Abstract
Objective
The aim of our study was to determine whether learning vector quantization neural networks could be used to predict liver metastases after a gastric cancer surgery.
Background
The prediction of tumor recurrence is invaluable for tailoring specific treatment and follow-up strategies for gastric cancer patients. At present, it is still impossible to make reliable predictions of tumor progression. The use of complex mathematical models such as neural networks has already been implemented for the study of various pathophysiological mechanisms, but to date they have never been used for predicting liver metastases after gastric cancer resection.
Methods
A total of 213 patients operated for gastric cancer between 1999 and 2005 were included in our study. They were stratified in a model development (140 patients) and validation group (73 patients). With the use of an auxiliary regression network, seven clinicopathological variables were selected to predict liver metastases.
Results
Forty-one patients developed liver metastases (19.2%). The longest follow-up was 2,754 days. Most liver metastases occurred in the first 799 days after discharge. All predictions were compared to actual recurrences with a two by two contingence table. The determined sensitivity and specificity for the development sample were 71 and 96.1%, respectively. The values for the test sample were 66.7 and 97.1%, respectively. The significance of the model was determined using various post-hoc tests, which all confirmed the effectiveness of our model.
Conclusion
The presented model exhibited a high negative predictive value and reasonable high sensitivity for liver metastases. To improve sensitivity, the inclusion of more patients and perhaps biological markers is still necessary.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The tremendous tendency for locoregional relapse, especially in serosa-positive tumors, is a major vice of gastric cancer. Those patients who survive the initial period disease-free are still at risk for hematogenous metastases. These are, paradoxically, most often seen in early stage gastric cancer, diminishing the potentially excellent long-term prognosis in T1 and T2 gastric cancer patients. In spite of all efforts to do so, it is still impossible to predict which patients might harbor dormant liver metastases, and, unfortunately, most of these cases are detected at a desperate stage. Consequently, a growing number of studies have been published emphasizing the importance of genetic markers in the genesis of gastric cancer recurrence [1–5], but even though encouraging results have been obtained, at present, further routine study along these lines would be too time- and cost-consuming. The use of indirect prognostic predictors could prove to be a simple time and cost-saving alternative.
Many papers have discussed this subject [6–10]. Although a plethora of mathematical models has been described, none of them are reliable enough to be used in clinical practice. Neural networks are useful tools in medicine [11]. When used, these usually implement Kohonen self-organizing neural networks. They have been successfully applied for classifying various forms of Parkinson’s disease [12], in diagnostic electromyography [13], and for studying breast cancer [14], but to our knowledge, they have never been used for prediction of liver metastases after gastric cancer resection. The aim of our study is to determine whether the use of learning vector quantization (LVQ) neural networks can be applied to predict liver metastases after gastric cancer resections with the analysis of indirect prognostic predictors.
Methods
Patients
From September 1999 to December 2005, 362 patients were operated on for gastric cancer at our institution. The data for each patient were collected and stored in our gastric cancer research database, comprising age, gender, ASA score, tumor size, location, Lauren, Ming and Borrman histological type, neo- and adjuvant chemo/radiotherapy data, type of resection, microscopically free margins, UICC and TNM stage, number of resected and positive lymph nodes, lymphocyte infiltration, vascular invasion, lymphangial invasion, tumor emboli to the lymph vessels, perineural invasion, and extranodal infiltration. During the 6-year period of data collection, some of the information was occasionally lost. If we used these missing values during the development of our model, the results would be corrupted, because the computer uses the average of the range of a predictor to fill the missing value. Consequently, only patients with no missing data (213 patients) were included in the study. Further inclusion criteria are histologically verified gastric cancer, resectable gastric cancer irrespective of level of curability (R0 and R2 resections), and patients with regular follow-up. Patients who died in the 60-day period following their operation were not included in the study. Age and gender distribution was balanced [gender: male 105 (49.3%), female 108 (50.7%), (P = 0.166); mean age: male 64.14 ± 11.69, female 64.86 ± 10.52, (P = 0.514)]. Patients from this primary group were randomly assigned into two subgroups. The size of the study group (development sample) used to develop the mathematical model was determined with the use of a nonlinear model. In this model, each predictor needed to be determined by a sufficient number of patients. Eventually, this group consisted of 140 patients. The validation group, used as a control to determine sensitivity and specificity of our model, consisted of 73 patients.
Surgery and Pathohistological Tumor Classification
The diagnosis of gastric adenocarcinoma was established endoscopically and confirmed with biopsy in all patients before the operation. The strategy was to achieve R0 resection (assuring 4–5-cm tumor-free safety margins in intestinal type; 8–10-cm tumor-free safety margins in diffuse type according to Lauren) accompanied by D2 lymphadenectomy. The surgical procedure was performed according to standards described elsewhere [15–17]. Resected specimens were examined according to standard pathohistological procedures and classified according to criteria described by the Japanese Research Society for Gastric Cancer [15–17]. The anatomical location, diameter of the tumor, lymphocyte invasion, vascular invasion, lymphangial invasion, extranodal invasion, and perineural invasion were all noted. The number of harvested and positive lymph nodes was also determined. Tumor invasion (T), lymph node (N) classification, and distant metastases (M) followed the UICC criteria [6]. Histological type was classified as intestinal, diffuse, or mixed, in accordance with Lauren’s classification. All patients were presented at an oncological-surgical board. The amenable patients were scheduled for adjuvant chemo/radiotherapy.
Data
Neural networks perform non-linear transformations between sets of variables. In principle, one could use raw input data for analysis, but this approach would yield poor results for a number of reasons. Thus, the data have to be first transformed into a new representation form before training a neural network. This was done in three steps: (1) standardization; (2) reduction of dimensionality; (3) incorporation of prior knowledge.
Pre-processing may take the form of a linear transformation of the input data. In the present application, we applied linear transformation by normalizing the input space with standardization. The transformed data have 0 mean and 1 SD. This step is necessary because we apply Eucledian distance—a method for determining the distance between two vectors—in the competitive layer. The input data are transformed into a vector form with its coordinates represented in a matrix. Wide ranges of variables would not be suitable for reproduction of vector coordinates, and the Eucledian distance would be very insensitive to such variables. The standardization itself was carried out with independent processing of each variable of the input space. For each of them (x i ) we calculated mean \( (\bar{x}_{i} ) \) and variance \( (\sigma_{i}^{2} ) \) using
where n = 1, … ,N labels the patterns (patience data). We defined a set of re-scaled variables given by
The next step was a reduction of dimensionality, which basically means that the number of input variables had to be reduced. The fact that such dimensionality reduction can lead to improved performance may appear paradoxical as it cannot increase the information content of the input data. The resolution is related to the curse of dimensionality (Masters 1995). As always, in practice, we are forced to work with a limited quantity of data. In such cases the inclusion of many variables into a model could lead to a point where the data are very sparse in relation to the number of predictors. Our model, and in fact any other mathematical model, would not be able to generalize, but would instead memorize the data. This would lead to pure classification results. In this application the reduction of dimensionality was guided by the application of an auxiliary regression network with a single neuron with a sigmoid activation function, which has very similar properties to logit regression. This procedure enabled us to reduce the input space to seven variables. The descriptive statistics for the selected variables and the dependent variable (representing the status) are presented in Tables 1 and 2.
Another important way to improve network performance is through the incorporation of prior knowledge referring to relevant information that might be used to develop a solution and that is additional to that provided by the development sample (training data). Prior knowledge can either be incorporated into the pre-processing and post-processing stages or into the network structure itself. It can also be used to modify the training process as in Jagric et al. [18]. In our case we incorporated the prior knowledge into the network structure by defining the number of neurons in the linear layer belonging to each outcome.
Learning Vector Quantization Network (LVQ)
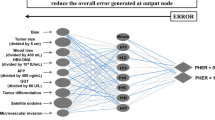
Learning Vector Quantization (LVQ) is a subtype of artificial neural networks introduced by Kohonen and colleagues [19–21]. The methodology has been described in detail elsewhere [22–34]. In brief, the LVQ network is a set of mathematical algorithms comparing the input data with a preexisting matrix of results and determining which part of this matrix has the most similar properties to the input data. This process is called the nearest prototype classification (NPC). In contrast to other types of neural networks, LVQ is a two-layer network, with one layer of synapses between them. A secondary linear layer transforms the results produced by a competitive layer into target results defined by the user (Fig. 1). This way the end result of learning is a certain number of outcomes (the so-called sub-classes) in the competitive layer and a defined number of outcomes (the so-called target classes) in the linear layer. The defined target classes in our case were binary results, i.e., liver metastases present or absent. According to the activity of the input neurons and the weights of their synapses, the activities of the output neurons are calculated. This process is a remnant of the data processing in the central nervous system, where multiple synapses pour information into a single neuron, which then transforms this input into an impulse in its axon. If the synapses are used frequently, they become consolidated and function more efficiently.
Results
Follow-Up
After hospital discharge, all patients had periodical follow-up examinations. At each examination standard laboratory, including tumor markers (CA 19-9, CEA), chest X-ray and abdominal ultrasound were performed. When a recurrence was suspected on the basis of elevation of tumor markers or imaging studies, the patient was scheduled for endoscopic examination and computer tomography. Where bone metastases were suspected, scintigraphy or positron emission tomography was scheduled. The type of recurrence was classified as liver metastasis (with or without locoregional relapse) or carcinoma progression that included locoregional recurrence or distant metastases.
Follow-up was closed in December 2007. From 213 patients included in our study, 41 (19.2%) had liver metastases, of which 15 (7%) were synchronous and 26 (12.2%) were metachronous. The mean follow-up for patients with liver metastases was 234 days (range, 0–1,849 days) and 1,061 days (range, 27–2,754 days) for patients without liver metastases. Most liver metastases occurred in the first 799 days (40 of 41, 97.56%), when the cumulative hazard was 20.3% ± 3%. The estimated risk of recurrence after 5 years (1,849 days) was 24.1 ± 5%.
Predictors
All 22 of the originally selected predictors were tested. As mentioned earlier, to improve the performance of our model, the number of predictors had to be reduced. This was achieved by means of the auxiliary regression network with a single neuron with a sigmoid activation curve. In this fashion, seven predictors were identified as significant, namely the size of the tumor, Lauren histological type, adjuvant chemo/radiotherapy, the TNM N status, the UICC stage, the number of positive lymph nodes, and the proportion of positive nodes of all resected nodes, which were then used in the final model.
We also compared patients with liver metastases and patients without liver metastases. Although there was no significant difference in the overall distribution of histological type (P = 0.279), there were clearly dissimilarities in the intestinal type distribution. The intestinal type was with 53.7%, the most prevalent histological type in the group with liver metastases (53.7% intestinal, 24.4% diffuse, 19.5% mixed), in contrast to patients without liver metastases where the histological types were equally distributed (39.5% intestinal, 37.2% diffuse, 20.3% mixed). There was an expected difference in the UICC stage distribution, because simultaneous liver metastases with the TNM stage and consequently a higher UICC stage were included. The comparison also identified the number of positive nodes and the fraction of positive nodes from all harvested nodes as significantly different (P < 0.0001 and P = 0.005, respectively). The number of all harvested nodes was similar in both groups. The size of the tumor was only insignificantly larger in the group with liver metastases (P = 0.451). In both groups, an equal part of patients received adjuvant chemo/radiotherapy (P = 0.474). The patient characteristics of patients with liver metastases and patients without liver metastases are shown in Table 3.
The Prediction of Liver Metastases
In this section we present the final model and its classification performance. By applying the procedures described in previous sections, we developed a model with nine neurons in the competitive layer and two neurons in the linear layer. The model is presented in Fig. 2 together with the matrices of weights for both layers.
The numbers in the IW1,1 matrix represent the weights of the predictors in the competitive layer. As described earlier, the values of the prognostic factors had to be standardized in order to use them in our model. Consequently, the weights cannot be interpreted as in other regression models. Because the relation to the actual values of the predictors is non-linear, the positive and negative values do not reflect a positive or a negative impact of a predictor on the outcome. The model exhibited the optimal performance with nine neurons; the first six predict no liver metastases, while the last three represent the patients with liver metastases. When we increased or lowered the number of neurons that predict liver metastases, the performance of our model either did not change at all or worsened. The final neurons of the linear layer are presented in the LW2,1 matrix. In this layer the final two neurons are binary. When a patient was analyzed, the values of his prognostic factors were transformed in a vector, which was analyzed by our model where it activated a neuron that was the closest in the network of the competitive layer. In the second layer, this neuron was transformed to present a binary result, i.e., metastases or no metastases. The result of our analysis is the prediction of two possible results, negative or positive (liver metastasis or no liver metastasis), against the objective measurement of the outcome, also measured dichotomously. The results of our predictions are presented in a two-by-two contingency table (Table 4) for classifying the number of times the model resulted in a correct positive prediction, a correct negative prediction, an incorrect positive prediction, and an incorrect negative prediction.
From 140 patients in the development sample, 38 had liver metastases. In the test sample, a total of 73 patients were included, and 3 among them had liver metastases. In the development sample, our model correctly predicted 27 liver metastases (71%) and incorrectly predicted 11 liver metastases (29%). The negative predictive value of the model for the development sample was 96% (98 from 102), whereas the false-negative value was 4%. The model developed was then used to predict metastases in the test sample. The positive predictive value was 66% (2 from 3), and the negative predictive value was 97% (68 from 70). The false positive and negative predictive values were 34 and 3%, respectively. Accordingly, the sensitivity of our model is 71% for the development sample and 66.7% for the test sample. The specificity of our model is 96.1 and 97.1% for the development and test sample.
To evaluate the effectiveness and significance of our model, various diagnostic post-hoc tests were used. The results of these and former tests and their confidence intervals are presented in Table 4. Kappa is a measure of agreement and takes on the value of zero if there is no more agreement between test and outcome than can be expected on the basis of chance. Kappa takes on the value of 1 if there is perfect agreement. It is considered that kappa values lower than 0.4 represent poor agreement. The kappa measure for the development sample is 0.712 and for the test sample 0.55. Positive likelihood indicates how much more likely it is to predict liver metastases in patients who actually have metastases than in the group of patients without such metastases. For the development sample, this value is 18.118 and for the test sample 23.333. A similar test indicates how likely it is to predict the absence of liver metastases in the group of patients without such metastases. Again, the values of 0.301 for the development sample and 0.343 for the test sample indicate that such prediction is more likely in the group without liver metastases. To determine the discriminative power of the test, the diagnostic odds ratio was used. This test has a value of one if the model does not discriminate between diseased and not diseased. Very high values, above one, mean that our model discriminates well. Values lower than one indicate that there is something wrong in the application of the test. For the development sample, the diagnostic odds ratio was 60.136, and for our test sample it was 68. A similar test to evaluate the overall performance of a model is Youden’s J. The J takes on the value of one if a diagnostic model discriminates perfectly without making any mistakes. For our development sample, this value is 0.671, and for the test sample 0.638. To access the diagnostic power of our model, a series of other tests were done, all of which are presented in Table 5.
Discussion
Gastric cancer is a disease with a strikingly high number of recurrences. Even a curative resection in many cases does not guarantee long-term survival. This implies the assumption that other factors besides microscopically free resection margins determine long-term survival and the occurrence of a potential relapse. Indeed, over the years many papers have been published describing such factors [4, 7, 9, 35, 36]. They are determined with routine pathohistological examination and molecular marker analysis, and have increasingly gained attention in recent years. The use of such predictors for determining a patient’s prognosis is of utmost importance in selecting the best individual treatment for a patient. In oncology, a large number of statistical models are used to predict recurrences with the application of prognostic variables [6, 8, 10, 37, 38].
In this study, with the use of LVQ neural networks we developed a model that would identify patients who will develop liver metastases after gastric resection. The final model distinguished between two groups of patients: patients who will develop liver metastases—isolated or in addition to gastric bed or peritoneal disease—and those who will not develop liver metastases. The latter group was defined as patients at risk for locoregional relapse, peritoneal dissemination, or without disease recurrence.
After surgical treatment, the patients were periodically followed up and examined according to a standard protocol. As described elsewhere [6, 39, 40], most of the recurrences occurred within the first 2 years. Liver metastases were observed in 19.2% of cases. In most cases, the recurrence was locoregional, in the form of peritoneal carcinomatosis. These incidences were similar to other studies in which liver metastases were found to be a rare mode of carcinomatous spread [7, 39, 41–43].
From all the selected prognostic factors included in this study, our model identified the size of the tumor, Lauren histological type, UICC stage, adjuvant chemo/radiotherapy, number of positive nodes, number of resected nodes, and the fraction of positive ones from all resected lymph nodes. Interestingly, these variables coincide with the findings of other authors studying the occurrence of liver metastases after gastric resection [7]. In their paper, Marrelli et al. [6] describe the preoperative tumor marker levels, Lauren’s intestinal histological type, and lymph node involvement as independent prognostic factors for liver metastases after radical surgical treatment of gastric cancer. In fact, many other studies confirm a significant association among intestinal histological type, high preoperative CEA values, lymph node involvement, and hematogenous metastases from gastric cancer [7, 39, 44, 45]. In our study, the comparison of tumor characteristic distribution between patients with and without liver metastases confirmed, as anticipated, a higher number of positive lymph nodes as well as a higher proportion of metastatic lymph nodes among harvested nodes in the liver metastases group. The comparison did, however, show some unexpected tumor characteristic differences between both groups. Even if the histological type distribution failed to show significant differences in favor of the intestinal type between both groups, the intestinal histological type was still the most prevalent in the group of patients with liver metastases. Surprisingly, the tumor size was also insignificantly different in the group with liver metastases; hence, there was no simple linear relationship between tumor progression, size of the tumor, and liver metastases. This mirrors the fact that the liver metastases producing tumors have diverse characteristics that cannot be appreciated by a simple statistical model. Our model, on the other hand, was able to identify even those patients who are considered to be at minor risk for liver metastases. This capability of our model accounts for its extraordinary value to make specific predictions of liver metastases.
The development of such a model requires accurate data, a suitable number of cases, and no missing values. The first two criteria are easily achieved, but as is usually the case, there were some patients with missing values. These cases could potentially endanger the analysis of the data and model development. In order to prevent this, we were forced to exclude these patients and risk selection bias of our data.
The sensitivity and specificity for our development sample were 71 and 96.1%, respectively. Similar values were obtained for the test sample, which had a sensitivity of 66.7% and a specificity of 97.1%. These results clearly demonstrate the high negative predictive value of our model. Even though the positive predictive value is somewhat lower, the model still correctly predicted most of the liver metastases. This result can mostly be accounted for by the low number of liver metastases in the sample. Thus, the sensitivity in the development sample in which 38 patients had liver metastases was 71% higher than in the test sample, in which only 3 out of 73 patients had such metastases. This means that the performance of the model and its sensitivity can be optimized simply by inclusion of a larger group of patients. The accuracy was tested with a range of post-hoc tests that all confirm the effectiveness of our model.
Many studies regarding the prediction of tumor recurrence with the use of mathematical models have been published [6, 8, 10, 37, 38]. They usually apply regression models [10], scoring systems [37], or tree algorithms [38] to stratify patients into risk groups according to their prognostic factors, but this and similar models present two key problems. The first drawback is that many patients are stratified in an intermediate risk group with an uncertain outcome. The model presented in our study divides the patients equally into two groups, one with a positive and the other with a negative prediction for a specific outcome. Such binary predictions are very important for therapeutic planning. In clinical practice, it is usually the intermediate risk group for which it is the most difficult to make decisions for the right oncological treatment. These clear-cut predictions, when made with reasonable accuracy, are of prime importance for improvement of the prognosis.
The second disadvantage of different models is their inability to predict the exact site of recurrence. Marrelli and colleagues [6] describe a model that, with reasonable sensitivity and specificity, predicts tumor recurrence after a curative resection, but their model fails to identify specific recurrence sites. The proponents of such models reason that with the use of more prognostic factors, precise predictions can be made. We suspect this might be only a part of the truth. Many statistical models do not acknowledge gastric cancer as a heterogenic disease; 1.4–6.4% of early cancers have been reported to recur [9, 46–48], and about half of them produce hematogenic metastases [9, 47, 49]. The morphological characteristics of these tumors are completely different from more advanced tumors with liver metastases. Our model is conceptualized to recognize a wide spectrum of morphological tumor characteristics that could produce the same outcome, and with simple modification a prediction of other specific recurrence sites could be made. The last novelty of this model is the use of adjuvant chemo/radiotherapy as a predictor. To our knowledge, none of the published models use this factor as one that can greatly alter the course of the disease for their predictions.
Despite the fact that gastric cancer is a disease with locoregional recurrence in over 80% of cases, modern adjuvant treatment aimed at locoregional control and better screening protocols improve patients’ survival. Consequently, patients who do not live long enough to show evidence of blood-borne metastases are now becoming more and more common. This positive trend might demand reconsideration in gastric cancer treatment strategies. Indeed, the greatest understanding of modern oncology is the individualization of cancer treatment. This can only be made possible by exact prediction of disease progression. To stratify patients, not only according to their survival prognosis, but also according to the recurrence site, will consequently alter the adjuvant treatment and follow-up strategy. Although the results presented in this study could greatly help in attaining this goal, they are still too optimistic to be relied upon. The inclusion of more patients along with the identification of different predictors, or even molecular markers, is still necessary to improve the efficiency of our model and to have it put to use in the seemingly hopeless war against cancer.
References
Vauhkonen M, Vauhkonen H, Sipponen P. Pathology and molecular biology of gastric cancer. Best Pract Res Clin Gastroenterol. 2006;20:651–674.
Cervantes A, Braun ER, Fidalgo AP, Gonzalez IC. Molecular biology of gastric cancer. Clin Transl Oncol. 2007;9:208–215.
Wright PA, Willians GT. Molecular biology and gastric carcinoma. 1993;34:145–147.
Dicken BJ, Bigam DL, Cass C, Mackey JR, Joy AA, Hamilton SM. Gastric adenocarcinoma. Review and considerations for future directions. Ann Surg. 2005;241:27–39.
Chan AOO, Chu KM, Lam SK, et al. Early prediction of tumor recurrence after curative resection of gastric carcinoma by measuring soluble E-cadherin. Cancer. 2005;104:740–745.
Marrelli D, De Stefano A, de Manzoni G, Morgagni P, Dileo A, Roviello F. Prediction of recurrence after radical surgery for gastric cancer. Ann Surg. 2005;241:247–255.
Marrelli D, Roviello F, De Stefano A, et al. Risk factors for liver metastases after curative surgical procedures for gastric cancer: a prospective study of 208 patients treated with surgical resection. J Am Coll Surg. 2004;198:51–58.
Bollschweiler E, Lubke T, Monig SP, Holscher AH. Evaluation of POSSUM scoring system in patients with gastric cancer undergoing D2-gastrectomy. 2005;5:2–7.
Lee HJ, Kim YH, Kim WH, et al. Clinicopathological analysis for recurrence of early gastric cancer. Jpn J Clin Oncol. 2003;33:209–214.
Yokota T, Saito T, Teshima S, et al. Early and late recurrences after gastrectomy for gastric cancer: a multiple logistic regression analysis. Upsala J Med Sci. 2002;107:17–22.
Gooi C, Mintchev M. Neural networks: a diagnostic tool for gastric electrical uncoupling? Inf Theor Appl. 2003;11:47–52.
Fritsch T, Kraus PH, Pruntek H, Tran-Gia P. Classification of Parkinson rating scale data using a self-organizing neural net. In: IEEE International conference on Neural Networks 1993;93–98.
Pattichis CS, Schizas CN, Middleton LT. Neural network models in EMG diagnosis. IEEE Trans Biomed Eng. 1995;486–496.
Allan R, Kinsner W. A study of microscopic images of human breast disease using competitive neural networks. Canadian Conference on Electrical and Computer Engineering 2001.
Grau JJ, Palmero R, Marmol M, et al. Time-related improvement of survival in resectable gastric cancer: the role of Japanese-style gastrectomy with D2 lymphadenectomy and adjuvant chemotherapy. World J Surg Oncol. 2006;4:53–62.
Kajitani T. Japanese Research Society for the Study of Gastric Cancer: the general rules for gastric cancer study in surgery and pathology. Jpn J Surg. 1981;11:127–145.
Nio Y, Tsubono M, Kawabata K, et al. Comparison of survival curves of gastric cancer patients after surgery according to the UICC stage classification and the general rules for gastric cancer study by the Japanese research society for gastric cancer. Ann Surg. 1993;218(1):47–53.
Jagric T. A nonlinear approach to forecasting with leading economic indicators, vol. 7. Stud Nonlinear Dyn Econom. 2003;7(2):1–18.
Masters T. Advanced Algorithms for Neural Networks: A C++ Sourcebook. NY: Wiley; 1995.
Kohonen T. Self Organization Maps. New York: Springer; 2001.
Kohonen T, Hynninen J, Kangas J, Laaksonen J, Torkkola K. Lvq pak: the learning vector quantization program package. 1995.
Pratt WK. Digital Image Processing. 2nd ed. NY: Wiley; 1991.
Rosenfeld A, Kak AC. Digital Picture Processing. London: Academic Press; 1982.
Gonzalez RC, Wints P. Digital Image Processing. London: Addison Wesley; 1987.
Handels H. Medizinische Bildverarbeitung. Stuttgart: BG Teubner; 2000.
Kohonen T. “The self-organizing map”. Proc IEEE. 1990;1464–1480.
Schmitz G, Krüger M, Ermert H. “Tissue characterization of the prostate using Kohonen maps”. Proc. 1994 IEEE Ultrasonics Symposium 1994;1487–1490.
Rychagov MN, Ilin SV, Masloboev YP. “Neural network tissue identification and characterization using multiplayer perception and Kohonen maps.” In: Humboldtian Conference “Biomedical Sciences-2001,” Moscow (Russia) 2001;25.
Ilin SV, Rychagov MN. “Segmentation of ultrasonic images by using neural networks of backward propagation”. In Proc. Nizhnij Novgorod Acoust. Scientific Session, Nizhnij Novgorod (Russia), pp. 404–406, 2002 (in Russian).
Shabalin A, Shabalin I, Zimbalov E. Project “SONO—2000”, http://www.sono.nino.ru 1318.
Kohonen T. Analysis of a simple self-organizing process. Biol Cybern. 1982;44(2):135–140.
Kohonen T. Self-organizing formation of topologically correct feature maps. Biol Cybern. 1982;43(1):59–69.
Kohonen T. Learning Vector Quantization. Technical Report. Otaniemi: Helsinki Univ. of Tech; 1986.
Kohonen T. Improved versions of learning vector quantization. Int Joint Conf Neural Netw. 1990;1:545–550.
Kooby DA, Suriawinata A, Klimstra DS, Brennan MF, Karpeh MS. Biologic predictors of survival in node-negative gastric cancer. Ann Surg. 2003;237:828–837.
Siewert JR, Böttcher K, Stien HJ, Roder JD, The German Gastric Carcinoma Study Group. Relevant prognostic factors in gastric cancer. Ten-Year Results German Gastric Cancer Study. 1998;228:449–461.
Grisaru DA, Covens A, Franssen E, et al. Histopathologic score predicts recurrence free survival after radical surgery in patients with stage IA2-IB1–2 cervical carcinoma. Cancer. 2003;97:1904–1908.
Radespiel-Tröger M, Hohenberger W, Reingruber B. Improved prediction of recurrence after curative resection of colon carcinoma using tree-based risk stratification. Cancer. 2004;100:958–967.
Yoo CH, Noh SH, Shin DW, et al. Recurrence following curative resection for gastric carcinoma. Br J Surg. 2000;87:236–242.
Shiraishi N, Inomata M, Osawa N, et al. Early and late recurrence after gastrectomy for gastric carcinoma: univariate and multivariate analyses. Cancer. 2000;89:255–261.
Koga S, Takebayashi M, Kaibara N, et al. Pathological characteristics of gastric cancer that develop hematogenous recurrence, with special reference to the site of recurrence. J Surg Oncol. 1987;36:239–242.
Maehara Y, Emi Y, Baba H, et al. Recurrences and related characteristics of gastric cancer. Br J Cancer. 1996;74:975–979.
Kasakura Y, Fujii M, Mochizuki F, et al. Is there a benefit of pancreaticosplenectomy with gastrectomy for advanced gastric cancer? Am J Surg. 2000;179:237–242.
Mori M, Sakaguchi H, Akazawa K, et al. Prognostic significance of CEA, Ca 19–9, and Ca 72–4 preoperative serum levels in gastric carcinoma. Oncology. 1999;57:55–62.
Ikeda Y, Mori M, Kajiyama K, et al. Indicative value of carcinoembryonic antigen (CEA) for liver recurrence following curative resection of stage II and III gastric cancer. Hepatogastroenterology. 1996;43:1281–1287.
Ichiyoshi Y, Toda T, Minamisona Y, Nagasaki S, Yakeishi Y, Sugimachi K. Recurrence in early gastric cancer. Surgery. 1990;107:489–495.
Shiozawa N, Kodama M, Chida T, Arakawa A, Tur GE, Koyama K. Recurrent death among early gastric cancer patients: 20-years’ experience. Hepatogastroenterology. 1994;41:244–247.
Sano T, Kobori O, Muto T. Lymph node metastasis for early gastric cancer. Br J Surg. 1992;79:241–244.
Folli S, Dente M, Dell’Amore D, et al. Early gastric cancer: prognostic factors in 223 patients. Br J Surg. 1995;82:952–956.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jagric, T., Potrc, S. & Jagric, T. Prediction of Liver Metastases After Gastric Cancer Resection with the Use of Learning Vector Quantization Neural Networks. Dig Dis Sci 55, 3252–3261 (2010). https://doi.org/10.1007/s10620-010-1155-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-010-1155-z