One new anthraquinone derivative, named 8-O-methylnidurufin (1), together with five known analogues (2–6), have been isolated from a gorgonian-derived fungus, Aspergillus sp. The structures were elucidated by combined spectroscopic methods including 1D and 2D NMR spectral data. All isolated metabolites were evaluated for their cytotoxic and antibacterial activities in vitro. Compound 2 showed significant cytotoxic activity against K562 and HL-60 cell lines with IC50 values of 0.87 and 1.46 μM, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Marine-derived fungi of the genus Aspergillus are well known as a prolific source of structurally unique and biologically active metabolites. A growing number of novel bioactive secondary metabolites isolated from this genus have been reported recently [1, 2]. As part of our continuing efforts to discover bioactive metabolites from marine-derived fungi in the South China Sea, we isolated several anthraquinone dimers, bisabolane sesquiterpenoids, and benzylazaphilone derivatives with potent cytotoxic activity [3–5]. Recently, we have investigated the chemical compositions of the fermentation broth of an Asperigillus sp. strain isolated from a gorgonian Dichotella gemmacea. One new anthraquinone derivative, named 8-O-methylnidurufin (1), together with five known analogues, nidurufin (2) [6], averufin (3) [7], 8-O-methylaverufin (4) [8], averufanin (5) [9], and 8-O-methylaverufanin (6) [10, 11], have been obtained from the fungal culture. Herein we describe the isolation, structure elucidation, and biological activity of these compounds.

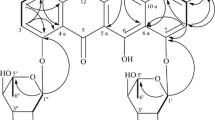
Compound 1 was isolated as an orange powder. Its molecular formula of C21H18O8 (13 degrees of unsaturation) was determined by HR-ESI-MS. Careful comparison of the 1H and 13C NMR data of 1 with those of nidurufin (2) [6] suggested that 1 was very similar to 2. The most obvious differences in their 1H NMR spectra were the presence of a singlet signal at δ 3.88 assignable to the methoxy group and a hydrogen-bonded hydroxy signal at δ 13.94 in 1 instead of two hydrogen-bonded hydroxy signals at δ 12.50 and 12.21 in 2. It can be deduced that 1 is the methylation derivative of 2. The methoxy group in 1 was also confirmed by the 13C NMR spectrum with a carbon signal at δ 56.2. In the HMBC spectrum, correlations from 8-OCH3 to C-8 showed that the methoxy group is attached to C-8. Detailed assignments of protons and carbons for 1 were unambiguously accomplished by analysis of the COSY, HMQC, and HMBC data (Table 1). The relative configuration of 1 was determined by combination of proton-proton coupling constant analysis and NOESY experiment.
The small coupling constant between H-1′ and H-2′ (J = 1.8 Hz) established the cis-configuration for these two protons. The NOESY correlation of H-2′/H3-6′ placed H-2′ and H3-6′ on the same face of the molecular. The absolute configuration of 1 could be determined based on biogenic considerations. Since 1 and 2 were produced in parallel by this fungus, the absolute configuration of 1 could be proposed as 1′R,2′S,5′S, identical to that of 2. Thus, the structure of 1 was named 8-O-methylnidurufin.
The structures of the known compounds 2–6 were identified as nidurufin (2) [6], averufin (3) [7], 8-O-methylaverufin (4) [8], averufanin (5) [9], and 8-O-methylaverufanin (6) [10, 11], respectively, on the basis of their NMR and ESI-MS data and by comparison with those previously reported in the literature. All of these known compounds (2–6) were involved in the aflatoxin biosynthetic pathway of Aspergillus spp. [12, 13]; hence, the co-occurring compound 1 could be presumed to be biosynthesized by the same pathway.
Compounds 1–6 were assessed for cytotoxic activity against human lung carcinoma K562 and human promyelocytic leukemia HL-60 cell lines by the MTT method as described previously [14]. Compound 2 displayed significant growth-inhibitory effects on K562 and HL-60 cell lines with IC50 values of 0.87 and 1.46 μM, respectively. Other compounds were found to be relatively noncytotoxic (IC50 > 10 μM).
The antibacterial activities of 1–6 were also evaluated by their minimum inhibitory concentration (MIC, μM) [15] against a panel of pathogenic bacteria including Gram-positive Staphylococcus aureus, S. albus, Bacillus cereus, Micrococcus tetragenus, and M. luteus, and Gram-negative Escherichia coli and Pseudomonas putida (Table 2). Compounds 1 and 4 displayed antibacterial activity against M. luteus with the same MIC values of 6.25 μM, more potent than those of 2 (MIC = 25 μM) and 3 (MIC = 50 μM), suggesting that the methoxy at C-8 may play a positive contribution. None of the compounds showed inhibitory effects against S. aureus, B. cereus, M. tetragenus, and P. putida.
Experimental
Melting points were determined on an X-6 micromelting point apparatus and are uncorrected. Optical rotations were measured on a Jasco P-1020 digital polarimeter. UV spectra were obtained on a Beckman DU 640 spectrophotometer. IR spectra were recorded on a Nicolet-Nexus-470 spectrometer using KBr pellets. NMR spectra were acquired using a JEOL JEM-ECP NMR spectrometer (600 MHz for 1H and 150 MHz for 13C) using TMS as internal standard. ESI-MS spectra were obtained from a Micromass Q-TOF spectrometer. Semipreparative HPLC was performed on a Waters 1525 system using a semipreparative C18 (Kromasil, 5 μm, 10 × 250 mm) column coupled with a Waters 2996 photodiode array detector. Silica gel (Qing Dao Hai Yang Chemical Group Co.; 200–300 mesh), Sephadex LH-20 (Amersham Biosciences), and octadecylsilyl silica gel (Unicorn; 45–60 μm) were used for column chromatography (CC). Precoated silica gel plates (Yan Tai Zi Fu Chemical Group Co.; G60, F-254) were used for thin-layer chromatography (TLC).
Fungal Material and Culture Conditions. The fungal strain of Aspergillus sp. was isolated from the inner part of a fresh tissue of gorgonian Dichotella gemmacea, which was collected from the South China Sea in December, 2009. The strain was deposited at the Key Laboratory of Marine Drugs, the Ministry of Education of China, School of Medicine and Pharmacy, Ocean University of China, Qingdao, P. R. China. The fungus was cultivated in 20 L liquid medium (10.0 g of glucose, 2.0 g of yeast extract, 2.0 g of peptone in 1 L of seawater, in 1 L Erlenmeyer flasks each containing 400 mL of culture broth) at 25°C without shaking for 30 days.
Extraction and Isolation . The fermentation broth (20 L) was extracted three times with an equal volume of EtOAc. The combined, filtered EtOAc solutions were evaporated to dryness, yielding 11.5 g of crude extract, which was subjected to silica gel column chromatography (CC) using step gradient elution with EtOAc–petroleum ether (0–100%) and then with MeOH–CHCl3 (0–100%) for separation into eight fractions (Fr.1–Fr.8). Fraction 2 was subjected to Sephadex LH-20 CC eluting with petroleum ether–CHCl3–MeOH (2:1:1, v/v/v) and then further purified on semipreparative HPLC (82%, MeOH–H2O) to give 3 (5.1 mg) and 5 (3.1 mg). Fraction 3 was separated by ODS CC eluted with 90% MeOH–H2O and further purified by HPLC (80%, MeOH–H2O) to give 4 (4.2 mg) and 6 (3.1 mg). Fraction 4 was applied to Sephadex LH-20 CC eluting with CHCl3–MeOH (1:1, v/v) and further purified by HPLC (85% MeOH–H2O) to afford 2 (1.8 mg). Fraction 5 was isolated by Sephadex LH-20 CC eluting with CHCl3–MeOH (1:1, v/v) and further purified by HPLC (78%, MeOH–H2O) to afford 1 (4.8 mg).
8- O -Methylnidurufin (1). Orange powder; mp 173–174°C; [α] 25D +47° (c 0.17, MeOH). UV (MeOH, λmax, nm) (log ε): 220 (2.88), 286 (2.60), 308 (2.24). IR (KBr, νmax, cm–1): 3428, 2926, 2852, 1609, 1436, 1382, 1265, 1206, 1127, 1028, 937, 836, 750. 1H NMR (600 MHz, DMSO-d6) and 13C NMR (150 MHz, DMSO-d6), see Table 1. ESI-MS m/z 397.1 [M – H]–; HR-ESI-MS m/z 397.0922 (calcd for C21H17O8, 397.0918).
Nidurufin (2). Orange powder. 1H NMR (600 MHz, DMSO-d6, δ, ppm, J/Hz): 12.50, 12.21 (each 1H, s, 1, 8-OH), 6.98 (2H, br.s, H-4, 5), 6.38 (1H, br.s, H-7), 5.03 (1H, d, J = 2.4, H-1′), 3.76 (1H, br.d, J = 2.4, H-2′), 2.15 (1H, m, Ha-4′), 1.81 (1H, m, Hb-4′), 1.55 (2H, m, H-3′), 1.53 (3H, s, H-6′). ESI-MS m/z 383.1 [M – H]–.
Averufin (3). Red powder. 1H NMR (600 MHz, acetone-d6, δ, ppm, J/Hz): 12.56, 12.20 (each 1H, s, 1, 8-OH), 7.23 (1H, br.s, H-5), 7.10 (1H, s, H-4), 6.64 (1H, br.s, H-7), 5.29 (1H, br.s, H-1′), 1.28–1.90 (6H, m, H-2′, 3′, 4′), 1.54 (3H, s, H-6′). ESI-MS m/z 369.1 [M + H]+.
8- O -Methylaverufin (4). Red powder. 1H NMR (600 MHz, acetone-d6, δ, ppm, J/Hz): 14.00 (1H, s, 1-OH), 7.34 (1H, d, J = 2.4, H-5), 7.05 (1H, s, H-4), 6.92 (1H, d, J = 2.4, H-7), 5.30 (1H, d, J = 2.4, H-1′), 3.97 (3H, s, 8-OCH3), 1.56–2.02 (6H, m, H-2′, 3′, 4′), 1.52 (3H, s, H-6′). ESI-MS m/z 383.1 [M + H]+, 787.4 [2M + H]+.
Averufanin (5). Orange powder. 1H NMR (600 MHz, acetone-d6, δ, ppm, J/Hz): 12.85, 12.20 (1H, s, 1, 8-OH), 7.25 (1H, d, J = 2.4, H-5), 7.12 (1H, s, H-4), 6.65 (1H, d, J = 2.4, H-7), 5.17 (1H, dd, J = 11.4, 1.8, H-1′), 3.84 (1H, m, H-5′), 1.47–1.96 (6H, m, H-2′, 3′, 4′), 1.32 (3H, d, J = 6.6, H-6′). ESI-MS m/z 371.2 [M + H]+, 393.3 [M + Na]+.
8- O -Methylaverufanin (6). Orange powder. 1H NMR (600 MHz, CDCl3, δ, ppm, J/Hz): 13.83 (1H, s, 1-OH), 9.83 (1H, s, 6-OH), 7.38 (1H, br.s, H-5), 7.20 (1H, s, H-4), 6.78 (1H, br.s, H-7), 5.17 (1H, dd, J = 11.4, 2.4, H-1′), 4.02 (3H, s, 8-OCH3), 3.75 (1H, m, H-5′), 1.39–1.96 (6H, m, H-2′, 3′, 4′), 1.30 (3H, d, J = 6.6, H-6′). ESI-MS m/z: 385.2 [M + H]+, 407.2 [M + Na]+, 423.2 [M + K]+, 791.5 [2M + Na]+.
Biological Assays. The cytotoxic activity against K562 and HL-60 cell lines were determined by the MTT method [14]. Adriamycin was used as a positive control.
The antibacterial activity against seven bacterial strains, including Gram-positive S. aureus (ATCC 27154), S. albus (ATCC 8799), B. cereus (ACCC 11077), M. tetragenus (ATCC 13623), M. luteus (ATCC 4698), and Gram-negative E. coli (ATCC 25922), and P. putida (ATCC 17848) were carried out as described previously [15].
References
J. W. Blunt, B. R. Coop, R. A. Keyzers, M. H. G. Munro, and M. R. Prinsep, Nat. Prod. Rep., 29, 144 (2012).
J. M. Finefield, J. C. Frisvad, D. H. Sherman, and R. M. Williams, J. Nat. Prod., 75, 812 (2012).
C. J. Zheng, C. L. Shao, Z. Y. Guo, J. F. Chen, D. S. Deng, K. L. Yang, Y. Y. Chen, X. M. Fu, Z. G. She, Y. C. Lin, and C. Y. Wang, J. Nat. Prod., 75, 189 (2012).
C. L. Shao, C. Y. Wang, M. Y. Wei, Y. C. Gu, Z. G. She, P. Y. Qian, and Y. C. Lin, Bioorg. Med. Chem. Lett., 21, 690 (2011).
L. L. Sun, C. L. Shao, J. F. Chen, Z. Y. Guo, X. M. Fu, M. Chen, Y. Y. Chen, R. Li, J. Nicole, Z. G. She, Y. C. Lin, and C. Y. Wang, Bioorg. Med. Chem. Lett., 22, 1326 (2012).
H. Ren and W. W. Liu, Arch. Pharm. Res., 34, 901 (2011).
Y. M. Lee, H. Li, J. Hong, H. Y. Cho, K. S. Bae, M. A. Kim, D. K. Kim, and J. H. Jung, Arch. Pharm. Res., 33, 231 (2010).
R. P. Maskey, I. G. Wollny, and H. Laatsch, J. Antibiot., 56, 459 (2003).
K. Sakai, S. Ohte, T. Ohshiro, D. Matsuda, R. Masuma, L. L. Rudel, and H. Tomoda, J. Antibiot., 61, 568 (2008).
J. S. E. Holkers, A. Kagall, J. Mulheirn, and P. M. White, Chem. Comm., 24, 911 (1966).
C. L. Shao, C. Y. Wang, M. Y. Wei, S. D. Li, Z. G. She, Y. C. Gu, and Y. C. Lin, Magn. Reson. Chem., 46, 886 (2008).
W. T. Shier, Y. Lao, T. W. J. Steele, and H. K. Abbas, Bioorg. Chem., 33, 426 (2005).
E. Sakuno, Y. Wen, H. Hatabayashi, H. Arai, C. Aoki, K. Yabe, and H. Nakajima, Appl. Environ. Microbiol., 71, 2999 (2005).
T. Mosmann, J. Immunol. Meth., 65, 55 (1983).
C. G. Pierce, P. Uppuluri, A. R. Teistan, J. F. L. Wormley, E. Mowat, G. Ramage, and J. L. Lopez-ribot, Nat. Protoc., 3, 1494 (2008).
Acknowledgment
This work was supported by the Key Program of National Natural Science Foundation of China (No. 41130858), the National Natural Science Foundation of China (Nos. 81172977; 41176121), the Program for New Century Excellent Talents in University, Ministry of Education of China (No. NCET-11-0472), and the Natural Science Foundation of Shandong Province (ZR2011DQ019; ZR2011HM085).
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in Khimiya Prirodnykh Soedinenii, No. 4, July–August, 2014, pp. 537–539.
Rights and permissions
About this article
Cite this article
Chen, M., Shao, CL., Kong, CJ. et al. A New Anthraquinone Derivative from a Gorgonian-Derived Fungus Aspergillus sp.. Chem Nat Compd 50, 617–620 (2014). https://doi.org/10.1007/s10600-014-1037-2
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10600-014-1037-2