Abstract
Purpose
DCBLD2 expression dysregulation has been reported in several types of human cancer. As yet, however, the role of DCBLD2 in colorectal cancer (CRC) is not known.
Methods
CRC tissues were obtained from patients undergoing surgery from February 2009 to May 2014 (n = 90). Tissue microarray construction and immunohistochemistry were carried out to determine DCBLD2 expression. In vivo studies were performed in 4-week-old BALB/c nude mice. In vitro studies were conducted using CRC-derived HT29 and HCT116 cell lines.
Results
DCBLD2 expression was found to be significantly increased in CRC tissues compared to adjacent normal tissues (p < 0.001). In addition, we found that DCBLD2 expression was positively correlated with the stage of the disease, the degree of differentiation and vascular invasion. High DCBLD2 expression was significantly associated with a poor overall survival. In vitro, DCBLD2 expression downregulation significantly reduced CRC cell proliferation and invasion. In a mouse xenograft model, DCBLD2 expression downregulation reduced lung metastasis and increased overall survival. Gene set enrichment analysis (GSEA) revealed that DCBLD2 overexpression induces epithelial–mesenchymal transition (EMT) and activates the JAK/STAT3 pathway.
Conclusions
We found that high DCBLD2 expression correlated with a poor clinical outcome, as well as tumorigenesis, invasion and metastasis of CRC cells. DCBLD2 may serve as a prognostic biomarker and a novel therapeutic target for CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Colorectal cancer (CRC) is an important cause of cancer-related morbidity and mortality in the world [1, 2]. Despite recent advances in diagnostic and therapeutic modalities for CRC, patient survival has not improved significantly [3, 4]. Also, the signaling pathways involved in CRC development are as yet poorly understood. A better understanding of CRC tumor biology using primary biopsy samples may facilitate the development of disease-specific procedures, and the identification of biomarkers that are sensitive and specific for CRC may help in the development of new diagnostic and therapeutic modalities [5].
The discoidin, CUB and LCCL domain-containing protein 2 (DCBLD2) cDNA has been cloned from human, mouse and rat coronary arterial endothelial and smooth muscle cells (CAECs and CASMCs, respectively) [6]. DCBLD2 is a type-I transmembrane protein [7], and its amino acid sequence is highly conserved across vertebrates, including the signal sequence and CUB, LCCL and coagulation factor V/VIII type-C (also discoidin) domains [8, 9]. The intracellular region contains several phosphorylation, acetylation and ubiquitylation sites without currently described molecular domains [10, 11]. The functions of the domains contained within DCBLD2 are as yet poorly understood. Based on knockout and knockdown results in mice, DCBLD2 has been found to exhibit positive effects on vascular repair and angiogenesis and negative effects on glucose uptake and thrombus formation [12,13,14].
DCBLD2 has been found to be overexpressed in lung cancer cells to participate in metastasis [7]. Furthermore, DCBLD2 has been found to be amplified in glioblastomas and to be required for epidermal growth factor receptor (EGFR)-stimulated carcinogenesis [15]. Conversely, DCBLD2 expression has been found to be downregulated in gastric and neuroendocrine cancers [16]. In gastric cancer DCBLD2 expression may be regulated by epigenetic modification, and the gene may act as a tumor suppressor in this cancer. Thus, DCBLD2 may play either positive or negative roles in different human cancer types.
In this study, we set out to assess DCBLD2 expression in primary CRC tissues compared to normal colonic tissues. We also evaluated the biological functions and putative clinical applications of DCBLD2.
2 Material and methods
2.1 Patient samples
For this study, tumor and normal colorectal tissues were collected from patients with CRC who underwent surgery at Guangzhou First Hospital (Guangzhou, China) from February 2009 to May 2014. The Ethics Committee of Guangzhou First Hospital approved the study protocol. The Institutional Animal Care and Use Committee approved the animal experimental procedures. All the patients included in this study provided written informed consent. All the research was carried out in accordance with the provisions of the Helsinki Declaration of 1975. None of the patients received preoperative radiotherapy or chemotherapy.
2.2 Cell culture
Human CRC-derived cell lines HT29, HCT116 and CaCo-2 were cultured as per instructions of the American Type Culture Collection (Manassas, VA, USA). RPMI-1640 medium (Invitrogen), supplemented with 10% fetal bovine serum (FBS), was used to culture the CRC cells in a humidified atmosphere containing 5% CO2 at 37 °C. For the cell proliferation assay (see below), culture medium with a low serum concentration (1.5% FBS-RPMI) was used.
2.3 Quantitative real-time PCR (qRT-PCR)
An Applied Biosystems 7900 quantitative PCR system was used to carry out quantitative real-time PCR. All the reactions were performed in triplicate using Brilliant® SYBR® Green QPCR Master Mix (Takara, Shiga, Japan) in a total volume of 20 μl. The amplified transcript level of each gene was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). All primer pairs used in this study are listed in Supplementary Table 1.
2.4 BrDU proliferation assay
A BrDU incorporation assay (Roche Molecular Biochemicals, Mannheim, Germany) was used to measure cell proliferation. In brief, cells were transfected with DCBLD2 or control siRNAs after culturing for 24 h. After the given time interval, BrDU labeling solution was added to the cells (10 μl/well), after which the cells were incubated for an additional 3 h. Next, the cells were fixed and the DNA was denatured using FixDenat (200 μl/well). Subsequently, anti-BrdU-POD solution (100 μl/well) was added, after which the cells were incubated for 90 min. Finally, the cells were washed with phosphate-buffered saline (PBS) three times, after which the immune complexes were identified by the substrate reaction and quantified by checking the absorbance at 370 nm (reference wavelength: approx. 492 nm).
2.5 Western blotting
Western blot analysis was conducted using standard procedures [17]. GAPDH (Kangchen, Shanghai, China) was used as a loading control. Antibodies directed against DCBLD2, p-JAK2, JAK2, p-STAT3, STAT3, E-cadherin, N-cadherin, vimentin, SNAI2 and GAPDH were purchased from Cell Signaling Technology Inc. (Beverly, MA, USA). Details regarding the antibodies and the doses used are shown in Supplementary Table 2.
2.6 Tissue microarray construction and immunohistochemical staining
Using pathologic examination of histology slides at Guangzhou First Hospital it was verified that the percentage of tumor cells in the CRC tissue sections was at least 70%. Next, tissue microarrays were constructed using standard techniques (Outdo Biotech, Shanghai, China) [18]. Expression analyses were carried out using a LSABt kit (DakoCytomation, Copenhagen, Denmark). Specific antibodies from Cell Signaling Technology (Beverly, MA, USA) were employed to detect DCBLD2, p-JAK2, JAK2, p-STAT3, STAT3, E-cadherin, N-cadherin, vimentin, SNAI2 and GAPDH expression. The resulting slides were inspected independently by two pathologists blinded to the clinical and the pathologic data. Quantification of protein expression was estimated using the method described by Xiong et al. [17]. Protein expression was evaluated according to the extent and intensity of staining (the percentage of positive cells was measured on a scale of 0–4: 0, 0–5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; 4, 76–100%; and the intensity of staining was measured on a scale of 0–3: 0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining). A final score was generated to determine the cut-off value for low- and high-expression groups by multiplying the extent-of-staining score by the intensity-of-staining score. Next, the protein expression levels were sorted into four categories: “-” for a score of 0–3, “+” for a score of 3–6, “++” for a score of 6–9, and “+++” for a score > 9; low expression was defined as a final score < 6, and high expression was defined as a final score ≥ 6.
2.7 shRNA construction, transfection and lentiviral transduction
A shRNA directed against DCBLD2 (GenBank accession number: NM_080927) and control shRNA/siRNA were purchased from Dharmacon RNA Technology (Lafayette, CO, USA). Transfection of the shRNAs/siRNAs was carried out as indicated in the manufacturer’s instructions. For lentivirus transfer, CRC cells were transfected using 2 μg of lenti-DCBLD2 shRNA-virus or lenti-control shRNA-virus. Lentivirus production and infection were carried out using standard procedures [19].
2.8 Transwell invasion assay
Transwell chambers containing 8-μm-pore polycarbonate membranes were coated with Matrigel on the upper side (BD Biosciences). Next, CRC cells were harvested and 1 × 105 cells were placed in serum-free medium in the upper chamber. In the lower chamber medium supplemented with 20% FBS (chemoattractant) was placed to induce invasion. After incubation for 48 h, the cells that had migrated to the bottom of the filter were stained with 0.1% crystal violet solution and counted.
2.9 In vivo experiments
To understand the effect of DCBLD2 in vivo, 4-week-old male BALB/c nude mice obtained from the Experimental Animal Centre of Shanghai Institutes for Biological Sciences (SIBS) were used. HCT116 cells (1.0 × 107) were injected into the right flank of these mice to develop subcutaneous CRC xenograft models. Starting at 10 days after subcutaneous inoculation, the mice were randomly divided into three groups (8 mice/group) and were injected with PBS, control shRNA lentiviruses, or DCBLD2 shRNA lentiviruses by multipoint intra-tumoral injection on alternate days for 14 days.
To additionally analyze the in vivo effect of DCBLD2 on tumor cell invasion, a CRC metastasis model was developed. To this end, HCT116 cells (2.5 × 106) were subcutaneously injected into the right flank of 4-week-old BALB/c nude mice. The mice were examined daily. The tumors were measured with calipers every 4 days. When the tumors reached a volume of ~25 mm3 (approximately 10 days after tumor cell inoculation) the animals were randomly assigned to one of three groups: intra-tumoral injection of PBS, lentiviruses encoding a control shRNA, or DCBLD2 shRNA lentiviruses (1 × 108 per tumor/mouse, twice a week for 13 weeks). The mice were sacrificed at week 13 to check for lung metastasis. The numbers of lung metastatic foci were determined in lung tissue sections stained with hematoxylin and eosin under a binocular microscope (Leica, DM 300).
2.10 Bioinformatics analysis
Human microarray expression data for CRC and normal adjacent tissues were obtained from the Gene Expression Omnibus (GEO). Dataset GSE9348 consisted of 65 pairs of CRC and normal adjacent tissues, while dataset GSE20842 consisted of 70 CRC and 12 normal adjacent tissues. After obtaining approval for this project from the consortium, RNA-Seq (STAD level 3) CRC data of The Cancer Genome Atlas (TCGA) and its corresponding clinical data were obtained from the TCGA website (https://tcga-data.nci.nih.gov/tcga/). Two additional cohorts (GSE14333 and GSE17536) were download from GEO for survival analyses.
To assess the pathogenesis of CRC through the DCBLD2 pathway, gene set enrichment analysis (GSEA) was performed on the TCGA dataset. The gene sets showing false discovery rates (FDR) of 0.25 were considered enriched. Hallmark (h.all.v5.1) and curated gene sets (c2.all.v5.1) were obtained from the Molecular Signatures Database. Gene sets containing a minimum of 15 genes were used.
2.11 Statistical analysis
SPSS 17.0 software (SPSS, Chicago, IL, USA) was used to perform statistical analyses. Continuous variables were measured as mean ± standard error and compared by Student’s t test. For categorical data, the chi-squared test or Fisher’s exact test (two sided) was used. Spearman’s correlation test was employed to correlate protein expression levels. Survival curves were constructed using the Kaplan–Meier method. The survival rates were compared using the log-rank test. A two-sided p value < 0.05 was considered statistically significant. Tumor volumes (mm3) were estimated using the formula: tumor volume (mm3) = (shorter diameter)2 × longer diameter / 2. Patient information for cohorts 1 and 2 is shown in Supplementary Figs. 3 and 4, respectively.
3 Results
3.1 DCBLD2 expression is upregulated in CRC
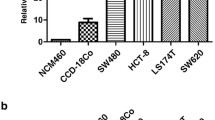
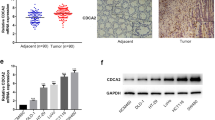
We found that the expression of DCBLD2 was increased in tumor tissues compared to normal colorectal epithelium in two independent microarray datasets obtained from the GEO database (p < 0.001, Fig. 1A and Supplementary Fig. 1). Subsequently, we analyzed DCBLD2 expression in another independent patient cohort from Guangzhou First People’s Hospital (cohort 1, n = 90). Using qRT-PCR, we found that the CRC tissues exhibited higher DCBLD2 mRNA expression levels than the normal adjacent tissues (p < 0.001, Fig. 1B). Also, the DCBLD2 protein expression in four fresh pairs of CRC and normal adjacent tissues as assessed by Western blotting was found to be significantly increased in the CRC tissues (Fig. 1C).
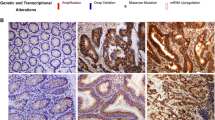
DCBLD2 expression in CRC tissues. a – b Analysis of DCBLD2 expression using the Wilcoxon test in dataset GSE9348, dataset GSE20842 and Guangzhou First People’s Hospital cohort 1 patients. c Western blot analysis of DCBLD2 protein expression in four paired tissue samples of patients from Guangzhou First People’s Hospital. d – e DCBLD2 protein levels as measured by immunohistochemistry in normal colorectal epithelial cells and CRC tissues from The Human Protein Atlas website and Guangzhou First People’s Hospital cohort 1
The Human Protein Atlas (https://www.proteinatlas.org/about/licence.) data indicate that DCBLD2 protein expression was present in most CRC tissues compared to normal colorectal epithelial tissues (patient ID: 2151, Fig. 1D). Additionally, immunohistochemical analysis of 90 pairs of clinical CRC samples (cohort 1), as well as representative images and summarized data of tissue microarray analysis of cohort 1 confirmed a higher DCBLD2 protein expression in tumor tissues compared to normal peritumoral tissues (p < 0.01, Fig. 1E).
3.2 DCBLD2 expression correlates with clinicopathological characteristics and prognosis
DCBLD2 expression was found to be significantly correlated with the degree of differentiation (p = 0.001) and the American Joint Committee on Cancer (AJCC) stage (p = 0.002) in the cohort 1 patients (Table 1). By Cox univariate analysis of the Guangzhou First People’s Hospital cohort 2, we found that a tumor size > 2 cm, the presence of lymph node metastasis, the presence of vascular invasion, and a high DCBLD2 expression correlated with a worse overall survival (Fig. 2A). In addition, we found by multivariate Cox regression analysis that DCBLD2 expression, lymphatic metastasis, tumor size, and vascular invasion served as independent predictors of overall survival after surgery (Fig. 2B).
Kaplan–Meier analysis revealed that DCBLD2 expression was significantly correlated with disease-free survival in GSE14333 (p = 0.026) and GSE17536 (p = 0.0003) as well as overall survival in cohort 2 patients (p < 0.0001) (Fig. 3A–C and Supplementary Fig. 2). The overall survival was better in CRC patients with a lower DCBLD2 expression. By receiver operating characteristic (ROC) curve analysis, we found that the sensitivity and specificity of DCBLD2 expression in predicting overall survival were 68% and 73%, respectively. The area under the curve (AUC) was 0.689 in Guangzhou cohort 2 (Fig. 3D).
Kaplan–Meier analysis of overall survival in CRC patients. a – c Associations between DCBLD2 expression and patient survival analyzed in the GSE14333 dataset, the GSE17536 dataset and Guangzhou First People’s Hospital cohort 2. d ROC curve analysis of DCBLD2 expression in Guangzhou First People’s Hospital cohort 2
3.3 DCBLD2 acts as an oncogene in CRC
To next investigate the role of DCBLD2 in CRC development, GSEA was performed on the two TCGA CRC datasets. We found that the gene sets related to positive regulation of cell proliferation and EMT were better correlated with patients exhibiting a high DCBLD2 expression than those with a low DCBLD2 expression in both datasets (Fig. 4A–B).
Association between DCBLD2 expression and in vitro CRC cell proliferation and invasion.a – b GSEA comparing the low-DCBLD2 expression group (blue) against the high-DCBLD2 expression group (red) in the TCGA CRC patient dataset (1% FDR, p < 0.005). c – d BrdU proliferation assays performed in HCT116 and HT29 CRC cells transfected with DCBLD2 siRNA1/2 (n = 3, Mann–Whitney test). e Transwell Matrigel invasion assay performed in HCT116 and HT29 cells transfected with control siRNA and DCBLD2 siRNA1/2, respectively (n = 3)
To verify the GSEA results, cell proliferation and transwell invasion assays were performed. The cell proliferation assay indicated that DCBLD2 expression downregulation significantly impaired the proliferation of HCT116 and HT29 cells (Fig. 4C – D). The transwell invasion assay indicated that DCBLD2 expression downregulation notably decreased the invasive capacities of HCT116 and HT29 cells (Fig. 4E). Conversely, we found that overexpression of DCBLD2 promoted the proliferation (Supplementary Fig. 4A) and invasion (Supplementary Fig. 4B) of Caco-2 cells exhibiting a low-DCBLD2 expression (Supplementary Fig. 3A).
Subsequent in vivo studies revealed that DCBLD2 expression downregulation considerably reduced HCT116 tumor growth (Fig. 5A–B) and tumor weight (Fig. 5C). In the CRC metastatic model, mice inoculated with DCBLD2 shRNA-expressing tumor cells showed fewer pulmonary metastases and longer overall survival times compared to the control group (Fig. 5D–F). Together, these data strongly suggest that DCBLD2 may promote CRC cell proliferation and metastasis both in vitro and in vivo.
Downregulation of DCBLD2 inhibits tumor cell growth and metastasis in a xenograft mouse model. a Tumor volumes after DCBLD2 shRNA1/2 adenovirus treatment (n = 8, *p < 0.05, Mann–Whitney test). b Tumors transfected with control shRNA adenovirus and DCBLD2 shRNA1/2 adenovirus (n = 8). c Tumor weights in mice after different treatments (n = 8, Mann–Whitney test). d Survival analysis of mice bearing CRC cell tumors transfected with DCBLD2 shRNA1/2 adenovirus or control shRNA adenovirus (n = 10). e – f Representative histologic images of tumor foci in lungs of nude mice and mice with/without metastases after injection with DCBLD2 shRNA1/2 adenovirus at 13 weeks (hematoxylin-eosin staining) (n = 10)
3.4 DCBLD2 overexpression activates the JAK-STAT pathway and EMT
The GSEA enrichment plots revealed that the Hallmark_IL6_JAK_STAT3_signaling gene signatures were more active in patients exhibiting high DCBLD2 expression levels (Fig. 6A). According to the GSEA previews, qRT-PCR revealed that DCBLD2 expression downregulation significantly increased E-cadherin expression and reduced N-cadherin, vimentin and Snai2 expression (Fig. 6B). It also showed opposite results in DCBLD2 overexpressing Caco-2 cells (Supplementary Fig. 5A). Moreover, Western blot analysis revealed that DCBLD2 expression downregulation upregulated E-cadherin expression and decreased the phosphorylation levels of JAK2, STAT3, and the EMT markers N-cadherin, vimentin and Snai2 in HT29 (Fig. 6C–D) and HCT116 (Fig. E–F) cells. Conversely, DCBLD2 overexpression decreased E-cadherin expression and increased the phosphorylation levels of JAK2 and STAT3, as well as the EMT markers N-cadherin, vimentin and Snai2 in Caco-2 cells (Supplementary Fig. 5B). These analyses suggest that DCBLD2 may play an oncogenic role in CRC and may control cancer progression through EMT and the JAK-STAT3 pathway.
Downregulation of DCBLD2 inhibits JAK/STAT3 pathway activation and EMT-related gene expression in CRC cells. a GSEA of dataset GSE28735 showing enrichment of the JAK/STAT3 pathway in samples with a high DCBLD2 expression. b mRNA levels of epithelial and mesenchymal markers as measured by qRT-PCR. c – d Protein levels of p-JAK2, JAK2, p-STAT3, STAT3 and EMT markers as measured by Western blotting in HT29 cells. GAPDH was used as an internal control. e – f Protein levels of p-JAK2, JAK2, p-STAT3, STAT3 and EMT markers as measured by Western blotting in HCT116 cells. GAPDH was used as an internal control. DCBLD2 siRNA Mix indicates DCBLD2 siRNA1:DCBLD2 siRNA2 = 1:1
4 Discussion
Several recent studies have suggested a role of DCBLD2 in various types of cancer [15, 20,21,22]. In this study, we provide evidence for a role of DCBLD2 in CRC. Specifically, we found that DCBLD2 was significantly overexpressed in CRC tissues compared to nontumor tissues and that this overexpression correlated with a higher AJCC grade, a poorer degree of histological differentiation, a higher incidence of vascular invasion, and a poorer overall survival of patients. These findings indicate that DCBLD2 may serve as a novel prognostic biomarker for CRC.
DCBLD2 is a known phospho-tyrosine target of EGFR and has been associated with EGFR-driven tumorigenesis and progression of gliomas and cervical and head and neck cancers [15, 23]. But the role of DCBLD2 in the development of CRC has remained unknown. Here, we found by GSEA of the TCGA dataset that positive regulation of cell proliferation and EMT pathways were positively correlated with a high DCBLD2 expression in CRC patients, which was further validated by a series of functional experiments. Expression downregulation of DCBLD2 in CRC cells significantly suppressed their growth and invasion in in vitro assays. The proliferation and metastasis-promoting effects of DCBLD2 were also observed in xenograft tumors in mice. These findings indicate that DCBLD2 may act as an oncogene in CRC. We also analyzed the molecular mechanism through which DCBLD2 overexpression promoted CRC cell invasion and metastasis. EMT is known to play an important role in the development of metastasis [1, 24,25,26,27]. Here, we found that DCBLD2 expression downregulation significantly upregulated epithelial markers and downregulated mesenchymal markers in CRC cells. Based on these results, we conclude that DCBLD2 overexpression may at least partly increase the invasiveness of CRC cells by promoting the EMT process. Further GSEA of the TCGA dataset revealed that the Hallmark_IL6_JAK_STAT3_signaling pathway was positively correlated with DCBLD2 expression. STAT3 activity has been reported to alter the architecture of the microtubule network, which in turn affects cell locomotion and focal adhesion turnover [28]. In addition to its function as a transcription factor, STAT3 can also interact with and destabilize microtubules, thereby promoting EMT [29]. STAT3 can also directly regulate EMT by modifying the transcription levels of EMT inducers and modulators [17, 30, 31]. We found that DCBLD2 expression downregulation significantly decreased the levels of p-JAK2, JAK, p-STAT3 and STAT3 in CRC cells. To the best of our knowledge, there are no previous reports on associations between DCBLD2 expression and the JAK/STAT signaling pathway and EMT. Further studies are, however, required to determine the direct regulatory role of DCBLD2 on STAT3 activation and EMT.
There are some limitations of this study that must be mentioned. First, this study is a single center study with a relatively small sample size. Second, as multiple hypotheses were tested in this study, there is an increased likelihood for type 1 errors. Third, all of the clinicopathological prognostic factors such as tumor budding and immune cell infiltration were not included in the univariate and multivariate survival analyses. Future multicenter studies with a larger sample size are required to validate the findings of this study.
In conclusion, we found that DCBLD2 is significantly overexpressed in CRC tissues. High DCBLD2 expression may serve as an independent adverse prognostic factor for CRC. Also, in vitro studies revealed that downregulation of DCBLD2 could inhibit CRC cell invasion via suppressing STAT3 activation and EMT. We conclude that DCBLD2 may serve as a biomarker and a novel therapeutic target for CRC. Additional studies are required to validate the findings of this study.
References
R.C. Bates, A.M. Mercurio, The epithelial-mesenchymal transition (EMT) and colorectal cancer progression. Cancer Biol. Ther. 4, 365–370 (2005). https://doi.org/10.4161/cbt.4.4.1655
S.D. Markowitz, D.M. Dawson, J. Willis, J.K. Willson, Focus on colon cancer. Cancer Cell 1, 233–236 (2002). https://doi.org/10.1016/S1535-6108(02)00053-3
L. Wei, X. Wang, L. Lv, Y. Zheng, N. Zhang, M. Yang, The emerging role of noncoding RNAs in colorectal cancer chemoresistance. Cell. Oncol. 42, 757–768 (2019). https://doi.org/10.1007/s13402-019-00466-8
D. Liu, Concomitant dysregulation of the estrogen receptor and BRAF/MEK signaling pathways is common in colorectal cancer and predicts a worse prognosis. Cell. Oncol. 42, 197–209 (2019). https://doi.org/10.1007/s13402-018-00420-0
M.S. Pepe, R. Etzioni, Z. Feng, J.D. Potter, M.L. Thompson, M. Thornquist, M. Winget, Y. Yasui, Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst. 93, 1054–1061 (2001). https://doi.org/10.1093/jnci/93.14.1054
K. Kobuke, Y. Furukawa, M. Sugai, K. Tanigaki, N. Ohashi, A. Matsumori, S. Sasayama, T. Honjo, K. Tashiro, ESDN, a novel neuropilin-like membrane protein cloned from vascular cells with the longest secretory signal sequence among eukaryotes, is up-regulated after vascular injury. J. Biol. Chem. 276, 34105–34114 (2001). https://doi.org/10.1074/jbc.M105293200
K. Koshikawa, H. Osada, K. Kozaki, H. Konishi, A. Masuda, Y. Tatematsu, T. Mitsudomi, A. Nakao, T. Takahashi, Significant up-regulation of a novel gene, CLCP1, in a highly metastatic lung cancer subline as well as in lung cancers in vivo. Oncogene 21, 2822–2828 (2002). https://doi.org/10.1038/sj.onc.1205405
M. Goujon, H. McWilliam, W. Li, F. Valentin, S. Squizzato, J. Paern, R. Lopez, A new bioinformatics analysis tools framework at EMBL-EBI. Nucl Acids Res. 38, W695–W699 (2010). https://doi.org/10.1093/nar/gkq313
H. Nielsen, Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73 (2017). https://doi.org/10.1007/978-1-4939-7015-5_6
T.M. Aten, M.M. Redmond, S.O. Weaver, C.C. Love, R.M. Joy, A.S. Lapp, O.D. Rivera, K.L. Hinkle, B.A. Ballif, Tyrosine phosphorylation of the orphan receptor ESDN/DCBLD2 serves as a scaffold for the signaling adaptor CrkL. FEBS Lett. 587, 2313–2318 (2013). https://doi.org/10.1016/j.febslet.2013.05.064
A.M. Schmoker, J.L. Weinert, K.J. Kellett, H.E. Johnson, R.M. Joy, M.E. Weir, A.M. Ebert, B.A. Ballif, Dynamic multi-site phosphorylation by Fyn and Abl drives the interaction between CRKL and the novel scaffolding receptors DCBLD1 and DCBLD2. Biochem J. 474, 3963–3984 (2017). https://doi.org/10.1042/bcj20170615
L. Nie, X. Guo, L. Esmailzadeh, J. Zhang, A. Asadi, M. Collinge, X. Li, J.D. Kim, M. Woolls, S.W. Jin, A. Dubrac, A. Eichmann, M. Simons, J.R. Bender, M.M. Sadeghi, Transmembrane protein ESDN promotes endothelial VEGF signaling and regulates angiogenesis. J. Clin. Invest. 123, 5082–5097 (2013). https://doi.org/10.1172/jci67752
X. Li, J.J. Jung, L. Nie, M. Razavian, J. Zhang, V. Samuel, M.M. Sadeghi, The neuropilin-like protein ESDN regulates insulin signaling and sensitivity. Am. J. Phys. Heart Circ. Phys. 310, H1184–H1193 (2016). https://doi.org/10.1152/ajpheart.00782.2015
M.N. O'Connor, A. Salles II, A. Cvejic, N.A. Watkins, A. Walker, S.F. Garner, C.I. Jones, I.C. Macaulay, M. Steward, J.J. Zwaginga, S.L. Bray, F. Dudbridge, B. de Bono, A.H. Goodall, H. Deckmyn, D.L. Stemple, W.H. Ouwehand, Functional genomics in zebrafish permits rapid characterization of novel platelet membrane proteins. Blood 113, 4754–4762 (2009). https://doi.org/10.1182/blood-2008-06-162693
H. Feng, G.Y. Lopez, C.K. Kim, A. Alvarez, C.G. Duncan, R. Nishikawa, M. Nagane, A.J. Su, P.E. Auron, M.L. Hedberg, L. Wang, J.J. Raizer, J.A. Kessler, A.T. Parsa, W.Q. Gao, S.H. Kim, M. Minata, I. Nakano, J.R. Grandis, R.E. McLendon, D.D. Bigner, H.K. Lin, F.B. Furnari, W.K. Cavenee, B. Hu, H. Yan, S.Y. Cheng, EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J. Clin. Invest. 124, 3741–3756 (2014). https://doi.org/10.1172/jci73093
E. Hofsli, T.E. Wheeler, M. Langaas, A. Laegreid, L. Thommesen, Identification of novel neuroendocrine-specific tumour genes. Br. J. Cancer 99, 1330–1339 (2008). https://doi.org/10.1038/sj.bjc.6604565
H. Xiong, J. Hong, W. Du, Y.W. Lin, L.L. Ren, Y.C. Wang, W.Y. Su, J.L. Wang, Y. Cui, Z.H. Wang, J.Y. Fang, Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J. Biol. Chem. 287, 5819–5832 (2012). https://doi.org/10.1074/jbc.M111.295964
T. Kato, S. Hayama, T. Yamabuki, N. Ishikawa, M. Miyamoto, T. Ito, E. Tsuchiya, S. Kondo, Y. Nakamura, Y. Daigo, Increased expression of insulin-like growth factor-II messenger RNA-binding protein 1 is associated with tumor progression in patients with lung cancer. Clin Cancer Res 13, 434–442 (2007). https://doi.org/10.1158/1078-0432.CCR-06-1297
Y.R. Weng, Y.N. Yu, L.L. Ren, Y. Cui, Y.Y. Lu, H.Y. Chen, X. Ma, W.X. Qin, W. Cao, J. Hong, J.Y. Fang, Role of C9orf140 in the promotion of colorectal cancer progression and mechanisms of its upregulation via activation of STAT5, beta-catenin and EZH2. Carcinogenesis 35, 1389–1398 (2014). https://doi.org/10.1093/carcin/bgu057
M. Kim, K.T. Lee, H.R. Jang, J.H. Kim, S.M. Noh, K.S. Song, J.S. Cho, H.Y. Jeong, S.Y. Kim, H.S. Yoo, Y.S. Kim, Epigenetic down-regulation and suppressive role of DCBLD2 in gastric cancer cell proliferation and invasion. Mol. Cancer Res. 6, 222–230 (2008). https://doi.org/10.1158/1541-7786.MCR-07-0142
I. Fukumoto, T. Kinoshita, T. Hanazawa, N. Kikkawa, T. Chiyomaru, H. Enokida, N. Yamamoto, Y. Goto, R. Nishikawa, M. Nakagawa, Y. Okamoto, N. Seki, Identification of tumour suppressive microRNA-451a in hypopharyngeal squamous cell carcinoma based on microRNA expression signature. Br. J. Cancer 111, 386–394 (2014). https://doi.org/10.1038/bjc.2014.293
S.M. Pagnotta, C. Laudanna, M. Pancione, L. Sabatino, C. Votino, A. Remo, L. Cerulo, P. Zoppoli, E. Manfrin, V. Colantuoni, M. Ceccarelli, Ensemble of gene signatures identifies novel biomarkers in colorectal cancer activated through PPARgamma and TNFalpha signaling. PLoS One 8, e72638 (2013). https://doi.org/10.1371/journal.pone.0072638
Y. Chen, T.Y. Low, L.Y. Choong, R.S. Ray, Y.L. Tan, W. Toy, Q. Lin, B.K. Ang, C.H. Wong, S. Lim, B. Li, C.L. Hew, N.S. Sze, B.J. Druker, Y.P. Lim, Phosphoproteomics identified Endofin, DCBLD2, and KIAA0582 as novel tyrosine phosphorylation targets of EGF signaling and Iressa in human cancer cells. Proteomics 7, 2384–2397 (2007). https://doi.org/10.1002/pmic.200600968
A. Loboda, M.V. Nebozhyn, J.W. Watters, C.A. Buser, P.M. Shaw, P.S. Huang, L. Van't Veer, R.A. Tollenaar, D.B. Jackson, D. Agrawal, H. Dai, T.J. Yeatman, EMT is the dominant program in human colon cancer. BMC Med. Genet. 4, 9 (2011). https://doi.org/10.1186/1755-8794-4-9
R. Kalluri, R.A. Weinberg, The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009). https://doi.org/10.1172/JCI39104
J.H. Tsai, J. Yang, Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 27, 2192–2206 (2013). https://doi.org/10.1101/gad.225334.113
S. Lamouille, J. Xu, R. Derynck, Molecular mechanisms of epithelial-mesenchymal transition. Nature Rev. Mol. Cell Biol. 15, 178–196 (2014). https://doi.org/10.1038/nrm3758
A. Ganguly, H. Yang, R. Sharma, K.D. Patel, F. Cabral, The role of microtubules and their dynamics in cell migration. J. Biol. Chem. 287, 43359–43369 (2012). https://doi.org/10.1074/jbc.M112.423905
D.C. Ng, B.H. Lin, C.P. Lim, G. Huang, T. Zhang, V. Poli, X. Cao, Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell. Biol. 172, 245–257 (2006). https://doi.org/10.1083/jcb.200503021
M. Colomiere, A.C. Ward, C. Riley, M.K. Trenerry, D. Cameron-Smith, J. Findlay, L. Ackland, N. Ahmed, Cross talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Brit. J. Cancer 100, 134–144 (2009). https://doi.org/10.1038/sj.bjc.6604794
M. Rokavec, M.G. Oner, H. Li, R. Jackstadt, L. Jiang, D. Lodygin, M. Kaller, D. Horst, P.K. Ziegler, S. Schwitalla, J. Slotta-Huspenina, F.G. Bader, F.R. Greten, H. Hermeking, IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 124, 1853–1867 (2014). https://doi.org/10.1172/JCI73531
Acknowledgments
This work was supported by the Natural Science Foundation of Guangdong Province, China (No. 2018A030313676), Guangdong Planned Project of Science and Technology (No.2017B020226001), Guangzhou Planned Project of Science and Technology (No. 201707010275), and the Fundamental Research Funds for the Central Universities (No. 2017ZD104).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
The Ethics Committee of Guangzhou First Hospital approved the study protocol. The Institutional Animal Care and Use Committee approved the experimental procedures. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Written informed consent was obtained from all individual participants included in this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 10260 kb)
Rights and permissions
About this article
Cite this article
He, J., Huang, H., Du, Y. et al. Association of DCBLD2 upregulation with tumor progression and poor survival in colorectal cancer. Cell Oncol. 43, 409–420 (2020). https://doi.org/10.1007/s13402-020-00495-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-020-00495-8