Abstract
Background
At present, the relationship between Nanog expression and the biological behavior and prognosis of colorectal cancer is still unclear.
Aim
The purpose of this study was to evaluate the expression and regulatory effects of Nanog in colorectal cancer and the correlation between Nanog protein expression and the prognosis of patients with colorectal cancer.
Materials and Methods
The differential expression of genes between CD133+ tumor cells and CD133− tumor cells were detected using RT2 Profiler™ PCR Array. The Nanog mRNA expression level was detected by RT-PCR and the protein level was detected using immunohistochemistry staining. The relationship between Nanog expression and clinicopathological parameters of colorectal cancer was determined.
Results
Nanog were expressed significantly higher in CD133+ tumor cells compared to CD133− tumor cells. It was observed that 72 (20.00 %) of the 360 cases positively expressed Nanog. Univariate analyses indicated that Nanog expression was related to histological grade, lymph node metastasis, TNM stage, and liver metastasis (P = 0.005, 0.001, 0.001 and 0.012, respectively). Spearman correlation analysis showed that Nanog expression has a linear correlation to liver metastasis (P = 0.001). After conducting multivariate analysis, histological grade, TNM stage, and Nanog were found to be related to liver metastasis (P = 0.020, 0.01 and 0.001, respectively). In the Cox regression test, the histological grade, Lymph node metastasis, TNM stage, liver metastasis, and Nanog were detected as the independent prognostic factors (P = 0.02, 0.045, 0.01, 0.001 and 0.001, respectively).
Conclusions
Nanog protein may be a potential biomarker for postoperative liver metastasis of colorectal cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, tumor stem cells (CSC) have been considered to be the culprits for tumor canceration, recurrence, and drug resistance [1, 2]. It has been reported as well that colorectal cancer stem cells may originate when normal intestinal epithelial stem cells mutate [3]. Other studies have reported that CD133+ tumor cells, which may be rich in colorectal cancer stem cells [4, 5], have the capacity to self-renew and have a strong ability to form solid tumors. Although cancer stem cell theory has gradually come to be understood, stem cell-related genes in malignant tumors have not gained much academic attention. More specifically, studies that have addressed the function and specific mechanism of stem cell-related genes in the biological behavior of colorectal cancer are sparse [6]. Although some identified molecule factors play important roles in the progress and metastasis of colorectal cancer, the mechanisms of colorectal stem cells are far from clear [7].
Several studies have reported that Oct-4, Nanog, nestin, and CD44, which are known as stem cell genes, are expressed higher in some solid tumor stem cells [8, 9]. These factors form a core regulatory network of self-renewal and differentiation in cancer stem cells [10]. The roles of these tumor self-renewal molecules in tumorigenesis, progression, and metastasis of malignant tumors, however, are still largely unknown.
Nanog is a transcription factor critically involved in the self-renewal of undifferentiated embryonic stem cells, which has been verified to play an important role in maintaining pluripotency [11]. Furthermore, Nanog has been reported to be related to the carcinogenesis of human cancers. The functional significance of Nanog in human colorectal cancer, however, remains unknown.
Currently, studies that have investigated the function and specific mechanisms of Nanog in colorectal cancer stem cells are rare [9]. Moreover, the relationship between Nanog expression and the biological behavior and prognosis of colorectal cancer is still unclear. In the present study, we investigated the clinical implications of Nanog protein in colorectal cancer. Our goal is to lay a foundation for managing colorectal cancer.
Materials and Methods
Patients and Tissue Specimens
For the present study, we selected 360 patients who had histologically confirmed colorectal cancer and underwent radical operations in Shengjing Hospital and Liaoning Province Tumor Hospital between January 2001 and January 2006 for immunohistochemistry staining test and prognosis analysis. TNM staging was determined by the American Joint Committee on Cancer (AJCC) Cancer Staging Manual [12]. The present study’s protocol was approved by the Ethics Committee of Shengjing Hospital and Liaoning Province Tumor Hospital.
Experimental Materials
CD2-FITC, CD3-APC, CD10-PE, CD16-FITC, CD18-APC, CD31-PE, CD326-FITC (EpCAM) and CD133-FITC were obtained from BD Pharmingen (BD Biosciences, USA). The FACSCalibur flow cytometer (BD Biosciences, USA) was obtained from BD Pharmingen. The RT2 Profiler™ PCR Array and the ABI PRISM 7700 Sequence Detection system (Applied Biosystems) were obtained from SABiosciences.
Experimental Methods
CD133+ Tumor Cell Sorting by Flow Cytometry
The clinical specimens were digested into single tumor cells using collagenase III. The tumor cells were suspended in 100 μl/106 cells of HBSS with 2 % HICS. The samples were then washed twice with HBSS/2 % HICS and suspended. Antibodies, including anti-CD2, -CD3, -CD10, -CD16, -CD18, -CD31, and -CD326 were added and incubated for 20 min on ice and then washed twice with HBSS/2 % HICS. Lineage+ cells were first eliminated by anti-CD2, -CD3, -CD10, -CD16, -CD18, -CD31, and -CD326 during flow cytometry. Dead cells were eliminated using the viability dye 7AAD. Then, CD133+ tumor cells were sorted by CD133 in flow cytometry.
PCR Microarray Experiment
A total of 106 CD133+ tumor cells and CD133− tumor cells were prepared from clinical specimens. Cells were lysed in TRIzol Reagent by repetitive pipetting. Several steps were taken to process the RNA, including phase separation, precipitation, washing the RNA, and removing the contaminating DNA from the prepared RNA. The RNA was assessed for yield and quality, after which real-time PCR could be performed using the ABI PRISM 7700 system. The ΔΔCt method was employed in the gene chip data analysis, whereby ΔCt (group 1) = average Ct–average of HK genes’ Ct for the group 1 array; ΔCt (group 2) = average Ct − average of HK genes’ Ct for the group 2 array; and ΔΔCt = ΔCt (group 2) − ΔCt (group 1).
RT-PCR
The RT-PCR was performed as described previously [13]. RNA was extracted from the cells with RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Semi-quantitative RT-PCR was carried out using the RT-PCR system (Promega, Madison, WI, USA). Primers were obtained from Invitrogen with the following sequences: for Nanog, forward 5′-CAGGAGTTTGAGGGTAGCTC-3′ and reverse 5′-CGGTTCATCATGGTACAGTC-3′ and the housekeeping gene GAPDH with primers 5′-CCA CCC ATG GCA AAT TCC CAT GGC A-3′ (forward primer) and 5′-TCT AGA CGG CAG GTC AGG TCC ACC-3′ (reverse primer), at annealing temperature of 54.5 °C for 30 cycles. The products were electrophoresed by 2.5 % agarose gel.
Western Blot Analysis
Total protein concentration was determined using the Bradford assay. After electrophoresis, proteins were transferred into a nitrocellulose membrane at a constant of 45 V for 1 h. After washing in TBST buffer with shaking at room temperature, the membrane was incubated overnight with the blocking solution (1 % bovine serum albumin). The membrane was incubated with anti-Nanog antibody (1:500 dilution, Sigma-Aldrich, St. Louis, MO, USA) for 2 h at room temperature. Subsequently, the membrane was incubated with anti-rabbit secondary antibody (1:500) at room temperature for 1 h, followed by washing with TBST three times and TBS twice. Images were taken using the Bio-Rad gel imaging system (Bio-Rad, USA).
Immunohistochemistry Experimental Procedures
Briefly, the specimen tissues were cut to a thickness of 4 μm. The sections were de-waxed with xylene, gradually hydrated with gradient alcohol, and then washed with PBS. Sections were incubated for 60 min with anti-Nanog antibody. After washing with PBS, sections were incubated for 30 min in the secondary antibody. Avidin Biotin Complex (1:1,000 dilution, Vector Laboratories, Burlingame, CA, USA) was then added to the sections for 1 h at room temperature. The immunoreactive products were visualized by catalysis of 3,3-diaminobenzidine (DAB) by horseradish peroxidase in the presence of H2O2, following extensive washing. Sections were then counterstained in Gill’s Hematoxylin and dehydrated in ascending grades of methanol before clearing in xylene, and mounting under a cover slip [14].
Nanog expression was classified semi-quantitatively according to the following criteria: 0 if <1 % of neoplastic cells discretely expressed Nanog in their cytoplasm and membrane; 1+ if ≥1 and <10 % of morphologically unequivocal neoplastic cells discretely expressed Nanog in their cytoplasm and membrane; and 2+ if ≥10 % of morphologically unequivocal neoplastic cells discretely expressed Nanog in their cytoplasm and membrane. We considered samples scored as 1+ or 2+ as positive.
Statistical Analysis
All data were analyzed using SPSS Statistical Software (version 13.0; SPSS Inc., Chicago, IL, USA). Relationships between tumor markers and other parameters were studied using the chi-square test, Fisher’s exact test, or independent t tests. Disease-specific survival was analyzed using the Kaplan–Meier method. The log-rank test was used to analyze survival differences. Multivariate analysis was performed using the Cox proportional hazards model selected in forward stepwise. A P value of less than 0.05 was considered statistically significant.
Results
Differential Gene Expression Detected Between CD133+ and CD133− Tumor Cells
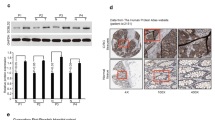
We compared the gene expression by PCR gene chip between CD133+ tumor cells and CD133− tumor cells, which were obtained from clinical specimens. Six differential genes, including transcription factors (GATA6 and Nanog), cytokines and growth factors (FGF5 and NODAL) and stem cell-differentiation/lineage markers (Nestin and OLIG2) were detected as varying significantly between CD133+ and CD133− tumor cells. RT-PCR and western blot analysis showed that Nanog mRNA and protein levels in CD133+ tumor cells were expressed significantly higher than the CD133− tumor cells (Fig. 1).
Nanog Expression in Colorectal Cancer and Its Relationship with Clinicopathological Characteristics
Immunohistochemical examination showed that Nanog was located in the cytoplasm and membrane of colorectal cancers (Fig. 2). In total, 72 (20.00 %) of the 360 cases positively expressed Nanog. Univariate analyses indicated that age, gender, and tumor size were not related to Nanog expression in colorectal cancer (P = 0.107, 0.407, and 0.079, respectively). Finally, Nanog expression was found to be related to histological grade, lymph node metastasis, TNM stage, and liver metastasis (P = 0.005, 0.001, 0.001, and 0.012, respectively) (Table 1). Furthermore, Spearman correlation analysis showed that Nanog expression has a line correlation to lymph node metastasis, TNM stage, and postoperative liver metastasis (P = 0.038, 0.01, and 0.001 respectively) (Table 2). After conducting multivariate analysis, histological grade, TNM stage, and Nanog were found to be related to liver metastasis (P = 0.020, 0.01 and 0.001, respectively) (Table 3).
Prognostic Analysis
Survival analysis revealed that the cases with positive Nanog expression exhibited a significantly higher postoperative liver metastasis rate compared to those without Nanog expression (38.89 vs. 6.94 %; P = 0.012). Survival analysis revealed that patients with Nanog expression experienced worse postoperative survival compared to those without Nanog expression (38.89 vs. 63.19 %; P = 0.001) (Fig. 3). Moreover, histological grade, lymph node metastasis, and TNM stage were associated with colorectal cancer-specific survival in all 360 cases (P = 0.032, 0.01, and 0.001, respectively; log-rank test; Fig. 3). In the Cox regression test, histological grade, lymph node metastasis, TNM stage, liver metastasis, and Nanog were detected as the independent prognostic factors (P = 0.02, 0.045, 0.01, 0.001 and 0.001, respectively) (Table 4).
Discussion
The concept of cancer stem cells (CSC) has been accepted gradually by most cancer researchers in recent years [15]. The CSC model is also a current focus of colorectal cancer research. According to the CSC concept, only a small minority of tumor cells have the ability to maintain the malignant population. In colorectal cancer, research has recently reported that CD133+ tumor cells were highly enriched in tumor-initiating colon CSCs that have the ability to self-renew and to recapitulate the bulk tumor population [16]. Studying the gene expression profiles of CSC, therefore, and selecting the potential targets for treating colorectal cancer are important.
In the present study, we selected CD133+ tumor cells from clinical specimens using flow cytometry and compared the gene expression using PCR gene chip between CD133+ tumor cells and CD133− tumor cells. Six differential genes were detected. RT-PCR and western blot analysis verified that Nanog was expressed significantly higher in mRNA and protein levels in CD133+ tumor cells compared to CD133− tumor cells. Nanog, a homeodomain transcription factor, is an essential regulator for promoting self-renewal of embryonic stem cells and inhibiting their differentiation [8]. In general, however, the clinical complications and the mechanism of Nanog in transforming and developing tumors are still unclear. Moreover, the role of Nanog in tumor development from different groups is still contradictory.
Nanog promotes dedifferentiation of p53-deficient mouse astrocytes into brain cancer stem-like cells [17]. In a recent study, Meng et al. [18] investigated the expression status and regulatory mechanism of the self-renewal molecule Nanog in colorectal cancer. They reported a significant correlation among over-expression of Nanog and poor prognosis, lymph node metastasis, and TNM classification of colorectal cancer [18].
In the present study, we studied the relationship between Nanog protein expression and pathological factors and prognosis in colorectal cancer. It was observed that 20.00 % of the 360 cases positively expressed Nanog, with Nanog expression being related to histological grade, lymph node metastasis, TNM stage, and liver metastasis. On the other hand, it was found that Nanog was linearly correlated to liver metastasis and was a prognostic factor of colorectal cancer. Nanog protein has been reported to be related to the poor prognosis and chemoresistance in other various cancers [19]. Hence, Nanog might provide a new potential target for chemotherapy in some tumors, although there is still no evidence of this for colorectal cancer. However, there also were some limitations in the study. All the enrolled cases underwent radical operations. More advanced cancers in which palliative surgery was performed were not studied in the study.
Many colorectal cancers are in an advanced stage when they are first diagnosed, and some are accompanied with distant organ metastases. The most effective therapy for advanced colorectal cancers with liver metastases [3, 7] is still controversial. Early screening of the distant metastasis in colorectal cancer, therefore, is very important. The outcome of the present study demonstrated that Nanog protein was related to liver metastasis of colorectal cancer, which may be used as a potential early liver metastasis screening factor in the colorectal cancer.
References
Li Y, Laterra J. Cancer stem cells: distinct entities or dynamically regulated phenotypes? Cancer Res. 2012;72:576–580.
Chu JE, Allan AL. The role of cancer stem cells in the organ tropism of breast cancer metastasis: a mechanistic balance between the “seed” and the “soil”? Int J Breast Cancer. 2012;2012:209748.
Galizia G, Gemei M, Del Vecchio L, et al. Combined CD133/CD44 expression as a prognostic indicator of disease-free survival in patients with colorectal cancer. Arch Surg. 2012;147:18–24.
Shimada R, Iinuma H, Akahane. Prognostic significance of CTCs and CSCs of tumor drainage vein blood in Dukes’ stage B and C colorectal cancer patients. Oncol Rep. 2012;27:947–953.
Gisina AM, Lupatov AY, Karalkin PA, et al. Detection of minor subpopulations of colorectal adenocarcinoma cells expressing cancer stem cell markers. Bull Exp Biol Med. 2011;151:234–238.
Lin L, Fuchs J, Li C, Olson V, Bekaii-Saab T, Lin J. STAT3 signaling pathway is necessary for cell survival and tumor sphere forming capacity in ALDH+/CD133+ stem cell-like human colon cancer cells. Biochem Biophys Res Commun. 2011;416:246–251.
Ding Q, Chang CJ, Xie X, et al. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. J Clin Invest. 2011;121:4526–4536.
Ezeh UI, Turek PJ, Reijo RA, Clark AT. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104:2255–2265.
Liu C, Cao X, Zhang Y, Xu H, Zhang R, Wu Y, Lu P, Jin F. Co-expression of Oct-4 and Nestin in human breast cancers. Mol Biol Rep. 2012;39:5875–5881
Liu CG, Lu Y, Wang BB, et al. Clinical implications of stem cell gene Oct-4 expression in breast cancer. Ann Surg. 2011;253:1165–1171.
Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49.
Ratto C, Ricci R. Potential pitfalls concerning colorectal cancer classification in the seventh edition of the AJCC cancer staging manual. Dis Colon Rectum. 2011;54:e232.
Khuda II, Koide N, Noman AS, et al. Astrocyte elevated gene-1 (AEG-1) is induced by lipopolysaccharide as toll-like receptor 4 (TLR4) ligand and regulates TLR4 signalling. Immunology. 2009;128:e700–e706.
He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, Li PY, Song YH, Lin JS. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2011. (Epub ahead of print). doi:10.1038/onc.2011.500.
Liu C, Chen B, Zhu J, et al. Clinical implications for nestin protein expression in breast cancer. Cancer Sci. 2010;101:815–819.
Vermeulen L, de Sousa E, Melo F, Richel DJ, Medema JP. The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol. 2012;13:e83–e89.
Moon JH, Kwon S, Jun EK, et al. Nanog-induced dedifferentiation of p53-deficient mouse astrocytes into brain cancer stem-like cells. Biochem Biophys Res Commun. 2011;412:175–181.
Meng HM, Zheng P, Wang XY, Liu C, Sui HM, Wu SJ, Zhou J, Ding YQ, Li JM. Overexpression of nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther. 2010;9:4.
Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–160.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, F., Dai, C., Zhang, R. et al. Nanog: A Potential Biomarker for Liver Metastasis of Colorectal Cancer. Dig Dis Sci 57, 2340–2346 (2012). https://doi.org/10.1007/s10620-012-2182-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-012-2182-8