Abstract
Surface soils from Guiyu, China (an intense e-waste processing center) were analyzed for persistent organic pollutants (POPs) and variations in composition of the resident bacterial communities. Denaturing Gradient Gel Electrophoresis analysis of bacterial 16S rRNA gene showed that e-waste pollution altered the bacterial community structure by promoting changes in species composition and species richness. Bacterial diversity was not decreased at e-waste open-burning sites, compared with a non e-waste site (reservoir site), due to flourishing of possible POPs-consuming bacterial cohorts. PAH-incubated experiments confirmed that different levels of PAHs might affect the bacterial community by suppressing or favoring certain groups of bacteria, for instance, uncultured Clostridium sp. and Massilia sp., respectively. Taxonomic analysis indicated β-proteobacteria and Firmicutes were abundant bacterial lineages in PAH-polluted soils. This study is the first reporting bacterial community structures at e-waste processing sites, and indicated that crude processing of e-waste has become a biohazard to the terrestrial environment warranting more extensive studies of microbial communities at e-waste polluted environments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Persistent toxic substances (PTS) that accumulate in the environment and in food webs result in long-term threats to public health and ecosystem stability (Opopol 2007). For example, polycyclic aromatic hydrocarbons (PAHs) are carcinogenic, mutagenic and teratogenic chemicals persistent in the environment (IARC 1983). PAHs mainly result from combustion in various industries, including automobile exhausts. PAHs enter food webs and accumulate in living organisms through lipid-rich cell membranes and adipose tissues, and eventually become a threat to human health. The extended range of PAHs distribution leads to broad harmful effects on the global ecosystem. In China, extensive investigations on PAHs (concentrations and distribution) have been carried out in the atmosphere (Lee et al. 2001), water (Zhou and Maskaoui 2003), sediment (Wu et al. 2003) and soil (Tao et al. 2004). Most PAHs are released from anthropogenic sources, and therefore most studies focused on PAHs from wastewater irrigation (Opopol 2007), vehicle exhausts (Chen et al. 2005), hydrocarbon spillage (Ou et al. 2004), industrial activities including coke ovens, gasworks petroleum refineries, wood conservation plants and power plants (Van Brummelen et al. 1996; Stalikas et al. 1997). However, information related to PAHs in electronic-waste (e-waste) processing areas is limited.
Guiyu, a village in southeast China, has been a booming e-waste processing center since 1995, and is a destination for e-waste illegally transported from developed countries. The extremely hazardous and dangerous e-waste “recycling” operations render the air, water and soil of Guiyu heavily polluted, which also pose a threat to the health of workers (who work with little or without any personal protective equipment) and local residents (Yu et al. 2006). Now, a number of papers (Deng et al. 2006; Leung et al. 2007; Wong et al. 2007a, b) have been published concerning the pollution of soil, air and water in Guiyu caused by heavy metals (e.g., lead and cadmium) and PTS, [i.e., PAHs, polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs), polybrominated diphenyl ethers (PBDEs)], from e-waste recycling using primitive techniques. The incomplete combustion of plastic waste at open burning sites was the main source of PAHs in the Guiyu soil (including both petrogenic and pyrogenic sources), reflected by the high dominance of phenanthrene (21.8%, as representative of low molecular weight PAHs) and pyrene (14%, as representative of high molecular weight PAHs) (Yu et al. 2006). The average and the highest concentrations of PAHs in the soil (582 and 3,206 μg/kg, respectively) were both the highest in southeast China. According to the criteria established by Maliszewska-Kordybach (1996), the Guiyu soil in general were identified as weakly contaminated (200–600 μg/kg), whereas the soils at the open burning area were classified as heavily contaminated (>1,000 μg/kg) (Yu et al. 2006).
Soil microbes play significant roles in recycling of plant nutrients, maintenance of soil structure, detoxification of noxious chemicals, and the control of plant pests and plant growth (Elsgaard et al. 2001; Filip 2002). Recent research further revealed microbial activities in the degradation of PAHs despite their stability (Gao et al. 2006). Thus, soil microbes may be even more important players in severe POPs contaminated areas, such as e-waste processing areas. Up to now, however, few studies have been conducted on the evaluation of microbial community changes at e-waste processing areas (Nakatsu et al. 2000).
It is commonly known that only about 1% of the microbes in environmental samples can be cultured in laboratory media (Torsvik et al. 2002). Molecular techniques, such as Denaturing Gradient Gel Electrophoresis (DGGE), allow for the determination of the presence of various microbes and the analysis of community structure (Fromin et al. 2002). The objectives of the present study were: (a) to study the bacterial community structures in Guiyu in situ soils by using DGGE analysis of 16S rRNA genes; (b) to design laboratory-incubated experiments to minimize the environmental complexity and to understand the bacterial response to certain POPs, based on the results from (a); and (c) to assess the potential effect of e-waste pollution on bacterial community structure in both in situ field and laboratory-incubated soil samples. This is the first study which shows the bacterial community profile at an e-waste open burning area.
Materials and methods
Study sites and soil sampling
The study sites were located at and near the e-waste burning area of Guiyu, Shantou, Guangdong Province, China (Fig. 1). Guiyu consists of a total area of 52 km2 and a population of 150,000. The soil was contaminated by persistent organic pollutants (PBDEs, PCDD/Fs, PAHs) (Yu et al. 2006; Leung et al. 2007) and heavy metals (Cd, Cr, Cu, Pb, Ni, Zn) (Leung et al. 2006, 2008; Wong et al. 2007a, b). Based on our previous studies, six soil samples (0–10 cm, n = 5) were collected along an e-waste pollution gradient in Guiyu including one sample collected at an adjacent reservoir (Ximei Reservoir (RS), no e-waste activity) in December 2004 (Yu et al. 2006). Approximately 500 g of each soil sample was collected with a stainless steel scoop. Each sample was comprised of a mixture of five subsamples collected at the four corners and the center of an area of about 10 × 10 m2. Each sample was divided into two parts and both parts were packed separately with aluminium foil and placed in polyethylene bags (Ziploc®). The first part of each sample was transported to the laboratory, air-dried at room temperature (Belkessam et al. 2005), sieved through a 2 mm sieve, and then stored at −20°C before physicochemical and PAHs, PCBs, PBDEs and PCDD/Fs analysis. The second part of each sample was put on dry ice and transported to the laboratory immediately, and then stored at −80°C for DNA extraction.
Physicochemical properties of Guiyu soil samples
Dissolved organic carbon (DOC) was measured using a TOC-VCPH Total Organic Carbon analyzer (SHIMADZU, USA). Soil organic matter (SOM) was determined by loss on ignition at 450°C until a constant weight was achieved (Howsam et al. 2000). The moisture content (MC) of the soil was determined after the soil was oven dried at 105°C for 24 h. Soil moisture content (MC) was calculated from: MC (%) = (1 − dry weight of soil/fresh weight of soil) × 100%. Soil pH was tested (soil:water = 1:5) by the Beckman pH meter (Beckman, USA).
Sample extraction, cleanup, and analysis of selected POPs
Samples of air-dried soil were extracted for 18 h with acetone and dichloromethane (v:v 1:1, 80 ml) in a Soxhlet apparatus according to the EPA Standard Method 3540C (USEPA 1996a). The concentrated extracts were purified using Florisil column clean-up (EPA Standard Method 3620B) (USEPA 1996c). The eluant was evaporated to less than 2 ml prior to analysis. Deuterated PAHs (acenaphthene-d 10, phenanthrene-d 10, chrysene-d 12 and perylene-d 12) were used as internal standards for quantification. GC–MS analysis was performed on a Hewlett Packard 6890 GC system equipped with a mass selective detector and a 30 m × 0.25 mm × 0.25 μm DB-5 capillary column (J & W Scientific Co. Ltd., USA). USEPA Standard Method 8270C (USEPA 1996b) was used to determine the PAH compounds. The concentrations of PCBs, PBDEs and PCDD/Fs in the soil samples had been previously measured and described (Wong et al. 2007b).
Laboratory experiments
The addition of PAHs to the soil from Ximei Reservoir under aerobic condition was designed to stimulate the growth of organisms capable of degrading PAHs and therefore enrich the population of these organisms in the samples. The soil samples (500 g) were air-dried at ambient temperature, crushed and sieved through 2 mm mesh with large pieces of plant materials, stones and soil animals removed. Different final concentrations (0, 10, 100, 500 and 1,000 μg/kg) of the mixtures of phenanthrene and pyrene (1:1) in acetone (purities higher than 98 and 96%, respectively; SIGMA, Germany) were spiked into triplicate soil samples according to the method described by Brinch et al. (2002). Control water content capacity was set as 40%. Samples were stored at −80°C for DNA extraction.
DNA extraction and PCR
Environmental bacterial DNA was extracted from 500 mg of both in situ Guiyu soils and laboratory-incubated soils using a FastPrep DNA isolation kit according to the manufacturer’s protocol (BIO 101, USA).
Bacterial 16S rRNA genes were amplified by touch-down PCR using the primer set 341F (5′-CCT ACG GGA GGC AGC AG-3′) and 907R (CCG TCA ATT CMT TTG AGT TT-3′) with GC-clamps (CGC CCG CCG CGC CCC GCG CCC GGC CC GCC GCC CCC GCC CC) attached to the forward primer (Muyzer et al. 1993, 1998). PCR was performed using the following reaction mixtures: 200 ng of DNA, 1× PCR buffer, 1 μM of each primer, 200 μM of each dNTPs, and 1 U Taq DNA polymerase (TaKaRa, Japan) to give a final volume of 50 μl. Thermal cycling was carried out under the following conditions: 95°C for 2 min, followed by 10 cycles of 95°C for 30 s, 65°C (decrease 1°C per cycle) for 1 min, and 72°C for 2 min; then followed by 15 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 5 min.
DGGE analysis
DGGE was performed using 8% polyacrylamide gel with a denaturing gradient of 40–80% (100% denaturant = 7 M urea, 40% (v/v) formamide) using Bio-Rad Protean II System for 18 h at a constant voltage of 125 V and a temperature of 60°C in 1× TAE buffer. After electrophoresis, the gels were stained for 20 min using SYBR Gold (1:1,000 dilutions, Invitrogen, USA) and photographed with UV transillumination (302 nm) with an Alpha Imager 2000 (Alpha-Innotech-Corporation, USA).
Digitized DGGE images were analyzed with Quantity One image analysis software (version 4.0, Bio-Rad, USA). DGGE band position and intensity were determined using a GELCOMPAR II software package (Applied Maths) and were manually modified. Band matching was performed with 1.00% position tolerance and 1.00% optimization. Cluster analysis was performed based on the Pearson similarity correlation and the Ward or Unweighted Pair Group Method with Arithmetic mean (UPGMA) dendrograming method.
The middle portion of each selected DGGE band was excised, washed with Milli-Q water, and incubated in 50 μl of Milli-Q water at room temperature for 4 h. After centrifugation at 11,000g for 60 s, the supernatant was transferred to a new tube, and 2 μl of it was used as the template for the PCR-DGGE analysis to check the band position and purity. Afterwards, PCR products were purified using the Wizard PCR prep DNA purification system (Promega, USA) and cloned into the vector with a TOPO TA Cloning Kit (Invitrogen, USA) according to the manufacturer’s instructions. The insertion of DNA fragments was confirmed by the same PCR-DGGE procedure. The 16S rRNA genes were sequenced from both ends with MegaBACE 500 (Amersham, USA). The nucleic acid sequences obtained were assembled using the Sequencher 4.2 (Gene Codes Corporation, USA). Affiliation of sequenced DGGE bands was determined using the BLASTN program available on the NCBI website (http://www.ncbi.nlm.nih.gov).
Statistical analysis
Shannon diversity index (H) is commonly used to measure biodiversity (Shannon and Weaver 1949). In the present study, Shannon diversity index was used to estimate bacterial diversity in both Guiyu and PAH-inoculated soils using the following equation: H = Σ (n ί /N) ln (n ί /N), where n ί is the peak height of the band ί, ί is the number of bands in each DGGE gel profile. N is the sum of peak heights in a given DGGE gel profile. Analysis of variance (ANOVA) was performed on experimental data and means were compared using the Duncan’s Multiple Range test with SPSS 11.5 (SPSS Inc. USA). Regression analysis was performed by SPSS 11.5 to investigate correlation between H and organic pollution. The significance level was P ≤ 0.05.
Nucleotide sequence accession numbers
The 16S rRNA gene sequences determined in this study were deposited in the GenBank with the following accession numbers: from EU561220 to EU561229.
Results and discussion
Distribution and concentration of POPs in Guiyu Soil
The basic physicochemical properties measured in this study are summarized in Table 1. Dissolved organic carbon (DOC) ranged considerably from 73 to 200 mg/kg among different sites. Soil organic carbon (SOC) was generally higher at open-burning and near open-burning sites than that at rice field and the reservoir site. The moisture contents (MC) at different sites were similar while pH values were generally lower at open-burning sites than at the other sampling sites.
The average total POPs (PAHs, PCBs, PBDEs, and PCDD/Fs) concentrations, Shannon diversity index (H) (deduced from DGGE profiles), and their correlations are presented in Table 2. In general, the range of various POPs (PAHs, PCBs, PBDEs, and PCDD/Fs) concentrations in soil decreased in the following order: open-burning sites > near open-burning site > rice field > reservoir area. The average PAH, PBDE, and PCDD/F concentrations at the open-burning sites were greater than that at the reservoir site by over 9,358, and 579 times, respectively. These POPs concentrations were statistically higher (P < 0.05) than all of the other sites (data not shown). The concentrations of PAHs in soil samples were generally higher near emission sources at OBSs, which may have been due to deposition of fly ash and emissions from e-waste open burning. The PBDE concentration at open-burning site (OBS2) was among the highest concentration reported in literature (36,215 μg/kg, dry wt). These data clearly indicated that the soils in Guiyu were heavily polluted by POPs at the open-burning sites, and that the ambient soils (at near open-burning sites or rice field) were also affected by the e-waste burning process. The POPs generated may travel in the air as fly ash (de Wit 2002), in the water, and settle in adjacent soils (Wong et al. 2007b; Leung et al. 2007). In addition, dumping of printed circuit boards and spent acid solution after acid stripping (to collect metals) may also transfer toxic chemicals, such as PBDEs, and many metals and affect soil properties (Leung et al. 2007).
Bacterial communities in field soil samples
Bacterial community structures of indigenous and laboratory-incubated soils were assessed by DGGE fingerprinting of bacterial 16S rRNA genes from soil-extracted DNA. Apparent differences of cluster analysis based on DGGE patterns were observed in the microbial communities among different soil samples in the Guiyu area, as shown in Fig. 2. Samples from rice field (RF) and near open-burning site (NOBS) were clustered together, and then grouped with the sample from the reservoir; whereas samples from different open-burning sites (OBSs) were loosely grouped (Fig. 2). The results indicated that the e-waste pollution and the emission of various POPs induced changes on bacterial activity and community composition in Guiyu soils.
Cluster analysis of DGGE gel pattern showing the amplified 16S rRNA gene fragments from field soil samples. The DGGE were performed for PCR products with templates combined from three replicates samples, which individually generated the same DGGE patterns (data not shown). Cluster analysis was performed based on the Pearson similarity correlation and the Ward dendrogramming method. For site abbreviations see Fig. 1
Subsequently, to preliminarily determine the relationship between e-waste pollution and soil bacterial community composition, gel band patterns of DGGE were interpreted by using the Shannon index and then compared with the concentrations of these POPs which were measured previously (Table 2). A higher Shannon index indicates a more diversified bacteria community. Results showed that the Shannon index varied from 0.98 to 1.21 at different sites. The most diversified bacterial community was determined to be present at OBS3 and the bacterial community with the lowest diversity index was found near at NOBS. No significant correlations were found between Shannon indices and different POPs, except for PAHs (r = 0.592, P < 0.05) (Table 2). Therefore, although e-waste contamination affected the general bacterial community structure, the relationship between various e-waste contamination and in situ bacterial community diversity index is still unclear in this study.
The effects of organic compounds on bacterial community structure in natural communities are complex. This complexity is reflected by the diverse physical, chemical, and biological factors (Battin et al. 2008). First of all, the activity and composition of soil bacterial community are closely related to soil fertility and environmental quality. The soils from Guiyu had distinct characteristics and quality. For example, most of the soil samples were contaminated by heavy metals and organic pollutants (Tables 1, 2). It has been demonstrated that the toxicity of heavy metals is an important factor affecting the composition of soil microbial community (Wang et al. 2006). Second, POPs entering the soil may affect bacterial communities not only by reducing the diversity, but they may also change the composition of the bacterial community by, for instance, enhancing certain POP-consuming cohorts (Gao et al. 2006). One of the advantages of the Shannon index is that it takes into account the number of species and the evenness of the given community (Shannon and Weaver 1949). Therefore, the increase in diversity index at the open-burning sites may have been caused by the presence of more unique POPs-consuming bacterial species or by greater species evenness among POPs-consuming bacteria, which equaled or exceeded the loss of POPs-sensitive bacteria. All these possible changes are not easy to identify, which is the reason why a correlation between diversity index and POPs concentration hard was to predict. Third, a molecular method based on DGGE of 16S rRNA genes was used to document the bacterial community and calculate the diversity index because this approach is a commonly employed technique used to describe the composition of complex microbial communities, and to gain a descriptive overview of possible differences among communities (Yakimov et al. 2005). However, molecular methods have biases, such as preferential amplification of certain sequences (Morasch et al. 2001) and different PCR amplification efficiencies of DNA from environmental samples (Chandler et al. 1997). In addition, DGGE reveals only major PCR amplified bacterial 16S rRNA genes and, therefore, underestimates the total diversity of bacterial assemblage due to relative low resolution of gel running and staining. These possible biases may influence fingerprinting results and have to be considered when interpreting data. Last, but not least, there are still many other factors that may change bacterial community in the field, such as effects of predators and lytic phages (Zhang et al. 2007). All of the above presents difficulties in analyzing the relationship between POPs in soil due to uncontrolled recycling of e-waste and in situ bacterial community. Therefore, to understand the bacterial response to POPs, and based on our results that in situ bacterial community compositions were only correlated with PAHs concentrations in soil, laboratory experiments were carried out to minimize the environmental complexity by using PAHs inoculated soils.
Bacterial communities in laboratory-incubated soil samples
All five replicates from each treatment showed the same DGGE pattern in preliminary experiments (data not shown). Two randomly selected replicates of each PAH-treated sample were used to compare changes in the bacterial community under different dosages of PAHs. Figure 3 shows the DGGE pattern and cluster analysis of PCR products amplified with DNA templates from different dosage treatments. Ward and UPGMA dendrogram methods generated the same topology of clustering. Cluster analysis of the DGGE profile indicated that the addition of PAHs had a significant impact on soil bacterial community structure. The samples clustered into two main groups consisting of non-acetone treated soils and acetone-treated soils. Within the acetone-treated cluster, samples contaminated with PAHs showed a similar bacterial community structure while the RS samples formed separate clusters (Fig. 3). Moreover, in the cluster of PAH-contaminated samples, samples were further grouped with respect to dosage of PAHs. For example, bacterial communities in soils inoculated with 1,000 and 500 mg/kg PAHs were similar members while bacterial communities in soils inoculated with 100 and 10 mg/kg PAHs clustered together. Based on culturing methods and physiological assays, a previous study showed that high concentrations of PAHs in soil may affect the microorganisms by reducing their diversity (Nakatsu et al. 2000). DNA fingerprinting suggests that the bacterial diversity was not simply reduced, but varied up and down according to different PAH dosage treatments. For instance, the presence of two specific phylotypes represented by DGGE bands-5 and -7 (Fig. 2; Table 3) were found to be enhanced by increasing PAHs concentration (indicated by band intensity). The bacterial community changed not only with regards to species richness (represented by number of bands) and diversity (indicated by Shannon index), but also in its composition (showed by cluster grouping; Fig. 2).
DGGE gel pattern and cluster analysis for samples from lab-incubation experiments. Cluster analysis was performed based on the Pearson similarity correlation and the Ward dendrogramming method. Initial concentrations of PAHs (phenanthrene: pyrene = 1:1) in treated soils (mg/kg, on a dry weight basis) are indicated. “U” represent unit as mg/kg; “A” in the bracket indicates using acetone as solvent. Numbers from 1 to 10 indicates the bands that sequenced in this study
DGGE analysis showed that bacterial communities in PAHs contaminated soils were significantly different from those in reservoir soil (Fig. 3). In general, the DGGE profile revealed 33 bands (identified using GELCOMPAR II; Applied Maths) with 1.00 % band position tolerance and 1.00% band optimization. Among the 33 bands, seven bands (21.1%) were identified in all 20 samples (Fig. 3). Ten of the 33 bands were sequenced (Fig. 3; Table 3). The DGGE pattern indicated that unique bacterial communities developed in soils with different dosages of PAH contaminations. Some of the bacteria (e.g., BAND-9) had zero or low tolerance to PAHs, while some of the bacteria (represented by BANDs-1, -2, -3,-5, -6, -8) were not dramatically affected by PAH contamination. The rest of the bacteria in the community (represented by BANDs-4, -7, -10) appeared to be present in higher numbers favored by different concentrations of PAHs. Whether or not this cohort of bacteria can be employed as bio-indicators or bio-tracers for PAHs or e-waste needs to be further investigated.
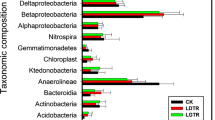
Taxonomic analysis based on top BLAST hits to the GenBank showed that all the DGGE bands sequenced were affiliated with β-proteobacteria (60%) and Firmicutes (40%) (Table 3). All Firmicute sequence clones were obtained from laboratory-incubated soil with a range of PAH concentrations. Most of them showed sequences that were similar with 16S rRNA genes from Clostridium sp. and with clones obtained from anoxic natural soils or petroleum-contaminated soils. Three DGGE band sequences retrieved top BLAST hits of sequences from Massilia sp. and one Acidivorax sp., both Betaproteobacteria and isolated from oxic soils or the oxic-anoxic interphase. Although three Massilia-like sequences shared high sequence similarity, their presence varied for the different ranges of PAHs concentration (Table 3).
The varying distribution of bacterial phylotypes in response to PAH treatment may indicate ongoing bacterial adaptation to environmental changes. It is hypothesized that varying PAH concentrations may provide additional metabolic niches for members of the Massilia spp., thereby facilitating ecotype speciation. This type of micro-diversification might also help Clostridium sp. to be one of the most successful bacteria to thrive in PAHs contaminated soil.
Conclusions
This study is the first to analyze bacterial community structures at e-waste processing sites. DGGE analysis suggests that the POPs in Guiyu soils altered the bacterial community by promoting changes in species composition and species richness. The diversity of in situ bacterial communities decreased at the near open-burning site (NOBS) (by suppressing POPs-sensitive species) and increased at the open-burning site (OBS) (by substantially favoring POPs-consuming species), compared with that at the reservoir (RS) site. Laboratory-incubation experiments showed that different PAH pollution levels affected the bacterial community by suppressing or favoring specific groups of bacteria. Taxonomic analysis indicated that β-proteobacteria and Firmicutes were the abundant bacterial lineages in PAH-polluted soils. The findings that differences in in situ bacterial community structures existed between e-waste processing and the reservoir site, and the different responses of bacterial community in laboratory-incubated soil to various dosages of PAHs calls for a more intensive study of microbial communities at e-waste polluted areas and a system to monitor PAHs or other POPs biohazards to microbial community in natural environments.
References
Battin TJ, Kaplan LA, Findlay S, Hopkinson CS, Marti E, Packman AI, Newbold JD, Sabater F (2008) Biophysical controls on organic carbon fluxes in fluvial networks. Nat Geosci 1:95–100
Belkessam L, Lecomte P, Milon V, Laboudigue A (2005) Influence of pre-treatment step on PAHs analyses in contaminated soils. Chemosphere 58:321–328
Brinch UC, Ekelund F, Jacobsen CS (2002) Method for spiking soil samples with organic compounds. Appl Environ Microb 68:1808–1816
Chandler DP, Fredrickson JK, Brockman FJ (1997) Effect of PCR template concentration on the composition and distribution of total community 16S rRNA gene clone libraries. Mol Ecol 6:475–482
Chen L, Ran Y, Xing B, Mai B, He J, Wei X, Fu J, Sheng G (2005) Contents and sources of polycyclic aromatic hydrocarbons and organochlorine pesticides in vegetable soils of Guangzhou, China. Chemosphere 60:879–890
de Wit CA (2002) An overview of brominated flame retardants in the environments. Chemosphere 46:583–624
Deng WJ, Louie PKK, Liu WK, Bi XH, Fu JM, Wong MH (2006) Atmospheric levels and cytotoxicity of PAHs and heavy metals in TSP and PM2.5 at an electronic waste recycling site in southeast China. Atmos Environ 40:6945–6955
Elsgaard L, Petersen SO, Debosz K (2001) Effects and risk assessment of linear alkylbenzene sulfonates in agricultural soil. 1. Short-term effects on soil microbiology. Environ Toxico Chem 20:1656–1663
Filip Z (2002) International approach to assessing soil quality by ecologically-related biological parameters. Agric Ecosyst Environ 88:169–174
Fromin N, Hamelin J, Tarnawski S, Roesti D, Jourdain-Miserez K, Forestier N, Teyssier-Cuvelle S, Gillet F, Aragno M, Rossi P (2002) Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ Microbiol 4:634–643
Gao Y, Yu XZ, Wu SC, Cheung KC, Tam NFY, Qian PY, Wong MH (2006) Interactions of rice (Oryza sativa L.) and PAH-degrading bacteria (Acinetobacter sp.) on enhanced dissipation of spiked phenanthrene and pyrene in waterlogged soil. Sci Total Environ 372:1–11
Howsam M, Jones KC, Ineson P (2000) PAHs in the soils of a mature, mixed-deciduous (quercus-fraxinus) woodland and the surrounding pasture. Water Air Soil Poll 121:379–398
IARC (1983) Monographs on the evaluation of the carcinogenic risk of chemicals to humans: polynuclear aromatic hydrocarbons, vol 32. WHO, Lyon
Lee SC, Ho KF, Chan LY, Zielinska B, Chow JC (2001) Polycyclic aromatic hydrocarbons (PAHs) and carbonyl compounds in urban atmosphere of Hong Kong. Atmos Environ 35:5949–5960
Leung A, Cai ZW, Wong MH (2006) Environmental contamination from electronic waste recycling at Guiyu, Southeast China. J Mater Cycl Waste Manag 8:21–33
Leung AOW, Luksemburg WJ, Wong AS, Wong MH (2007) Spatial distribution of polybrominated diphenyl ethers and polychlorinated dibenzo-p-dioxins and dibenzofurans in soil and combusted residue at Guiyu, an electronic waste recycling site in southeast China. Environ Sci Tech 41:2730–2737
Leung AOW, Duzgoren-Aydin NS, Cheung KC, Wong MH (2008) Heavy metals concentrations of surface dust from e-waste recycling and its human health implications in southeast China. Environ Sci Tech 42:2674–2680
Maliszewska-Kordybach B (1996) Polycyclic aromatic hydrocarbons in agricultural soils in Poland: preliminary proposals for criteria to evaluate the level of soil contamination. Appl Geochem 11:121–127
Morasch B, Richnow HH, Schink B, Meckenstock RU (2001) Stable hydrogen and carbon isotope fractionation during microbial toluene degradation: mechanistic and environmental aspects. Appl Environ Microbiol 67:4842–4849
Muyzer G, Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes encoding for 16S rRNA. Appl Environ Microbiol 59:695–700
Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C (1998) Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans ADL, van Elsas JD, de Bruijn FJ (eds) Molecular microbial ecology Manual, Suppl 3, 3.3.4, Kluwer, Dordrecht, pp 1–27
Nakatsu CH, Torsvik V, Øvreås L (2000) Soil community analysis using DGGE of 16S rRNA gene polymerase chain reaction products. Soil Sci Soc Am J 64:1382–1388
Opopol N (2007) Impact of pops on the republic of moldova environment and public health. In: The fate of persistent organic pollutants in the environment, pp 405–424. http://www.springerlink.com/content/p80g75628u3566jm
Ou SM, Zheng JH, Zheng JS, Richardson BJ, Lam PKS (2004) Petroleum hydrocarbons and polycyclic aromatic hydrocarbons in the surficial sediments of Xiamen Harbour and Yuan Dan Lake, China. Chemosphere 56:107–112
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Stalikas CD, Chaidou CI, Pilidis GA (1997) Enrichment of PAHs and heavy metals in soils in the vicinity of the lignite-fired power plants of West Macedonia (Greece). Sci Total Environ 204:135–146
Tao S, Cui YH, Xu FL, Li BG, Cao J, Liu WX, Schmitt G, Wang XJ, Shen WR, Qing BP, Sun R (2004) Polycyclic aromatic hydrocarbons (PAHs) in agricultural soil and vegetables from Tianjin. Sci Total Environ 320:11–24
Torsvik V, Ovreas L, Thingstad TF (2002) Prokaryotic diversity magnitude dynamics, and controlling factors. Science 296:1064–1066
USEPA (1996) Method 3540C Revision 3, Soxhlet Extraction. In: Test methods for evaluating solid waste, physical/chemical methods; SW-846, Washington, DC
USEPA (1996) Method 8270C, Semivolatile organic compounds by gas chromatography/mass spectrometry (GC/MS). In: Test methods for evaluating solid waste, physical/chemical methods; SW-846, Washington, DC
USEPA (1996) Method 3620B, Florisil Cleanup. In: Test methods for evaluating solid waste, physical/chemical methods; SW-846, Washington, DC
Van Brummelen TC, Verweij SA, Wedzinga SA, Van Gestel CAM (1996) Enrichment of polycyclic aromatic hydrocarbons in forest soils near a blast furnace plant. Chemosphere 32:293–314
Wang YP, Yan J, Wang SH, Lin Q, Chen XC, Chen YX (2006) The influence of soil heavy metals pollution on soil microbial biomass, enzyme activity, and community composition near a copper smelter. Ecotox Environ Saf 67:75–81
Wong CSC, Duzgoren-Aydin NS, Aydin A, Wong MH (2007a) Evidence of excessive releases of metals from primitive e-waste processing in Guiyu, China. Environ Pollut 148:62–72
Wong MH, Wu S, Deng WJ, Yu XZ, Luo Q, Leung AOW, Wong CSC, Luksemburg WJ, Wong AS (2007b) Export of toxic chemicals—a review on the case of uncontrolled electronic waste recycling. Environ Pollut 149:131–140
Wu Y, Zhang J, Zhu Z (2003) Polycyclic aromatic hydrocarbons in the sediments of the Yalujiang Estuary, North China. Mar Pollut Bull 46:619–625
Yakimov MM, Denaro R, Genovese M, Cappello S, D’Auria G, Chernikova TN, Timmis KN, Golyshin PN, Giluliano L (2005) Natural microbial diversity in superficial sediments of Milazzo Harbor (Sicily) and community successions during microcosm enrichment with various hydrocarbons. Environ Microbiol 7:1426–1441
Yu XZ, Gao Y, Wu SC, Zhang HB, Cheung KC, Wong MH (2006) Distribution of polycyclic aromatic hydrocarbons in soils at Guiyu area of China, affected by recycling of electronic waste using primitive technologies. Chemosphere 65:1500–1509
Zhang R, Weinbauer MG, Qian PY (2007) Viruses and flagellates sustain richness and reduce biomass accumulation of bacterioplankton in coastal marine waters. Environ Microbiol 9:3008–3018
Zhou JL, Maskaoui K (2003) Distribution of polycyclic aromatic hydrocarbons in water and surface sediments from Daya Bay, China. Environ Pollut 121:269–281
Acknowledgments
The authors thank Y. M. Tam for assistance in sequencing work. Financial support from The Research Grants Council of the University Grants Committee of Hong Kong (Central Allocation Group Research Project HKBU 1/03C) and a private donation is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, W., Wang, H., Zhang, R. et al. Bacterial communities in PAH contaminated soils at an electronic-waste processing center in China. Ecotoxicology 19, 96–104 (2010). https://doi.org/10.1007/s10646-009-0393-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-009-0393-3