nhe/Msc and pHA-Nhe/Aat, bearing respectively the ‘a + b’ and ‘a’ region of HA 5-1, caused Type II symptoms (Figs. 2B, 3A, II), characterized by mild mosaic and slight leaf distortion. The ‘a’ region (HA 5-1) shared by pHA-Nhe/Msc and pHA-Nhe/Aat corresponded to the C-terminal half of P1 protein (Fig. 2B). The recombinant viruses derived from pHA-Aat/Mlu, pHA-Aat/Msc, and pHA-Msc/Mlu, with the ‘b + c’, ‘ b’, and ‘c’ fragments (i.e., the HC-Pro gene) of HA 5-1, respectively, induced Type III symptoms displayed as mild mottling on leaves coupled with normal-appearing young leaves (Figs. 2B, 3A, III). The recombinant viruses derived from pHA-Nhe/Bsiw, p51-Mlu/Not, and pHA-Nhe/Mlu induced Type IV mildest symptoms of near-normal appearance, similar to that induced by HA 5-1 (Fig. 3A, IV). Differing from the Type I symptom-causing recombinants, which bore the genomic region of nt 950–3261 from HA, the Type IV symptom-causing recombinants bore the corresponding region from the mild mutant HA 5-1. Notably, the plants inoculated with the transcripts from pHA-NA/MM (with ‘b’ region from HA) or p51-Msc/Mlu (with ‘c’ region from HA) also showed Type IV symptoms (Figs. 2B, 3A, IV). However, the clone pHA-NA/MM possessed the ‘a + c’ region from HA 5-1, while p51-Msc/Mlu was identical to HA 5-1, except its possession of the ‘c' region of HA (Fig. 2B, 3A, IV).
Correlation of pathogenicity and hypersensitive reaction
After initial inoculation onto papaya plants and subsequent transfer to C. metuliferus, the recombinant viruses were mechanically transferred from the latter to the local lesion host, C. quinoa. The results of the assays on C. quinoa are summarized in Fig. 2. The recombinant viruses that caused Type I and Type II symptoms on papaya were able to induce local lesions on C. quinoa plants (Fig. 4A, C). Also, the recombinant virus derived from pHA-Aat/Msc with the ‘b’ region from HA 5-1, which induced Type III symptoms on papaya was able to induce local lesions on C. quinoa. However, the recombinant viruses derived from pHA-Aat/Mlu (with ‘b + c’ region from HA 5-1) and pHA-Msc/Mlu (with ‘c’ region from HA 5-1), which induced Type III symptoms on papaya were unable to induce local lesions on C. quinoa (Figs. 2B, 4D). Also, the pHA-NA/MM- and p51-Msc/Mlu-derived Type IV recombinants, which did not induce conspicuous symptoms on papaya, were unable to induce HR reaction on C. quinoa (Figs. 2B, 4B, D). However, the results of ELISA evidenced the accumulation of the chimeric viruses in the leaves of C. quinoa (data not shown).
Relatedness of PRSV pathogenicity on the systemic host papaya and the local lesion host Chenopodium quinoa. Similar to the severe parental strain HA, recombinant viruses derived from pHA-Mlu/Not, p51-Nhe/Not, p51-Nhe/Mlu caused Type I symptoms of severe mosaic and leaf distortion on papaya (A). Similar to the parental mild strain HA 5-1, the recombinants derived from pHA-Nhe/Bsiw, p51-Mlu/Not, pHA-Nhe/Mlu, pHA-NA/MM, and p51-Msc/Mlu induced Type IV infection without conspicuous symptoms on papaya (B). Recombinant viruses inducing severe Type I symptoms on papaya and the viruses derived from pHA-Nhe/Msc, pHA-Nhe/Aat, and pHA-Aat/Msc caused local lesions on C. quinoa (C). Recombinant viruses causing mild Type IV symptoms on papaya and viruses derived from pHA-Aat/Mlu and pHA-Msc/Mlu infected C. quinoa without local lesions (D)
To summarize, the recombinant viruses derived from pHA-Nhe/Bsiw, p51-Mlu/Not, pHA-Nhe/Mlu, and pHA-NA/MM with the common segment of ‘a + c’ from HA 5-1 displayed diminished pathogenicity on papaya plants. The recombinant viruses from pHA-Nhe/Bsiw, p51-Mlu/Not, pHA-Nhe/Mlu, pHA-Aat/Mlu, pHA-NA/MM, and pHA-Msc/Mlu with the common ‘c’ region from HA 5-1 were unable to induce local lesion on C. quinoa. Altogether, our results suggested the criticality of the ‘c’ region from HA genome in the induction of severe symptoms on papaya plants, as well as the elicitation of the HR reaction on the local lesion host C. quinoa. However, the recombinant virus derived from p51-Msc/Mlu (the complementary construct of pHA-Msc/Mlu), possessing ‘c’ segment from HA genome and the rest of the genome from HA 5-1, was not able to induce conspicuous symptoms on papaya and to elicit HR on the local lesion host, C. quinoa (Fig. 2).
Verification of attenuated infection
Papaya plants that manifested Type I, II, and III symptoms reacted positively with PRSV antiserum, when assayed by ELISA three weeks after inoculation. Recombinant virus was also detected in those plants that showed Type IV symptoms. However, lower ELISA values were recorded from the plants displaying Type II, III or IV symptoms, in comparison to that recorded from the plants displaying Type I symptoms (Fig. 2). The ELISA-positive plants with milder symptoms were further analyzed by immunoblotting. The results evidenced the detection of a 35 kDa antigen corresponding to the CP of PRSV from the infected plants displaying Type II, III, and IV symptoms (Fig. 3B). SSEM also visualized numerous PRSV antiserum-decorated filamentous particles of 700–800 nm in the upper non-inoculated leaves of the infected (ELISA-positive) plants with mild symptoms, including the plants of Type IV symptoms (data not shown). Moreover, the results of RT-PCR with specific primers and subsequent restriction analysis of the RT-PCR products in pursuit of the EcoRI site unique to HA 5-1 genome (nt 2940–2945) confirmed the identity of the recombinants accumulated in plants of papaya and C. quinoa (data not shown). These results confirmed the Type IV symptoms as a manifestation of a highly attenuated PRSV infection of papaya and C. quinoa plants.
Sequence analyses of the genomic region responsible for symptom attenuation
The genomic region (nt 950–3261) of pHA51 (Fig. 5A, showing cDNA version), implicated in the symptom attenuation, was sequenced to locate possible sequence variations from that of HA. A comparison of the sequence with the corresponding stretch from HA revealed discrepancies in eighteen nucleotides (Fig. 5B). The six changed nucleotides in the P1 gene caused three amino acid substitutions in the protein, I309→ S, E351→ K, and K481→ Q (HA→ HA 5-1). Alignment of potential P1 translates from HA 5-1, HA and seven other severe PRSV strains identified isoleucine or leucine residue at the position 309 of P1 of the severe PRSV strains and its substitution by the polar serine in HA 5-1 (Fig. 6A). Hence, this change was considered critical. Moreover, at the position 481, lysine residue was found conserved among the eight severe strains, while it was mutated to glutamine residue in HA 5-1 (Fig. 6A). At position 351, most of the PRSV severe strains possessed the acidic glutamic acid residue, while in HA, it was the basic lysine residue (Fig. 5B).
Sequence variation in the genomic region nt 950–3261 of the PRSV mild mutant HA 5-1 in comparison with its parental severe strain HA. (A) Organization of PRSV polyprotein. The black shaded area represents the region determining the pathogenicity on papaya for HA isolate. (B) Schematic representation of the sequence variations in HA 5-1 (in comparison to HA) in the genomic region (nt 950–3261) corresponding to the C-terminal half of P1, complete HC-Pro and N-terminal portion of P3. The corresponding amino acid changes are indicated. ‘:’ represents structurally similar residues, ‘▴’ represents critical changes. ‘△’ represents less critical changes when compared to the conserved domain from other potyviruses. The character of ‘a’, ‘b’, and ‘c’ represent the regions, nt 950–1653, 1653–2860, 2860–3261, respectively
Alignment of the amino acid sequences of P1 and HC-Pro of the mild mutant PRSV HA 5-1 with those of the parental strain PRSV HA and other PRSV strains (A); alignment of the same with those of other PRSV strains and potyviruses (B). Identical amino acids are indicated as ‘−’. Arrows indicate the differences between HA and HA 5-1. GenBank accession numbers are indicated in Table 2. Numbers reflect the amino acid positions in the polyprotein
The genomic region analyzed (nt 950–3261) also encompassed seven of the changed nucleotides located in the HC-Pro gene. These mutations resulted in four amino acid substitutions, F753→ L, V940→ I, D944→ N, and Q952→ R (HA → HA 5-1). The change of F753 → L in the central region of HC-Pro was considered critical, since it represented a replacement of an amino acid residue with an aromatic ring side chain by a residue with a linear side chain, though there was no change in hydrophobicity. The critical nature of F753 in PRSV HA was strongly suggested by the conservation of the residue among the potyviruses (Fig. 6B). The D944→ N replacement may also be important, since it was a case of replacement of a highly acidic residue by a less acidic one. It was found that in the corresponding position, most of the compared potyviruses, including HA, had a highly acidic residue, either aspartic acid or glutamic acid (Fig. 6B). Therefore, it appeared that the existence of a negatively charged amino acid at that position in HC-Pro was critical. The mutation Q952 → R that changed the amino acid from slightly acidic to highly basic one may influence the functioning of HC-Pro. The arginine residue at the position 952 of HC-Pro in HA 5-1 may represent a back-mutation regaining its conservation among potyviruses (Fig. 6B). Therefore, the change was considered less critical.
The change of V940→ I in HC-Pro was considered less critical, since this was a substitution of the original hydrophobic amino acid by another hydrophobic amino acid, comparable also in structure (Fig. 5B). The same inference could be applicable to the other amino acid substitutions (HA to HA 5-1, I1038 →V, V1057 →I) in P3 of HA 5-1 (Fig. 5B).
Discussion
Identification and characterization of viral genetic determinants of plant disease symptom phenotypes would help to generate useful attenuated strains helpful in viral disease control. In the present study, the genomic regions of PRSV that influence the severity of disease symptoms on papaya were identified by analyzing a series of recombinant viruses constructed from the in vitro infectious clones of a Hawaii severe strain HA and its mild mutant, HA 5-1. Generated from the severe strain HA, by nitrous acid mutagenesis (Yeh and Gonsalves 1984), the milder mutant, HA 5-1 was expected to bear virulence-decreasing mutations interspersed throughout the genome. The sequence analyses of the genomic region of nt 950–3261 revealed the presence of eighteen point mutations of unclustered distribution (Fig. 5). Genetic analysis of the recombinants implicated the C-terminal portion of P1 (nt 950–1653) of severe strain HA and the C-terminal portion of HC-Pro (nt 2860–3261) of mild mutant HA 5-1, respectively, in determining the severe and apparently symptomless infections of PRSV on papaya. The correlation of attenuated infection of papaya to mutations in the C-terminal portion of HC-Pro (nt 2860–3261) of HA 5-1 did not seem to be host-specific, as evidenced by non-induction of local lesions in C. quinoa by certain PRSV recombinants that shared the same region with HA 5-1. Thus, the results implied that the PRSV determinants of the symptom severity in papaya also underlie the elicitation of hypersensitive reaction in C. quinoa.
Papaya plants inoculated with the recombinant transcript from pHA-Nhe/Msc showed mosaic and leaf distortion symptoms of Type II (Fig. 3, II). The clone, pHA-Nhe/Msc was identical to severe strain HA, but for its possession of the ‘a + b’ region of the milder mutant HA 5-1, which differed from HA at four amino acid residues (Fig. 5B). The potential translate of the transcript from pHA-Nhe/Msc was identical to that of the recombinant derived from pHA-Nhe/Aat, but for single amino acid substitution (F753 → L) in the ‘b’ region. These two constructs possessed the same ‘a’ segment, which corresponded to the C-terminal half of the P1 from HA 5-1 and induced milder symptoms of Type II on papaya plants. Sequence comparison with P1 protease conserved domains showed that the three amino acid substitutions in P1 (i.e., I309→ S, E351→ K and K481→ Q) were not located at the protease activity motifs, such as the H456, D465, S499, and FIVRG519–523 (Verchot et al. 1992). Conservation of I309 and K481 among the compared PSRV strains (Fig. 6A) and the mutationally changed polarity and acidic property, respectively, of these positions implied their criticality. P1 protein’s involvement in genome amplification has been suggested for Tobacco etch virus (TEV) (Verchot and Carrington 1995) and Potato virus Y (PVY) (Arbatova et al. 1998). Its RNA-binding activity has been attributed to its specific structure and ionic interaction with the phosphate backbone of viral RNA (Brantley and Hunt 1993; Merits et al. 1998). The present three amino acid substitutions in P1 that reduce symptom severity to Type II may operate by interfering with the interaction of P1 with other proteins in PRSV replication or by altering P1 structure and ionic charge, thereby reducing its RNA binding ability.
The recombinant transcripts from pHA-Aat/Mlu, pHA-Aat/Msc, and pHA-Msc/Mlu, which induced Type III symptoms in papaya (Fig. 3A) were identical to that of PRSV HA in all the regions, except in the regions ‘b + c’, ‘b’, or ‘c’ derived from PRSV HA 5-1 (Figs. 2B, 5B). The fact that the recombinant transcript from pHA-Nhe/Msc with the ‘a + b’ region of PRSV HA 5-1 caused more severe symptoms (Type II) on papaya than that (Type III) induced by the recombinant virus from pHA-Aat/Msc with only ‘b’ region of HA 5-1 (Fig. 2B) implied possible interaction of the C-terminal portion of the P1 with HC-Pro. The four amino acid changes (PRSV HA to HA 5-1, F753→ L, V940→ I, D944→ N, and Q952→ R) in the ‘b + c’ region corresponding to the HC-Pro gene lead to the symptoms changing from Type I to Type III, as revealed by the milder symptoms induced by the transcript derived from pHA-Aat/Msc, as against the wild-type symptoms induced by the transcript from pT3-HAG.
Correlation of symptom Types II–IV and recombinant virus accumulation (as assessed by ELISA) in the host was suggestive of the influence of mutations in HC-Pro on the accumulation of PRSV in papaya plants. In TVMV, the N-terminal and central regions of HC-Pro are involved in genome amplification and pathogenicity (Klein et al. 1994). A mutation in TEV, in the central region of HC-Pro was found to suppress virus amplification and render the virus defective in long-distance movement (Kasschau et al. 1997).
The presently analyzed mutations in HC-Pro were found to be out of the conserved motifs of the HC-Pro functional domains involved in aphid transmission: KITC (nt 598–601) and PTK (nt 856–858); symptom expression: FRNK (nt 727–730); genome amplification: IGN (nt 797–799); systemic movement: CCC (nt 838–840); protease activity: GYCY (nt 888–891) and H (nt 963) (Plisson et al. 2003). However, considering the mutationally induced local changes in the amino acid positions and the conserved residues among the compared sequences of the potyviruses, we propose that the changes of F753 → L and D944 → N in HC-Pro have important effects on symptom severity, while the change of Q952→ R is less important since the latter mutation restored the conserved residue at the position (Figs. 5B, and 6B).
The recombinant transcripts from pHA-Nhe/Bsiw, p51-Mlu/Not, pHA-Nhe/Mlu, pHA-NA/MM, and p51-Msc/Mlu induced Type IV mild symptoms similar to that induced by PRSV HA 5-1 (Fig. 2). Except for the one derived from p51-Msc/Mlu, all the recombinants shared the common region of ‘a + c’ of PRSV HA 5-1, in which the region ‘c’ represented the C-terminal region of HC-Pro (nt 2860–3261). Altogether, the results suggested that in HA 5-1, the mutations in the C-terminal region of P1 together with those in HC-Pro decrease the severity of symptoms (from Type I–IV). Based on sequence analysis, the mutation of D944→ N in the C-terminal region of HC-Pro was considered critical for this interaction (Figs. 5, 6). The recombinant transcripts of p51-Msc/Mlu contained most of the sequence from PRSV HA 5-1 but with the ‘c’ region (nt 2860–3261) from PRSV HA induced Type IV infection (Figs. 2B, 5B), indicating that the replacement of the region nt 2860–3261 of PRSV HA 5-1 by the corresponding region from HA was inadequate to restore the pathogenicity of HA 5-1 on papaya plants. Our results suggest that the homologous combination of the C-terminal region of P1 (‘a’ region) and the C-terminal region of HC-Pro (‘c’ region) from the severe HA is critical for the induction of severe symptoms in papaya plant. Conversely, the homologous combination of the corresponding regions from the mild HA 5-1 decreases the symptom severity to apparently symtomless Type IV.
The recombinant viruses that caused Type I and Type II symptoms on papaya were also able to induce local lesions on C. quinoa (Fig. 2A, B). However, the recombinants causing Type IV symptoms on papaya failed to induce conspicuous symptoms on plants of C. metuliferus (results not shown) and local lesions on C. quinoa plants, suggesting possible common (or contiguously arranged) genetic determinants for symptom severity in the systemic hosts of papaya and C. metuliferus and HR reaction in the local lesion host, C. quinoa.
Among the recombinant viruses that induced Type III symptoms on papaya, the recombinant from pHA-Aat/Msc was able to induce local lesions on C. quinoa, while those from pHA-Aat/Mlu and pHA-Msc/Mlu failed to do so. Except for the recombinant from p51-Msc/Mlu, all others containing the C-terminal portion of HC-Pro (‘c’ region) from HA were able to induce local lesions on C. quinoa plants. On the contrary, the recombinants possessing the corresponding region from HA 5-1 infected C. quinoa plants without HR reaction. Similarly, the recombinant from p51-Msc/Mlu also infected C. quinoa without HR reaction, though it possessed the ‘c’ segment of PRSV HA. Thus, our results suggest the C-terminal portion of HC-Pro as the major elicitor for the induction of HR reaction in C. quinoa. However, its interaction with the N-terminal portion of HC-Pro and the C-terminal region of P1 could modulate the disease symptom on C. quinoa. The host-dependent determination of symptom phenotype by the potyvirus gene silencer HC-Pro resembles the ability of its counterpart P19 in Tomato bushy stunt virus, which induces lethal systemic necrosis in Nicotiana benthamiana and HR (manifested as necrotic lesions) in N. tabaccum (Scholthof et al. 1995).
Interactions of pathogen and plant host genes result either in systemic infection or local lesion development characteristic of the host’s hypersensitive reaction. Multifunctionality of single proteins such as HC-Pro and multiprotein involvement in a single function, as exemplified by HC-Pro, CI and CP cooperation in viral movement cell-to-cell (Urcuqui-Inchima et al. 2001; Ballut et al. 2005) render potyviral pathogenicity a complex phenomenon to explain. Owing to the multiplicity of symptom determinants (operating differentially through viral protein variants) and interacting host factors, the disease symptom patterns are diverse. Natural recombination or laboratory dissection of co-operative genetic determinants of disease symptom phenotype from a genomic region (Chu et al. 2000) and exchange between two strains of a virus (representing contrasting extremes of disease symptom trait) can produce different shades of symptom pattern, which may not be distinguishable by simple morphological observation of the diseased plants. This phenomenon, which is important in the construction of mild strains, was evidenced in the present study as the individual differences in the ability of recombinants of Type I category to amplify in the host (as interpreted from ELISA data) and those of Type III category to elicit HR in C. quinoa. Moreover, since large number of host proteins are known to interact directly with viral proteins (Leonard et al. 2000; Ruffel et al. 2002; Yoshii et al. 2004; Brizard et al. 2006; Canto et al. 2006), genetic variations in viral and/or host factors have the potential to alter disease symptomology in a viral strain- and/or host-specific manner. The symptomology can be modulated further by host-specific RNA determinants (Omarov et al. 2004; Krause-Sakate et al. 2005), the variety of which may exceed their protein counterparts.
To summarize, in this study, by genetic analysis of a highly virulent strain of PRSV HA and its mild mutant HA 5-1, we have identified a set of amino acids in P1 and HC-Pro critical for determining PRSV systemic infection of papaya and HR in C. quinoa. The infection of papaya with highly attenuated symptoms of near-normal appearance by the mild strain HA 5-1 and loss of its ability to elicit hypersensitive reaction in C. quinoa could be attributed to mutations of these critical amino acids. Genetic analysis of recombinants between HA and HA 5-1 revealed that the symptom attenuation of HA 5-1 on papaya was caused by the interaction of several mutations in P1 and HC-Pro. HA 5-1 is a stable mild strain successfully employed in cross-protection for control of PRSV for several years (Yeh and Gonsalves 1984). Its stability may be attributed to the possible coordination of its multiple mutated residues. The present data on symptom modulation by critical mutations in the interacting regions of P1 and HC-Pro encourage future construction of agriculturally relevant novel mild strains of PRSV for controlling papaya ringspot disease in different geographic areas. Moreover, the present information may also be valuable for the construction of attenuated potyvirus strains which are useful to express heterologous proteins in host plants, as exemplified by an attenuated ZYMV strain employed to express human interferon-alpha 2 in cucurbits (Arazi et al. 2001).
References
Akira, K., Itaru, O., Kayoko, A., Yutaka, C., Shuu, H., Yoshiko, N. N., Akiko, I., & Yoshio, E. (1999). One amino acid change in cucumber mosaic virus RNA polymerase determines virulent/avirulent phenotypes on cowpea. Phytopathology, 89, 186–1192.
Arazi, T., Slutsky, S. G., Shiboleth, Y. M., Wang, Y., Rubinstein, M., Barak, S., Yang, J., & Gal-On, A. (2001). Engineering zucchini yellow mosaic potyvirus as a non-pathogenic vector for expression of heterologous proteins in cucurbits. Journal of Biotechnology, 87, 67–82.
Arbatova, J., Lehto, K., Pehu, E., & Pehu, T. (1998). Localization of the P1 protein of potato Y potyvirus in association with cytoplasmic inclusion bodies and in the cytoplasm of infected cells. Journal of General Virology, 79, 2319–2323.
Atreya, C. D., Atreya, P. L., Thornbury, D. W., & Pirone, T. P. (1992). Site-directed mutations in the potyvirus HC-Pro gene affect helper component activity, virus accumulation, and symptom expression in infected tobacco plants. Virology, 191, 106–111.
Atreya, C. D., & Pirone, T. P. (1993). Mutational analysis of the helper component-proteinase gene of a potyvirus: effects of amino acid substitutions, deletions, and gene replacement on virulence and aphid transmissibility. Proceedings of National Academy of Sciences USA, 90, 11919–11923.
Ballut, L., Drucker, M., Pugniere, M., Cambon, F., Blanc, S., Roquet, F., Candresse, T., Schmid, H. P., Nicolas, P., Gall, O. L., & Badaoui, S. (2005). HcPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymic activities. Journal of General Virology, 86, 2595–2603.
Brantley, J. D., & Hunt, A. G. (1993). The N-terminal protein of the polyprotein encoded by the potyvirus tobacco vein mottling virus is an RNA-binding protein. Journal of General Virology, 74, 1157–1162.
Brizard, J. P., Carapito, C., Delalande, F., Dorsselaer, A. V., & Brugidou, C. (2006). Proteome analysis of plant-virus interactome. Molecular and Cellular Proteomics, 5, 2279–2297.
Canto, T., Uhrig, J. F., Swanson, M., Wright, K. M., & MacFarlane, S. A. (2006). Translocaiton of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. Journal of Virology, 90, 9064–9072.
Chiang, C. H., & Yeh, S. D. (1997). Infectivity assays of in vitro and in vivo transcripts of papaya ringspot potyvirus. Botanical Bulletin of Academia Sinica, 38, 153–163.
Chu, M., Desvoyes, B., Turina, M., Noad, R., & Scholthof, H. B. (2000). Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion. Virology, 266, 79–87.
Chu, M., Lopez-Moya, J. J., Llave-Correas, C., & Pirone, T. P. (1997). Two separate regions in the genome of the tobacco etch virus contain determinants of the wilting response of Tabasco pepper. Molecular Plant-Microbe Interactions, 10, 472–480.
Fellers, J. P., Tremblay, D., Handest, M. F., & Lommel, S. A. (2002). The Potato virus Y MSNR NIb-replicase is the elicitor of a veinal necrosis-hypersensitive response in root knot nematode resistant tobacco. Molecular Plant Pathology, 3, 145–152.
Gal-On, A. (2000). A point mutation in the FRNK motif of the potyvirus helper component-protease gene alters symptom expression in cucurbits and elicits protection against the severe homologous virus. Phytopathology, 90, 467–473.
Gonsalves, D., & Ishii, M. (1980). Purification and serology of papaya ringspot virus. Phytopathology, 70, 1028–1032.
Jenner, C. E., Tomimura, K., Ohshima, K., Hughes, S. L., & Walsh, J. A. (2002). Mutations in Turnip mosaic virus P3 and cylindrical inclusion proteins are separately required to overcome two Brassica napus resistance genes. Virology, 300, 50–59.
Jenner, C. E., Wang, X., Tomimura, K., Ohshima, K., Ponz, F., & Walsh, J. A. (2003). The dual role of the potyvirus P3 protein of turnip mosaic virus as a symptom and avirulence determinant in brassicas. Molecular Plant-Microbe Interactions, 16, 777–784.
Kasschau, K. D., Cronin, S., & Carrington, J. C. (1997). Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology, 228, 251–262.
Klein, P. G., Klein, R. R., Rodriguez-Cerezo, E., Hunt, A. G., & Shaw, J. G. (1994). Mutational analysis of the tobacco vein mottling virus genome. Virology, 204, 759–769.
Krause-Sakate, R., Redondo, E., Richard-Forget, F., Jadão, A. S., Houvenaghel, M. C., German-Retana, S., Pavan, M. A., Candresse, T., Zerbini, F. M., & Gall, O. L. (2005). Molecular mapping of the viral determinants of systemic wilting induced by a Lettuce mosaic virus (LMV) isolate in some lettuce cultivars. Virus Research, 109, 175–180.
Leonard, S., Plante, D., Wittmann, S., Daigneault, N., Fortin, M. G., & Laliberte, J. F. (2000). Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlated with virus infectivity. Journal of Virology, 74, 7730–7737.
Masuta, C., Nishimura, M., Morishita, H., & Hataya, T. (1999). A single amino acid change in viral genome-associated protein of potato virus Y correlates with resistance breaking in ‘Virgin A mutant’ tobacco. Phytopathology, 89, 118–123.
Merits, A., Guo, D., & Saarma, M. (1998). VPg, coat protein and five non-structural proteins of potato A potyvirus bind RNA in a sequence-unspecific manner. Journal of General Virology, 79, 3123–3127.
Omarov, R. T., Rezende, J. A., & Scholthof, H. B. (2004). Host-specific generation and maintenance of Tomato bushy stunt virus defective interfering RNAs. Molecular Plant-Microbe Interactions, 17, 195–201.
Paalme, V., Gammelgård, E., Järvekülg, L., & Valkonen, J. P. T. (2004). In vitro recombinants of two nearly identical potyviral isolates express novel virulence and symptom phenotypes in plants. Journal of General Virology, 85, 739–747.
Plisson, C., Drucker, M., Blanc, S., German-Retana, S., Le Gall, O., Thomas, D., & Bron, P. (2003). Structural characterization of HC-Pro, a plant virus multifunctional protein. Journal of Biological Chemistry, 278, 23753–23761.
Purcifull D. E., Edwardson J. R., Hiebert E., & Gonsalves, D. (1984). Papaya ringspot virus. Kew, UK: CMI/AAB Descriptions of Plant Viruses.
Redondo, E., Krause-Sakate, R., Yang, S. J., Lot, H., Le Gall, O., & Candresse, T. (2001). Lettuce mosaic virus pathogenicity determinants in susceptible and tolerant lettuce cultivars map to different regions of the viral genome. Molecular Plant-Microbe Interactions, 14, 804–810.
Rodriguez-Cerezo, E., Klein, P. G., & Shaw, J. G. (1991). A determinant of disease symptom severity is located in the 3′-terminal noncoding region of the RNA of a plant virus. Proceedings of National Academy of Sciences USA, 88, 9863–9867.
Ruffel, S., Dussault, M. H., Palloix, A., Moury, B., Bendahmane, A., Robaglia, C., & Caranta, C. (2002). A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant Journal, 32, 1067–1075.
Saenz, P., Cervera, M. T., Dallot, S., Quiot, L., Quiot, J. B., Riechmann, J. L., & Garcia, J. A. (2000). Identification of a pathogenicity determinant of Plum pox virus in the sequence encoding the C-terminal region of protein P3 + 6K(1). Journal of General Virology, 81, 557–566.
Saenz, P., Quiot, L., Quiot, J. B., Candresse, T., & Garcia, J. A. (2001). Pathogenicity determinants in the complex virus population of a Plum pox virus isolate. Molecular Plant-Microbe Interactions, 14, 278–287.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: A laboratory manual, 2nd ed. NY: Cold Spring Harbor Laboratory.
Scholthof, H. B., Scholthof, K., Kikkert, M., & Jackson, A. O. (1995). Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology, 213, 425–438.
Simon-Buela, L., Guo, H. S., & Garcia, J. A. (1997). Long sequences in the 5′ noncoding region of plum pox virus are not necessary for viral infectivity but contribute to viral competitiveness and pathogenesis. Virology, 233, 157–162.
Suehiro, N., Natsuaki, T., Watanabe, T., & Okuda, S. (2004). An important determinant of the ability of Turnip mosaic virus to infect Brassica spp. and/or Raphanus sativus is in its P3 protein. Journal of General Virology, 85, 2087–2098.
Tribodet, M., Glais, L., Kerlan, C., & Jacquot, E. (2005). Characterization of Potato virus Y (PVY) molecular determinants involved in the vein necrosis symptom induced by PVYN isolates in infected Nicotiana tabacum cv. Xanthi. Journal of General Virology, 86, 2101–2105.
Ullah, Z., & Grumet, R. (2002). Localization of Zucchini yellow mosaic virus to the veinal regions and role of viral coat protein in veinal chlorosis conditioned by the zym potyvirus resistance locus in cucumber. Physiological and Molecular Plant Pathology, 60, 79–89.
Urcuqui-Inchima, S., Haenni, A. L., & Bernardi, F. (2001). Potyvirus proteins: a wealth of functions. Virus Research, 74, 157–175.
Verchot, J., & Carrington, J. C. (1995). Debilitation of plant potyvirus infectivity by P1 proteinase-inactivating mutations and restoration by second-site modifications. Journal of Virology, 69, 1582–1590.
Verchot, J., Herndon, K. L., & Carrington, J. C. (1992). Mutational analysis of the tobacco etch potyviral 35-kDa proteinase: identification of essential residues and requirements for autoproteolysis. Virology 190, 298–306.
Wang, C. H., & Yeh, S. D. (1992). Nucleotide sequence comparison of the 3′-terminal regions of severe, mild, and non-papaya infecting strains of papaya ringspot virus. Archives of Virology, 127, 345–354.
Wang, H. L., Wang, C. C., Chiu, R. J., & Sun, M. H. (1978). Preliminary study on papaya ringspot virus in Taiwan. Plant Protection Bulletin, 20, 133–140.
Yeh, S. D., & Gonsalves, D. (1984). Evaluation of induced mutants of papaya ringspot virus for control by cross protection. Phytopathology, 74, 1081–1085.
Yeh, S. D., Gonsalves, D., & Provvidenti, R. (1984). Comparative studies on host range and serology of papaya ringspot virus and watermelon mosaic virus 1. Phytopathology, 74, 1081–1085.
Yeh, S. D., Gonsalves, D., Wang, H. L., Nanba, R., & Chiu, R. J. (1988). Control of papaya ringspot virus by cross protection. Plant Disease, 72, 375–380.
Yeh, S. D., Jan, F. J., Chiang, C. H., Doong, T. J., Chen, M. C., Chung, P. H., & Bau, H. J. (1992). Complete nucleotide sequence and genetic organization of papaya ringspot virus RNA. Journal of General Virology, 73, 2531–2541.
Yoshii, M., Nishikiori, M., Tomita, K., Yoshioka, N., Kozuka, R., Naito, S., & Ishikawa, M. (2004). The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. Journal of Virology, 78, 6102–6111.
Acknowledgements
The authors thank Drs. M. J. Chen, S. T. Hsu, and H. T. Hsu for their encouragement and advice. This research was partly supported by the grants NSC 87-2312-B-005-005 and NSC 89-2321-B-005-001 from the National Science Council of the Republic of China on Taiwan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chiang, CH., Lee, CY., Wang, CH. et al. Genetic analysis of an attenuated Papaya ringspot virus strain applied for cross-protection. Eur J Plant Pathol 118, 333–348 (2007). https://doi.org/10.1007/s10658-007-9130-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-007-9130-z