Abstract
Background
Several lines of evidence have indicated that growth arrest-specific transcript 5 (GAS5) functions as a tumor suppressor and is aberrantly expressed in multiple cancers. GAS5 was found to be downregulated in gastric cancer (GC) tissues, and ectopic expression of GAS5 inhibited GC cell proliferation.
Aims
The present study aimed to explore the underlying mechanisms of GAS5 involved in GC cell proliferation.
Methods
GAS5 and miR-222 expressions in GC cell lines were estimated by quantitative real-time polymerase chain reaction. The effects of GAS5 and miR-222 on GC cell proliferation were assessed by MTT assay and 5-bromo-2-deoxyuridine (BrdU) incorporation assays. The interaction between GAS5 and miR-222 was confirmed by luciferase reporter assay and RNA immunoprecipitation assay. The protein levels of the phosphatase and tensin homolog (PTEN), phosphorylated protein kinase B (Akt) (p-Akt), Akt, phosphorylated mammalian target of rapamycin (mTOR) (p-mTOR), and mTOR were determined by western blot.
Results
GAS5 was downregulated and miR-222 was upregulated in GC cells. GAS5 directly targeted and suppressed miR-222 expression. GAS5 overexpression and miR-222 inhibition suppressed cell proliferation, increased PTEN protein level and decreased p-Akt and p-mTOR protein levels in GC cells while GAS5 knockdown and miR-222 overexpression exhibited the opposite effects. Moreover, mechanistic analyses revealed that GAS5 regulated GC cell proliferation through the PTEN/Akt/mTOR pathway by negatively regulating miR-222.
Conclusions
GAS5/miR-222 axis regulated proliferation of GC cells through the PTEN/Akt/mTOR pathway, which facilitated the development of lncRNA-directed therapy against this deadly disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) ranks the fourth most prevalent malignant tumor and the second leading cause of cancer-associated mortality worldwide, accounting for 700,000 deaths annually [1]. Although great improvements have been made in the diagnosis and therapy of GC in recent years, the clinical outcome of GC patients remains unsatisfactory, with the 5-year overall survival rate of GC patients lower than 30% [2]. More than 80% of GC patients are diagnosed at an advanced stage which is accompanied by malignant proliferation, extensive invasion and distant metastasis, thus leading to a poor prognosis [3]. Therefore, it is urgently needed to identify effective therapeutic targets for GC patients.
In the past few years, increasing evidence has highlighted the important roles of noncoding RNAs (ncRNAs), including the well-known microRNA (miRNA) and the recently acknowledged long noncoding RNA (lncRNA), in the pathogenesis of multiple human cancers [4, 5]. miRNAs are a group of highly conserved small ncRNAs with 20–25 nucleotides in length, which play a regulatory role in gene expression at the posttranscriptional level by directly targeting the 3′-untranslated region (3′-UTR) of target mRNA [6]. An increasing number of miRNAs have been revealed to take part in diverse physiological and pathological processes, including cell proliferation, differentiation, invasion, metastasis, and apoptosis [7]. Studies have increasingly indicated that aberrantly expressed miRNAs function as oncogenes or tumor suppressors in various malignant tumors and contribute to carcinogenesis and progression of numerous cancers, including GC [8]. miR-222, known as a member of oncomiRs, has been shown to promote the oncogenesis of several tumors including GC [9,10,11]. LncRNAs, a novel class of ncRNAs long than 200 nucleotides, have received increasing attentions in cancer research [12]. Amounting functional lncRNAs have been shown to be involved in the regulation on gene expression at various levels, including chromatin modification, transcription and posttranscriptional processing [13]. Dysregulated lncRNAs play oncogenic or tumor-suppressive roles in the pathogenesis of multiple cancers [14, 15]. LncRNA growth arrest-specific transcript 5 (GAS5), localized at chromosome 1q25, was originally isolated from a subtraction cDNA library [16]. Several lines of evidence have indicated that GAS5 functions as a tumor suppressor and is aberrantly expressed in multiple cancers [17]. Interestingly, GAS5 was found to be downregulated in GC tissues, and ectopic expression of GAS5 inhibited GC cell proliferation [18, 19]. Numerous evidences previously proposed the competing endogenous RNA (ceRNA) hypothesis that lncRNAs function as ceRNAs or molecular sponges of miRNAs in regulating its expression and biological functions [20]. It is well established that lncRNA GAS5 serves as a ceRNA in the occurrence and development of tumors [21]. For example, GAS5 exerted tumor-suppressive functions by targeting miR-222 in human glioma [22]. However, whether GAS5 could regulate the development of GC by reducing the expression of miR-222 remains to be further investigated.
In the present study, we demonstrated that GAS5 was downregulated and miR-222 was upregulated in GC cell lines. Furthermore, GAS5 functioned as an endogenous sponge by directly binding to miR-222 and suppressed miR-222 expression. Importantly, mechanistic analysis demonstrated that GAS5/miR-222 axis regulated GC cell proliferation through the PTEN/Akt/mTOR pathway. Therefore, the GAS5/miR-222/PTEN/Akt/mTOR pathway may be a novel potential therapeutic target for GC.
Materials and Methods
Cell Culture and Transfection
Human GC cell lines (SGC-7901, MKN-28, MKN-45, and MGC-803) and normal gastric epithelial cell line GES-1 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin (Sigma-Aldrich, St. Louis, MO, USA), and 100 μg/ml streptomycin (Sigma-Aldrich) at 37 °C in humidified atmosphere with 5% CO2, and the medium was replaced every 2 days.
pcDNA3.1-GAS5 (GAS5), pcDNA3.1 empty vector (Vector), siRNA against GAS5 (si-GAS5), negative control siRNA (si-NC), miR-222 mimics (miR-222), negative control miRNA (miR-NC), miR-222 inhibitor (miR-222), and miRNA inhibitor negative control (anti-miR-NC) were synthesized and purchased from GenePharma Co., Ltd. (Shanghai, China). Transfection with oligonucleotides or plasmids was performed with Lipofectamine 2000 reagents (Invitrogen) according to the manufacturer’s instructions. Cells were collected 48 h after transfection, and the expression of GAS5 and miR-222 was monitored using quantitative real-time PCR (qRT-PCR).
RNA Extraction and qRT-PCR
Total RNA was extracted from cells using Trizol reagent (Invitrogen). For qPCR, 1 µg of total RNA was reversely transcribed into cDNA using PrimeScript RT-polymerase (Takara, Dalian, China). The expression of GAS5 was evaluated using a SYBR Premix Ex Taq™ kit (Takara), and GAPDH was used as the internal control. The miR-222 expression was quantified using TaqMan microRNA assays (Ambion, Austin, TX, USA), and U6 small nuclear RNA (snRNA) was used as an endogenous control. qRT-PCR was carried out using the Applied Biosystems 7900 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The relative gene expression was calculated using the 2−ΔΔCt method. The primers used were synthesized by Sangon Biotech Co. Ltd. (Shanghai, China), and the primer sequences were as follows: GAS5, forward 5′-AGC TGG AAG TTG AAA TGG -3′, reverse 5′-CAA GCC GAC TCT CCA TAC C-3′; GAPDH, forward 5′-AAC GTG TCA GTG GTG GAC CTG-3′, reverse 5′-AGT GGG TGT CGC TGT TGA AGT-3′; miR-222, forward 5′-AGC TAC ATC TGG CTA CTG GGT AAA A-3′, reverse 5′-GTC GTC AAC GAT ACG CTA CGT AAC G-3′; U6, forward 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′, reverse 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′.
Western Blot
For western blot analysis, cells were lysed using RIPA buffer (Sigma-Aldrich). Protein lysates were resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After blocking with 5% skimmed milk in tris-buffered saline (TBS) containing 0.1% Tween-20 for 2 h at room temperature, the PVDF membranes were probed overnight at 4 °C with the primary antibodies PTEN (Abcam, Cambridge, MA, USA), phosphorylated-Akt (p-Akt) (Ser473; Santa Cruz Biotech, Santa Cruz, CA, USA), Akt (Santa Cruz Biotech), phosphorylated-mTOR (p-mTOR) (Ser2448; Santa Cruz Biotech), mTOR (Santa Cruz Biotech), β-actin (Santa Cruz Biotech) at 4 °C overnight, followed by incubation with horseradish peroxidase-labeled second antibody goat anti-rabbit IgG (Santa Cruz Biotech) at 37 °C for 2 h. The protein was visualized using an ECL system (Amersham Pharmacia, Piscataway, NJ, USA). Protein bands were quantified using Quantity One software (Bio-Rad, Hercules, CA, USA).
MTT Assay
Briefly, the transfected SGC-7901 and MGC-803 cells were seeded in 96-well plates at 1.5 × 103 cells/well and cultured at 37 °C with 5% CO2 for 0, 24, 48, and 72 h. Subsequently, 20 µl of MTT solution (5 mg/ml; Sigma-Aldrich) was added to each well, followed by another incubation of 4 h. The medium was then removed, and 150 µl dimethyl sulfoxide (DMSO) was added to each well to dissolve blue formazan crystals. The absorbance at 570 nm was measured using a microplate reader (Bio-Tek, Winooski, VT, USA).
5-Bromo-2-Deoxyuridine (BrdU) Incorporation Assay
Cell proliferation enzyme-linked immunosorbent assay-BrdU kit (Millipore) was used to analyze cell proliferation following the manufacturer’s protocols. Briefly, the transfected SGC-7901 and MGC-803 cells (1.5 × 103 cells/well) were seeded in 96-well plates at a density of 2 × 103 cells/well and 10 μM of BrdU was added to each well. Following incubation for 0, 24 h, 48 h, and 72 h at 37 °C with 5% CO2, cells were washed with PBS, fixed by 4% paraformaldehyde (PFA) for 20 min and treated with 0.3% Triton X-100 for 10 min. After blocking with 10% goat serum for 1 h, cells were labeled with peroxidase-coupled BrdU primary antibody (Santa Cruz Biotechnology) overnight at 4 °C. After washing with PBS, cells were then incubated with peroxidase substrate (tetramethylbenzidine) for 30 min. The absorbance at 450 nm was determined by a microplate reader (Bio-Tek).
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted using the EZ-Magna RIP kit (Millipore) following the manufacturer’s protocol. Briefly, cells at 80% confluency were scraped off and lysed in complete RIPA lysis buffer (Sigma-Aldrich) containing 10% proteinase inhibitor cocktail (Sigma-Aldrich) and 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich). Then, 100 μl of whole cell extract was immunoprecipitated with RIP wash buffer containing A + G magnetic beads conjugated with human Argonaute2 (Ago2) antibody (Millipore) or mouse immunoglobulin G (IgG) (Millipore) as a negative control. Proteinase K was used to digest the protein in the samples, and the immunoprecipitated RNA was isolated. Purified RNA was then subjected to qRT-PCR analysis to demonstrate the presence of the binding targets between GAS5 and miR-222.
Luciferase Reporter Assay
To construct dual-luciferase reporter plasmids, the predicted binding sequence of miR-222 in the wild-type of GAS5 gene and the corresponding mutant type were synthesized and inserted into the downstream of firefly luciferase gene in pGL3 basic luciferase reporter vector (Promega Corp., Madison, WI, USA), named as GAS5-Wt and GAS5-Mut. SGC-7901 and MGC-803 cells were cotransfected with 0.4 μg of GAS5-Wt and GAS5-Mut, along with 0.02 μg of the pRL-TK control vector and 50 nM miR-222 or miR-NC using Lipofectamine 2000 reagents (Invitrogen). Cells were collected 48 h after transfection, the luciferase activities were measured through Dual-Luciferase reporter assay system (Promega), and renilla luciferase activity was used as the normalization.
Statistical Analysis
All data were analyzed with the SPSS software 19.0 (SPSS Inc., Chicago, IL, USA). The Student’s t test or one-way analysis of variance (ANOVA) was used to determine the statistical significance between two or more groups. The results were considered as statistically significant when P value is less than 0.05.
Results
GAS5 Was Downregulated While miR-222 Was Upregulated in GC Cells
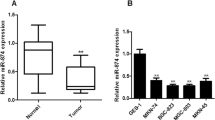
As previous reported, GAS5 expression was lower and miR-222 was higher in GC [19, 23]. Similarly, the expression of GAS5 was markedly reduced (Fig. 1a) and miR-222 was significantly elevated (Fig. 1b) in GC cell lines SGC-7901, MKN-28, MKN-45, and MGC-803 compared with the normal gastric epithelial cell line GES-1, especially in SGC-7901 and MGC-803 cells, as demonstrated by qRT-PCR. Therefore, SGC-7901 and MGC-803 cells were selected for gain- and loss-of-function analyses of GAS5 and miR-222. SGC-7901 and MGC-803 cells were introduced with GAS5, si-GAS5, miR-222, anti-miR-222, or matched controls and qRT-PCR was carried out to check the transfection efficiency. As shown in Fig. 1c, d, transfection of GAS5 remarkably improved GAS5 expression while si-GAS5 introduction effectively suppressed GAS5 expression in both SGC-7901 and MGC-803 cells. Meanwhile, the expression level of miR-222 was dramatically increased by miR-222 transfection and significantly decreased in anti-miR-222-treated SGC-7901 and MGC-803 cells (Fig. 1e, f).
Expressions of GAS5 and miR-222 in GC cells. a, b qRT-PCR analysis of GAS5 and miR-222 expressions in GC cell lines (SGC-7901, MKN-28, MKN-45, and MGC-803) and the normal gastric epithelial cell line GES-1. c, d qRT-PCR analysis of GAS5 expression in SGC-7901 and MGC-803 cells transfected with GAS5, si-GAS5, or respective controls. e, f qRT-PCR analysis of miR-222 expression in SGC-7901 and MGC-803 cells treated with miR-222, anti-miR-222, or matched controls. Data are presented as means ± standard deviation (SD), n = 3. *P < 0.05
GAS5 Directly Targeted and Negatively Regulated the Expression of miR-222 in GC Cells
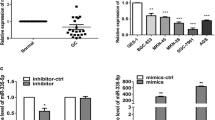
In view of the inverse expression change of GAS5 and miR-222 in GC cells, we supposed that GAS5 functioned as a molecular sponge of miR-222 in GC cells. Through online bioinformatics analysis by StarBase v2.0 program (http://starbase.sysu.edu.cn/), we found that miR-222 contained the complementary bases pairing with GAS5 (Fig. 2a). Since miRNAs exerted their gene silencing functions through a RNA-induced silencing complex (RISC) [24], RIP assay was performed in SGC-7901 cell extracts using antibody against Ago2, a key component of RISC, to confirm whether GAS5 and miR-222 were in the same RISC complex. RNA levels in the immunoprecipitates were examined by qRT-PCR. As illustrated in Fig. 2b, GAS5 and miR-222 were enriched in Ago2 immunoprecipitates compared with IgG control group. To verify the direct binding between GAS5 and miR-222, the wild-type or mutated GAS5 fragments containing the potential miR-222 binding sites were cloned into luciferase reporter plasmids. The luciferase reporter assay indicated that miR-222 overexpression restrained the luciferase activity of GAS5-WT compared with control group while it exhibited no influence on the luciferase activity of GAS5-MUT in SGC-7901 (Fig. 2c) and MGC-803 (Fig. 2d) cells. The regulatory association between GAS5 and miR-222 was further explored by determining the expression of miR-222 in SGC-7901 and MGC-803 cells transfected with GAS5, si-GAS5, or matched controls by qRT-PCR. The results demonstrated that ectopic expression of GAS5 reduced miR-222 expression in contrast to Vector group in SGC-7901 and MGC-803 cells (Fig. 2e). On the contrary, GAS5 knockdown improved the expression of miR-222 relative to si-NC group in SGC-7901 and MGC-803 cells (Fig. 2f). Collectively, these results suggested that GAS5 reduced the expression of miR-222 through directly targeting miR-222 in GC cells.
GAS5 functioned as a ceRNA and negatively regulated the expression of miR-222 in GC cells. a Schematic of wild-type or mutated GAS5 sequences containing the potential miR-222 binding sites. b Association between GAS5 and miR-222 with Ago2. RIP assay was performed in SGC-7901 cell extracts using antibody against Ago2. GAS5 and miR-222 levels in the immunoprecipitates were examined by qRT-PCR. c, d The relative luciferase activity was measured by luciferase reporter assay in SGC-7901 and MGC-803 cells cotransfected with GAS5-WT or GAS5-MUT and miR-222 or miR-NC. e, f The expression of miR-222 was detected by qRT-PCR in SGC-7901 and MGC-803 cells transfected with GAS5, si-GAS5, or corresponding controls. Data are presented as means ± SD, n = 3. *P < 0.05
GAS5 Overexpression Suppressed Cell Proliferation in GC Cells
We next performed gain- and loss-of-function experiments to confirm the biological function f GAS5 in GC. The effect of GAS5 on cell proliferation at 0, 24, 48, and 72 h was evaluated by MTT and BrdU incorporation assays. As a result, SGC-7901 cells with GAS5 transfection showed a marked inhibition of cell proliferation compared with that in Vector-transfected cells, as demonstrated by MTT (Fig. 3a) and BrdU incorporation (Fig. 3b) assays. Conversely, the results of MTT (Fig. 3c) and BrdU incorporation (Fig. 3d) assays also revealed that cell proliferation was promoted in GAS5 knockdown-MGC-803 cells in comparison with si-NC group. Therefore, we concluded that GAS5 overexpression suppressed cell proliferation in GC cells.
GAS5 overexpression suppressed cell proliferation in GC cells. Cell proliferation at 0, 24, 48, and 72 h of SGC-7901 cells following transfection with GAS5 or Vector and MGC-803 cells after transfection with si-GAS5 or si-NC was assessed by MTT (a, c) and BrdU incorporation (b, d) assays. Data are presented as means ± standard error (SEM), n = 3. *P < 0.05
miR-222 Inhibition Repressed Cell Proliferation in GC Cells
Similarly, the effect of miR-222 on GC cell proliferation was also evaluated by MTT and BrdU incorporation assays. MTT (Fig. 4a, c) and BrdU incorporation (Fig. 4b, d) assays both proved that elevated miR-222 expression promoted cell proliferation of SGC-7901 cells versus miR-NC group, while miR-222 inhibition by anti-miR-222 restrained cell proliferation in MGC-803 cells relative to anti-miR-NC group. Together, these data uncovered that miR-222 inhibition repressed cell proliferation in GC cells.
miR-222 inhibition repressed cell proliferation in GC cells. MTT (a, c) and BrdU incorporation (b, d) assays were performed to detect cell proliferation at 0, 24, 48, and 72 h after SGC-7901 cells were transfected with miR-222 or miR-NC and MGC-803 cells were transfected with anti-miR-222 or anti-miR-NC. Data are presented as means ± SEM, n = 3. *P < 0.05
GAS5 Regulated GC Cell Proliferation Through Negatively Regulating miR-222
To explore the effect of interaction between GAS5 and miR-222 on GC cell proliferation, rescue experiments were performed in SGC-7901 cells transfected with Vector, GAS5, or along with miR-222 or miR-NC and MGC-803 cells transfected with si-NC, si-GAS5, or combined with anti-miR-222 or anti-miR-NC. MTT (Fig. 5a, c) and BrdU incorporation (Fig. 5b, d) assays demonstrated that cell proliferation was repressed by exogenous expression of GAS5 in SGC-7901 cells compared to Vector transfection group, while forced expression of miR-222 recuperated the inhibitory effect of GAS5 on cell proliferation. In contrast, GAS5 knockdown led to a promotion of cell proliferation in MGC-803 cells compared with si-NC group, which was partially reversed by miR-222 suppression. Taken together, these results revealed that GAS5 regulated GC cell proliferation through negatively regulating miR-222.
GAS5 inhibited GC cell proliferation through negatively regulating miR-222. MTT (a, c) and BrdU incorporation (b, d) assays were carried out to estimate cell proliferation at 0, 24, 48, and 72 h after SGC-7901 cells were transfected with Vector, GAS5, or along with miR-222 or miR-NC and MGC-803 cells were transfected with si-NC, si-GAS5, or combined with anti-miR-222 or anti-miR-NC. *P < 0.05
Both GAS5 Overexpression and miR-222 Downregulation Modulated the PTEN/Akt/mTOR Pathway
Since documents have reported that GAS5 inhibited cell proliferation and progression of prostate cancer through the Akt/mTOR pathway [25], and miR-222 could regulate PTEN/Akt/mTOR pathway [26, 27], we explored the association between GAS5, miR-222 and the PTEN/Akt/mTOR pathway in GC cells. Western blot was performed to detect the protein levels of PTEN and phosphorylation of Akt and mTOR in SGC-7901 cells transfected with GAS5, miR-222 or matched controls and MGC-803 cells introduced with si-GAS5, anti-miR-222 or corresponding controls. The results uncovered that SGC-7901 cells with restoration of GAS5 expression showed a promotion in the PTEN level and a suppression in the p-Akt and p-mTOR (Fig. 6a) while MGC-803 cells with GAS5 knockdown exhibited the opposite effects on the protein levels of PTEN, p-Akt, and p-mTOR (Fig. 6b). Furthermore, forced expression of miR-222 decreased PTEN level and increased the levels of p-Akt and p-mTOR, but not the total amounts of Akt and mTOR in SGC-7901 cells compared with miR-NC group (Fig. 6c). Conversely, miR-222 inhibition improved PTEN level and reduced the phosphorylation of Akt and mTOR in MGC-803 cells. Collectively, these findings demonstrated that GAS5 overexpression and miR-222 inhibition regulated the PTEN/Akt/mTOR pathway.
GAS5 overexpression and miR-222 downregulation suppressed the PTEN/Akt/mTOR pathway. The protein levels of PTEN, p-Akt, Akt, p-mTOR, and mTOR in SGC-7901 cells transfected with GAS5, miR-222 or matched controls (a, c) and MGC-803 cells introduced with si-GAS5, anti-miR-222 or corresponding controls (b, d). Data are presented as means ± SD, n = 3. *P < 0.05
GAS5/miR-222 Axis Regulated GC Cell Proliferation Through the PTEN/Akt/mTOR Pathway
To further investigate the potential mechanism of GAS5 involved in GC cell proliferation, we analyzed the protein levels of PTEN, p-Akt, Akt, p-mTOR, and mTOR in SGC-7901 cells cotransfected with GAS5 and miR-222 and MGC-803 cells cotransfected with si-GAS5 and anti-miR-222. As compared with the Vector, the protein level of PTEN was increased and phosphorylation of Akt and mTOR were reduced in GAS5-transfected SGC-7901 cells, while the effect of GAS5 on the protein levels PTEN, p-Akt, and p-mTOR was overturned in SGC-7901 cells cotransfected with GAS5 and miR-222 (Fig. 7a). In addition, GAS5 knockdown decreased the protein level of PTEN and increased the protein levels of p-Akt and p-mTOR compared with si-NC group in MGC-803 cells, which was partially reversed by cotreatment with GAS5 knockdown and miR-222 inhibition (Fig. 7b). Since the tumor suppressor PTEN negatively regulates the PI3 K/Akt/mTOR pathway, which is a prototypic survival pathway that plays a crucial role in cell proliferation, growth, and survival [28, 29], we concluded that GAS5/miR-222 axis regulated GC cell proliferation through the PTEN/Akt/mTOR pathway.
GAS5/miR-222 axis regulated GC cell proliferation through PTEN/Akt/mTOR pathway. The protein levels of PTEN, p-Akt, Akt, p-mTOR, and mTOR from SGC-7901 cells treated with Vector, GAS5, GAS5 + miR-222, or GAS5 + miR-NC (a) and MGC-803 transfected with si-GAS5, si-NC, si-GAS5 + anti-miR-NC, or si-GAS5 + anti-miR-222 (b) were evaluated by western blot. Data are presented as means ± SD, n = 3. *P < 0.05
Discussion
Recent studies have indicated that lncRNAs play an important role in many biological progression and the abnormally expressed lncRNAs participate in the pathogenesis and development of GC [30]. For example, lncRNA prostate cancer-associated transcript 1 (PCAT-1) was demonstrated to be upregulated in GC tissues and cells and play an oncogenic role in GC progression [31]. LncRNA the actin filament-associated protein 1 antisense RNA 1 (AFAP1-AS1) was reported to be highly expressed in GC and AFAP1-AS1 knockdown suppressed cell viability and promoted apoptosis by the PTEN/p-AKT pathway [32]. Moreover, the upregulated lncRNA colon cancer-associated transcript 2 (CCAT2) was found to indicate poor prognosis of GC and promoted GC cell proliferation and invasion [33]. Therefore, lncRNAs have been widely emerged as potential therapeutic targets for cancers [34]. Notably, several studies have reported the tumor-suppressive role of GAS5 in GC cells [18, 19]. Consistently, our study confirmed the downregulation of GAS5 in GC cells. Furthermore, we demonstrated that GAS5 overexpression inhibited GC cell proliferation, while GAS5 knockdown promoted GC cell proliferation.
Although GAS5 has been shown to be deregulated and play a tumor-suppressive role in GC cells, the detailed molecular mechanisms by which GAS5 regulated GC cell proliferation remain to be elucidated. Recently, GAS5 has been found functioning as a miRNA sponge to be involved in tumor progression. For example, Xue et al. [35] reported that GAS5 inhibited tumorigenesis and enhanced radiosensitivity in vitro and in vivo by suppressing miR-135b expression in non-small cell lung cancer. Guo et al. [36] revealed that GAS5 enhanced the expression of PTEN to promote endometrial cancer cell apoptosis by inhibiting miR-103 expression. Hu et al. [37] discovered that GAS5 suppressed cell migration and invasion of hepatocellular carcinoma cells by negative regulation of miR-21. Our study indicated that GAS5 acted as an endogenous sponge to inhibit miR-222 expression, as demonstrated by luciferase reporter assay, RIP assay, and qRT-PCR analysis. miR-222, belonging to the miR-221/miR-222 family, is located on human chromosome Xp11.3, with the same seed sequences with its homologous miRNA, miR-221 [38]. Several studies have suggested that miR-222 was highly expressed in multiple cancer types and reported the role of miR-222 in cancer progression as an oncomiRNA by promoting the proliferation, migration and invasion of cancer cells [39, 40]. Our study also proved that miR-222 expression was elevated in GC cells. Ectopic expression of miR-222 promoted cell proliferation and miR-222 inhibition impeded cell proliferation in GC cells. More importantly, forced expression of miR-222 recuperated the inhibitory effect of GAS5 restoration on GC cell proliferation, while miR-222 inhibition attenuated the promotive effect of GAS5 knockdown on GC cell proliferation, suggesting that GAS5 suppressed GC cell proliferation by sponging miR-222. Similarly, GAS5 was found to suppress tumor malignancy by targeting miR-222 in glioma cells [22]. GAS5 also increased p27 expression by functioning as a competing endogenous RNA for miR-222, thereby inhibiting liver fibrosis progression [41].
The Akt/mTOR pathway is a key signaling pathway that plays a crucial role in various physiological and pathological processes, such as proliferation, growth, survival, apoptosis and metabolism, and is constitutively activated in many malignant human tumors [42, 43]. Besides, abnormal activation of Akt/mTOR signaling pathway is generally attributed to PTEN dysregulation, which is one of the commonly lost or mutated tumor suppressor that acts as a central negative regulator of this pathway [44]. Thus, targeting the PTEN/Akt/mTOR signaling pathway has been considered to be a potential strategy against cancer treatment [45]. A previous study has reported that GAS5 could inactivate the Akt/mTOR pathway by targeting miR-103 in prostate cancer [25]. Additionally, it was reported that the Akt/mTOR pathway was activated in miR-222-overexpressing bladder cancer cells and blockade of the Akt/mTOR pathway prevented miR-222-induced cell proliferation [27]. In line with these previous studies, we found that the PTEN/Akt/mTOR pathway was regulated by GAS5 overexpression and miR-222 inhibition in GC cells. Moreover, the present study further uncovered that GAS5 overexpression could suppress cell proliferation of GC cells through the PTEN/Akt/mTOR pathway by negatively regulating miR-222.
In conclusion, our study provided the evidence that GAS5 overexpression could suppress cell proliferation of GC cells through the PTEN/Akt/mTOR pathway by negatively regulating miR-222, which enhanced our understanding of the pathogenesis and development of GC. Therefore, our study may be important to facilitate the development of lncRNA-directed therapy against GC.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29.
Kagawa S, Shigeyasu K, Ishida M, et al. Molecular diagnosis and therapy for occult peritoneal metastasis in gastric cancer patients. World J Gastroenterol. 2014;20:17796–17803.
Liu HS, Xiao HS. MicroRNAs as potential biomarkers for gastric cancer. World J Gastroenterol. 2014;20:12007–12017.
Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181.
Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166.
Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610.
Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108.
Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432–10439.
le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708.
Zhang C, Kang C, You Y, et al. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol. 2009;34:1653–1660.
Chun-Zhi Z, Lei H, An-Ling Z, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367.
Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–1910.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159.
Ma C, Shi X, Zhu Q, et al. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37:1437–1444.
Pickard MR, Williams GT. Molecular and cellular mechanisms of action of tumour suppressor GAS5 lncRNA. Genes. 2015;6:484–499.
Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793.
Pickard MR, Williams GT. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget. 2016;7:10104–10116.
Sun M, Jin FY, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319.
Guo X, Deng K, Wang H, et al. GAS5 inhibits gastric cancer cell proliferation partly by modulating CDK6. Oncol Res Treat. 2015;38:362–366.
Liu D, Yu X, Wang S, et al. The gain and loss of long noncoding RNA associated-competing endogenous RNAs in prostate cancer. Oncotarget. 2016;7:57228–57238.
Ye K, Wang S, Zhang H, et al. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem. 2017. https://doi.org/10.1002/jcb.26145.
Zhao X, Wang P, Liu J, et al. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Mol Ther. 2015;23:1899–1911.
Li N, Tang B, Zhu ED, et al. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS Lett. 2012;586:722–728.
Gregory RI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640.
Xue D, Zhou C, Lu H, et al. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumour Biol. 2016;37:16187–16197.
Li B, Lu Y, Wang H, et al. miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother. 2016;79:93–101.
Zeng LP, Hu ZM, Li K, et al. miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR axis in bladder cancer cells. J Cell Mol Med. 2016;20:559–567.
Blanco-Aparicio C, Renner O, Leal JF, et al. PTEN, more than the AKT pathway. Carcinogenesis. 2007;28:1379–1386.
Bai X, Jiang Y. Key factors in mTOR regulation. Cell Mol Life Sci. 2010;67:239–253.
Fang XY, Pan HF, Leng RX, et al. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356:357–366.
Bi M, Yu H, Huang B, et al. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in gastric cancer cells through regulating CDKN1A. Gene. 2017;626:337–343.
Guo JQ, Li SJ, Guo GX. Long noncoding RNA AFAP1-AS1 promotes cell proliferation and apoptosis of gastric cancer cells via PTEN/p-AKT pathway. Dig Dis Sci. 2017;62:2004–2010.
Wu SW, Hao YP, Qiu JH, et al. High expression of long non-coding RNA CCAT2 indicates poor prognosis of gastric cancer and promotes cell proliferation and invasion. Minerva Med. 2017;108:317–323.
Zhou S, Wang J, Zhang Z. An emerging understanding of long noncoding RNAs in kidney cancer. J Cancer Res Clin Oncol. 2014;140:1989–1995.
Xue Y, Ni T, Jiang Y, et al. LncRNA GAS5 inhibits tumorigenesis and enhances radiosensitivity by suppressing miR-135b expression in non-small cell lung cancer. Oncol Res. 2017. https://doi.org/10.3727/096504017X14850182723737.
Guo C, Song WQ, Sun P, et al. LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J Biomed Sci. 2015;22:100.
Hu L, Ye H, Huang G, et al. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691–2702.
Garofalo M, Quintavalle C, Romano G, et al. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33.
Sun Q, Jiang CW, Tan ZH, et al. MiR-222 promotes proliferation, migration and invasion of lung adenocarcinoma cells by targeting ETS1. Eur Rev Med Parmacol Sci. 2017;21:2385–2391.
Fu X, Li Y, Alvero A, et al. MicroRNA-222-3p/GNAI2/AKT axis inhibits epithelial ovarian cancer cell growth and associates with good overall survival. Oncotarget. 2016;7:80633–80654.
Yu F, Zheng J, Mao Y, et al. Long non-coding RNA growth arrest-specific transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism of competing endogenous RNA. J Biol Chem. 2015;290:28286–28298.
Bauer TM, Patel MR, Infante JR. Targeting PI3 kinase in cancer. Pharmacol Ther. 2015;146:53–60.
Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318.
Zhang XC, Piccini A, Myers MP, et al. Functional analysis of the protein phosphatase activity of PTEN. Biochem J. 2012;444:457–464.
Hennessy BT, Smith DL, Ram PT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004.
Acknowledgments
This study was supported by grants from the Science & Technology Department of Henan Province (9412016Y1403).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No potential conflicts of interest were disclosed.
Rights and permissions
About this article
Cite this article
Li, Y., Gu, J. & Lu, H. The GAS5/miR-222 Axis Regulates Proliferation of Gastric Cancer Cells Through the PTEN/Akt/mTOR Pathway. Dig Dis Sci 62, 3426–3437 (2017). https://doi.org/10.1007/s10620-017-4831-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-017-4831-4