Abstract
Shark fins are among the most highly prized seafood products in the world with massive consumption in Asia over the past several decades. The demand for shark fins is a major driver of the enormous population declines of elasmobranchs that are generally vulnerable to overexploitation. This study aims to better understand the species composition of shark fin products in Thailand and their conservation statuses by using DNA-based species identification. Various types and sizes of shark fins were collected from 4 locations in Thailand. DNA barcoding method based on a fragment of the cytochrome c oxidase subunit I (COI) gene was applied to species identification. Fins from at least 15 shark species were found from Thailand’s markets. The spottail shark (Carcharhinus sorrah) and the night shark (Carcharhinus signatus) were the two dominant species presented in this study. 34% of identifiable samples are the species that have not been record in this region. 62% of species detected from the fin samples are categorized under the threatened categories of IUCN Red List. Species composition reported in shark fin products potentially helps indicate the appropriate conservation action and increases awareness from monitoring the trade in elasmobranch products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shark fins are one of the most valuable fish products and their demand is a major driver for shark fisheries in the world’s oceans (Clarke et al. 2006b; Wong et al. 2009; Fields et al. 2015; Feitosa et al. 2018), where it was estimated that between 26 and 73 million sharks were killed for the global fin trade annually worldwide (Clarke et al. 2006b). Based on the Food and Agriculture Organization (FAO) report, the number of fins in the global trades had continuously experienced substantial declines from the peak of the trade in 2003, where the global exports and imports decreased by 12.6% and 7.7% respectively by 2011 (Dent and Clarke 2015), suggesting the unsustainability of the trades for this shark product and concur to the declines of sharks populations in many parts of the world.
Sharks and other chondrichthyans are now considered to be facing higher extinction risks than most other vertebrate groups with the current rate of overexploitation (Dulvy et al. 2014; Pacoureau et al. 2021), as they are intrinsically susceptible to fishing pressure due to their slow life history characteristics, i.e., slow growth rates, late sexual maturity, low fecundity, long inter-birth interval, long gestation periods (Hoenig and Gruber 1990; Cortés 2000). With large declines of sharks observed throughout the world’s oceans that has grown to be an international concern (Stevens et al. 2000), the fin trade could keep driving the fisheries of elasmobranch species further toward more unsustainable catch levels and even extinction of some species in the near future (Dulvy and Forrest 2010). Moreover, the species composition of sharks in the fin trade could also shift from larger and more valuable species to smaller and more-resilient ones as the slower-growing species are gradually fished out, reflecting the changes in community structures as observed in some heavily fished regions where the shark fin markets continue growing (Lam and Sadovy de Mitcheson 2011; Arunrugstichai et al. 2018).
The shark population in Thailand has undergone a severe decline. According to national fisheries statistics of shark landings in Thailand from last 10 years, a 2,873 metric tons of sharks landed from all fishing gear in commercial fisheries was reported in 2012, and dramatically dropped to 628 metric tons in 2016 (Department of Fisheries 2014, 2018). These revealing figures accounted for a 78% decline in the catch for sharks. After The Royal Ordinance on Fisheries, B.E. 2558 was launched to prevent IUU fishing in 2015, shark landings continuously declined to lower catch levels, with 545 metric tons reported for 2020 (Department of Fisheries 2021). The catch reports could be underestimated since sharks were categorized as bycatch in commercial fisheries, with limited monitoring capacity, while landings are also becoming more difficult to record as traders often hide the catch from the public eye with the growing awareness of shark conservation among Thai citizens. Moreover, shark catch data are not taken from the small-scale artisanal fisheries. In Thailand, shark commodities are utilized in several ways. Shark meat can be freshly consumed or processed to salted fish, sweetened fish, and fish balls in both local and industrial scale processors. Shark liver oil is utilized in the cosmetic industry. Putrid shark and other shark by-products are utilized as raw material for animal feeds. Shark fin is dried and used as raw material for shark fin soup (Department of Fisheries 2020).
In the case of shark fin, Thailand was the world’s second-largest quantity exporter listed from the Food and Agriculture Organization (FAO) (Dent and Clarke 2015). Due to production of shark fin products for the global trade, many shark species in Thailand might have been overexploited to the point of collapse long before monitoring began, as shown in the massive decline of Sphyrna lewini along with large carcharhinid landings within a decade 2005–2015 (Arunrugstichai et al. 2018). Moreover, it was reported that the Thailand market is focused on small low-value shark fins (Dent and Clarke 2015), which are obviously obtained from small sized sharks or immature individuals. Immature sharks might be overexploited to supply the market and their population might suffer from overfishing. Classification of species in each size class of fin product is almost globally absent even though it can be useful information. Details of species in each fin size could potentially reveal the species which undergo overfishing. However, the species of elasmobranchs in the fin trades has never been investigated in Thailand.
Species identification of sharks can be done from external morphology of the first dorsal or pectoral using appropriate identification guidelines (Abercrombie et al. 2013; Marshall and Barone 2016) however, it is hard for less distinctive fin types such as pelvic fin and anal fin. Classification of shark species from shark fin products is more challenging in curly dried fins or it is rather impossible in processed fin which diagnostic characters are chemically damaged during the process to remove the skin. For this reason, DNA-based approaches with advances in nucleotide sequencing, species-specific primers selection, real-time polymerase chain reaction and establishment of global sequence libraries have been developed to identify shark species at lowest taxonomic level (Clarke et al. 2006a; Wong et al. 2009; Fields et al. 2015; Cardenosa et al. 2017, 2018; Abercrombie et al. 2018; Domingues et al. 2021). Moreover, DNA-based approaches are one of the main effective methods to identify CITES-listed species and other threatened species in processed fin products (Fields et al. 2015, 2018; Cardenosa et al. 2018). The mitochondrial cytochrome c oxidase subunit 1 (COI) barcoding approach using BLAST taxonomy databases are a global identification system for animals and provide an ability to identify larger numbers of shark recently (Ward et al. 2005; Fields et al. 2015, 2018; Bhattacharya et al. 2016; Cardenosa et al. 2017, 2020; Abercrombie et al. 2018; Bingpeng et al. 2018). Implementation of two or more markers produce more accuracy in taxonomic study (Domingues et al. 2021) however, it requires greater cost and effort for the molecular method.
The management of shark resources is necessary to have information of shark species and their conservation status on the fin trade to assess questions of sustainable use. Recently, the Department of Fisheries of Thailand (DoF) as the main policy maker for managing sharks stocks in Thailand have completed Thailand National Plan of Action for the Conservation and Management of Sharks (NPOA-Sharks, Thailand) to support International Plan of Action for the Conservation and Management of Sharks (IPOA-Sharks) (Department of Fisheries 2020) and the establishment of DNA barcode reference libraries and taxonomy study are also included as species identification of elasmobranch product, as the basic information to make a proper conservation and management plan for shark and other relatives. Nonetheless, there are limited data to assess species of shark fin trade in Thailand which is an obstacle to develop appropriate management strategy. The main aim of this study is to identify species from shark fin products in Thailand with respect to their conservation status as listed on the International Union for Conservation of Nature Red List (IUCN Red List), CITES to understand species composition and their conservation status in Thailand’s market. Sizes of shark fin samples are also classified to investigate whether size-species specification remains in samples. This study is the first DNA-based species identification of shark fin products in Thailand, which could provide important conservation messages to consumers, in addition to valuable information to help policymakers establish a sustainable management plan, launch appropriate law enforcement, and strengthen trade monitoring efforts for this keystone species.
Method
Sample collection
We collected samples from available sources in the supply chain of shark fin products in partnership with the Department of Fisheries of Thailand (DoF) from 4 sampling locations; Bangkok and vicinity, Chonburi, Songkhla and Pattani. Fins samples were randomly collected from retail markets, restaurants, warehouses, seaports, and fishing ports. Fin samples were classified into 4 types including (1) frozen fin: unprocessed fins kept on freezers or cold storage; (2) dried fin: unprocessed fin that contain both skin and cartilage; (3) dried processed fin: fins that have been chemically treated to remove their skin, turn to yellow or golden color and keep in dried condition; and (4) wet processed fins: ready-to-cook fin that their texture is pliable after chemically treated. Details of numbers and sample type in each collecting place cannot be disclosed due to owner agreements. Number of samples is reported in terms of percentage in each location and percentage of each fin type in total. Length of fins was measured from the fin baseline to the tip of the fin in a perpendicular line (Marshall and Barone 2016). Size of fin was categorized as small (S: <10 cm), medium (M: 10–15 cm), large (L: 16–30 cm) and extra-large (XL: >30 cm). Each fin tissue sample was clipped and put in individual microcentrifuge tubes for DNA extraction.
DNA based species identification
A sliced part of the tissues approximately 1 cm2 were overnight incubated in 10% SDS (sodium dodecyl sulfate) lysis buffer and Proteinase K. Tissues in the lysis buffer were homogenized. Genomic DNA was extracted by phenol-chloroform extraction (Sambrook et al. 1989). A cytochrome c oxidase I (COI) amplicon was amplified using 2 processes to obtain high yield product PCR (a clear band of PCR product were checked through agarose gel electrophoresis) for DNA sequencing. Initially, a multiplex PCR assay modified from Cardenosa et al. (2017) was performed with minor change of primers volume and some chemical reagents. Briefly, each 25 µl of PCR master mix consist of 0.5 µl of extracted DNA, 12.5 µl of GoTaq® Master Mixes (Promega), 1.5 µl of VF2-tl primer (5’-TGTAAAACGACGGCCAGTCAACCAACCACAAAGACATTG GCAC-3’), 0.8 µl of FishR1_tl and FishR2_tl primer (5’-CAGGAAACAGCTATGACACTTCAGGGTGACCGAAGAATCAGAA-3’ and 5’-CAGGAAACAG CTATGACACCTCAGGGTGTCCGAARAAYCARAA-3’), 0.8 µl of shark150R primer (5’-AAGATTACAAA AGCGTGG GC-3’) and 0.2 µl of Shark474F primer (5’-CHATTTCCCAATATCAAACACC-3’). The thermal cycle consisted of a cycle of 2 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 52 °C, and 1 min at 72 °C, followed by a final extension step of 72 °C for 10 min. Multiplex PCRs were visualized using Midori green Advanced DNA stain (Nippon Genetic Europe GmbH, Germany) and checked on a 3% agarose gel to verify successful amplification. PCR product was purified and sequenced by Macrogen, Inc. (South Korea) using the M13 forward primer and the M13 reverse primer as describe by Cardenosa et al. (2017). A multiplex PCR assay yield 650, 200 or 150 bp (Cardenosa et al. 2017) depending on quality of DNA. If the first protocol did not yield a good quality of PCR product, another protocol described by Fields et al. (2015) were performed subsequently and yield an amplicon of 130 bp (Fields et al. 2015). A pair of primers (FishF2_t1 5’-TGTAAAACGACGGCCAGTCGACTAATC ATAAAGATATCGGCAC-3’ and Shark COI-MINIR 5’-AAGATTACAA AAGCGTGGGC-3’) was tested against a wide range of annealing temperatures (50°–60 °C) to find appropriate annealing temperatures for PCR assay. Each 25 µl of PCR master mix consist of 0.5 µl of extracted DNA, 12.5 µl of GoTaq® Master Mixes (Promega), 1 µl of FishF2_t1 and Shark COI-MINIR primer. The thermal cycle consisted of a cycle of 2 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 52 °C, and 1 min at 72 °C, followed by a final extension step of 72 °C for 10 min. PCR products were visualized as above and checked on a 2% agarose gel. PCR product was purified and sequenced by Macrogen, Inc. (South Korea). Nucleotide sequences were checked and trimmed using MEGA6 (Tamura et al. 2013) and BioEdit Software (Hall 2011). All sequences were entered in Barcode of Life Data System (BOLD: https://v3.boldsystems.org/index.php/databases) using BOLD database and the Basic Local Alignment Search Tool (BLAST: https://blast.ncbi.nlm.nih.gov/Blast.cgi) using Genbank database (Altschul et al. 1997) to identify them to the lowest taxonomic level. Species-level identification was made for sequences with 100% similarity to both BLAST and BOLD database. Generic name was assigned when sequence was identified to the same species with a 100% match in a database and ≥ 99% in another database. Sequences that possess lower 99% similarity to one of two databases were eliminated from analysis. The conservation status of DNA sequences where it were possible to identify species were obtained from IUCN Red List of Threatened Species website (https://www.iucnredlist.org/) and define to Thailand Red Data (Krajangdara et al. 2019; Department of Fisheries 2020). Species listed in Convention on International Trade in Endangered Species (CITES) Appendix were checked.
Result
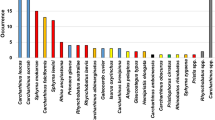
A total of 206 fin samples were collected from four locations. The majority samples were collected from Bangkok and vicinity (52%) followed by Songkhla (39%), Chonburi (6%), and Pattani (3%). Majority of the fins collected were small size (50.0%) followed by large size (32.0%) (Table 1). We successfully amplified DNA from 166 to 206 fins. Four types of fins categorized as frozen fin, dry fin, processed dry fin, and processed wet fin were collected at the sampling sites and the percentage of success DNA-amplification of each fin type was 100%, 86%, 64%, and 50%, respectively. 10% of amplified samples was cut off after BOLD and BLAST searches. Another part of amplified samples (14%) was reported for generic names. Total amplified sample which 100% match in BOLD and BLAST searches was 75%. Species identification of shark fin product through DNA-based techniques confirms the presence of at least 15 shark species (Table 2). Two species, Loxodon macrorhinus and Triaenodon obesus have 99% similarity in one of databases, however; species name was reported instead of only the generic name since there is a species in these two genera. The contribution to the total identified samples from each shark family are as follow, Carcharhinidae (85%), Triakidae (7%), Sphyrnidae (5%), and other (3%) (Fig. 1). Spottail shark (Carcharhinus sorrah) and the night shark (Carcharhinus signatus) are the two dominant species presented in this study (Fig. 1). Most of species can be found in Thai waters, while 34% of identifiable species including Carcharhinus leiodon, C. signatus, Mustelus canis, and Mustelus schmitti are not known to occur in this region (Table 2). A large portion of the species detected from the fin samples are categorized as Near Threatened and Endangered in IUCN Red List (Table 2). Most of the identifiable species, 62%, were assessed under the threatened categories of IUCN Red List (Table 2). The conservation status of the highest dominant species, C. sorrah is Near Threatened under IUCN Red List but record as Vulnerable in Thailand Red Data. The two species of the hammerheads (Sphyrna lewini and Sphyrna mokarran) identified in this study were listed as Critically Endangered in both IUCN red list and Thailand Red Data and are globally regulated by CITES and listed in Appendix II. Most of the samples from Bangkok and vicinity consist of Near Threatened (38%) and Vulnerable species (38%) listed in IUCN Red List followed by Critically Endangered (16%) (Fig. 2). Shark fin products from Chonburi are categorized as Endangered (67%) and Near Threatened species (33%). Large proportions of fins from Songkhla are from Endangered (52%) and Near Threatened species (36%). All samples from Pattani are from Near Threatened species. Two species of Carcharhinus spp. (C. signatus and C. sorrah) are found in all sizes of fin and the other two species (Table 2). Seven species have been found only in small fin including Carcharhinus limbatus, Carcharhinus obscurus, Carcharhinus plumbeus, L. macrorhinus, Rhizoprionodon acutus, M. canis, and Chiloscyllium punctatum, Species which are mostly detected in large fin size are C. amblyrhynchoides, C. signatus and S. mokarran. Tree species, C. amboinensis, Carcharhinus brevipinna and C. leiodon are mostly identified from extra-large fin size. All sequences from processed wet fin sample were unable to match to any species with high possibility. Dry fin and processed dry fin are two main fin types that can be identify to species level. Some species found in only a fin type; M. schmitti was found from frozen fin, C. amblyrhynchoides, C. limbatus, C. signatus, L. macrorhinus, S. mokarran, and C. punctatum were found from dry fin; C. leiodon, C. plumbeus, R. acutus and M. canis were found from processed dry fin. Four species which have not been recorded in Thailand were found from all three fin types.
Discussion
This is the first study to identify the species contributing to the shark fin trade in Thailand. The number of assessed shark fins is quite low compared to the number of fins being traded because market accesses were limited; however, we identified 15 shark species of which two are CITES regulated. A large portion of the samples were sharks that cannot be found in Thai waters. The native distribution of C. signatus and M. canis, M. schmitti are in the Atlantic Ocean. The Smooth tooth blacktip shark, C. leiodon is record from Western Indian Ocean (www.fishbase.org). This finding is concordant with the trade records obtained from The Customs Department in Thailand, where shark fin products (HS-03039200 and HS-03057100) are imported from many countries such as Argentina, Norway, United States, Indonesia, Singapore, China and Taiwan (The Customs Department 2021). The geographical origin of shark fins in Thailand’s market is hard to trace due to the obscure nature of the trade of this lucrative and occasionally illegal product, where re-export is also common in many countries before being imported into Thailand and commodity coding revisions from The Customs Department (Dent and Clarke 2015). A large proportion of shark species recorded in Thailand for this study could not be determined that they were captured in Thai waters since some species are widespread (Dulvy et al. 2014). Moreover, the large percentage of shark species that occur outside of Thai waters provides evidence that the fins traded in Thai markets rely heavily on sources from outside the country, which maybe imported to satisfy the local demand or re-export as a trading hub. Preserved method of fin didn’t reveal specific international transport of identifiable species that didn’t recorded in Thailand since those species can be found from all fin types. Lack of traceability shark fin trade and the interlink of source and sink for globally threatened species are still a barrier to understand the trade route of shark fin in Thailand. The traceability system should be established both domestically and internationally.
Over half of species reported in this study are listed under threatened categories listed in IUCN Red List and Thailand Red data. There are 6 Critically Endangered species, 16 Endangered species and 22 Vulnerable species of sharks that can be found in Thai water listed in IUCN Red List (Krajangdara 2021). Two Critically Endangered species, 2 Endangered species and 6 Vulnerable species from the IUCN Red List are presented in this study. The record of these 10 species reveals utilization of threatened species of Thailand in fin trade. The highest proportion of C. sorrah infer to the large volume of this species being exploited to support Thailand’s small low-value shark fins market. This species is one of the dominant shark species in Thai fisheries (Krajangdara 2021) and is frequently captured in Southeast Asia (Fahmi and Sumadhiharga 2007; Moore et al. 2012) and Southwest coast of India (Akhilesh et al. 2011). The C. sorrah is classified as Vulnerable in Thailand Red data, but their status is Near Threatened listed in IUCN Red List (Krajangdara et al. 2019). The recent status on IUCN Red List of this species is based on the biological data of Australian samples (Giles et al. 2014). Its status listed in IUCN Red List should be revised due to the prevalence of the species in the marketplace. Moreover, the stock of C. sorrah should be assessed for each regions for fisheries management since there is genetic variation in C. sorrah populations across Indo–West Pacific region (Giles et al. 2014) and its landings were dominated by immature size in Southeast Asia (Fahmi and Sumadhiharga 2007; Moore et al. 2012). Two species listed in CITES appendix were found among the samples and many Critically Endangered species from IUCN Red List were detected, highlighting the utilization of these species in the global shark fin trade, while there is currently no law to prohibit capturing them in Thailand. Monitoring of these species should be prioritized to understand their conservation status and sources of fin trade.
Threatened species are recorded in all locations except Pattani. Various proportion of all threatened categories found in samples collected from Bangkok and vicinity reflect that it is the center of Thailand’s shark fin market which obtains product from many sources, while the markets outside of Bangkok probably obtain shark fin products from the local sources or one of dealers in Bangkok and vicinity. Moreover, it was noted that our surveys around out of town markets were done during COVID-19 pandemic when most of the shops were closed due to low number of travelers. The types of sample sources are limited, which might fail to detect some other species present in fins trade in Thai market, however the record of threatened species from each location presents useful information for further investigation.
Many identifiable species that are classified as Critically Endangered, Endangered, and Vulnerable were found in small size class. The fins commonly marketed and exported from Thailand are small low-value fins (Dent and Clarke 2015) which results in being the majority size class of fin samples collected in this study. It was difficult to determine maturity of sharks from shark fins alone since there is limited information on the relationships between fin length and size at maturity of different shark species. However, we considered that all small size samples, except smaller sharks Chiloscyllium spp. and Squalus sp., are immature because it was estimated that the most commonly traded fins i.e. dorsal, pectoral or caudal fin of many mature shark species such as C. brevipinna, C. plumbeus, C. limbatus, C. sorrah, C. amboinensis etc. are larger than 10 cm (Al-Qasmi 1994; Oktaviyani et al. 2020). We could not accurately determine which fin type our sample consisted of, however, the small size of fin samples in this study are considerably smaller than all fin types of mature sharks in general (Al-Qasmi 1994; Oktaviyani et al. 2020). Harvesting immature sharks can affect the recruitment process of shark population, and hinder the recovery potential of the stock from exploitation (Fahmi and Sumadhiharga 2007). This finding should raise awareness to shark management policymakers and consumers. Study of relationships between fin length and mature size of shark will indicate the minimum size of shark fins that should be allowed in the trade for sustainable fishery in the case that the fisheries were targeted for sharks. However, given the critical status of shark species globally, the consumed shark fins could be globally threatened and illegal. Traceability of shark fin trade should be mandated.
The quality of DNA template directly affects the success rate of amplification in PCR. In case of shark fin product in this study, the range of DNA quality from excellent to bad condition that yield from different types of fins can be found. Frozen fins which are generally well-preserved in cold storage and dried fins which are chemically untreated provide higher success rate of amplification. Processed dried fins and processed wet fins should be avoided from further sample collection for this kind of study since it was difficult to amplify. Effective approach modified from Cardenosa et al. (2017) and Fields et al. (2015) were applied to utilize when various quality of DNA template was an obstacle.
This study emphasizes conservation concerns, particularly for the large volume of elasmobranchs categorized as Critically Endangered assessed by the IUCN Shark Specialist Group. In addition, the majority Vulnerable and Endangered species identified from shark fin product indicated that we need more law enforcement and trade monitoring for those threatened species before their populations decline even further. Species identification using external morphological examination can be a first step to identify CITES-listed species or regulated species (Marshall and Barone 2016), however, missing key diagnostic characteristics of processed fin or even in dried fins remains problematic (Ferrette et al. 2019; Villate-Moreno et al. 2021), therefore we recommend DNA-based identification methods to detect threatened species and CITES-listed species in shark fin trade. A standard molecular diagnostic technique is another step to enhance species identification accuracy (Van Houtan et al. 2020; Villate‐Moreno et al. 2021). This approach is also more convenient for the trade monitoring staffs since it does not require extensive training in taxonomy, which is especially in shortage in Thailand and DNA based approach can be performed by outsources. DoF should be empowered and equipped with all necessary resources to be able to fulfil CITES obligation. This should be prioritized in Thailand’s NPOA-Sharks. A random molecular test of subsamples of shark products in regular period will provide invaluable information for future evaluations for the threatened and CITES listed species, aid in better management for shark resources in this region, and also help in enforcement on the trades of illegal shark species within Thailand as one of the hub of fins trade in the world.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Abercrombie DL, Cardeñosa D, Chapman DD (2018) Genetic approaches for identifying Shark Fins and Other Products: a Tool for International Trade monitoring and enforcement. Abercrombie & Fish, Marine Biological Consulting, Suffolk County, NY, p 13
Abercrombie DL, Chapman DD, Gulak SJB, Carlson JK (2013) Visual Identification of Fins from Common Elasmobranchs in the Northwest Atlantic Ocean., NMFS-SEFSC-643, pp 51
Akhilesh KV, Ganga U, Pillai NGK, Vivekanandan E, Bineesh KK, Shanis CPR, Hashim M (2011) Deep-sea fishing for chondrichthyan resources and sustainability concerns—a case study from southwest coast of India. Indian J Geo-Mar Sci 40:347–355
Al-Qasmi AM (1994) Physico-Chemical characterization of Shark-Fins. University of Rhode Island
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Arunrugstichai S, True JD, White WT (2018) Catch composition and aspects of the biology of sharks caught by thai commercial fisheries in the Andaman Sea. J Fish Biol 92:1487–1504. https://doi.org/10.1111/jfb.13605
Bhattacharya M, Sharma AR, Patra BC, Sharma G, Seo EM, Nam JS, Chakraborty C, Lee SS (2016) DNA barcoding to fishes: current status and future directions. Mitochondrial DNA Part A 27:2744–2752. https://doi.org/10.3109/19401736.2015.1046175
Bingpeng X, Heshan L, Zhilan Z, Chunguang W, Yanguo W, Jianjun W (2018) DNA barcoding for identification of fish species in the Taiwan Strait. PLoS ONE 13:e0198109. https://doi.org/10.1371/journal.pone.0198109
Cardenosa D, Fields A, Abercrombie D, Feldheim K, Shea SKH, Chapman DD (2017) A multiplex PCR mini-barcode assay to identify processed shark products in the global trade. PLoS ONE 12:e0185368. https://doi.org/10.1371/journal.pone.0185368
Cardenosa D, Fields AT, Babcock EA, Shea SKH, Feldheim KA, Chapman DD (2020) Species composition of the largest shark fin retail-market in mainland China. Sci Rep 10:12914. https://doi.org/10.1038/s41598-020-69555-1
Cardenosa D, Quinlan J, Shea KH, Chapman DD (2018) Multiplex real-time PCR assay to detect illegal trade of CITES-listed shark species. Sci Rep 8:16313. https://doi.org/10.1038/s41598-018-34663-6
Clarke SC, Magnussen JE, Abercrombie DL, McAllister MK, Shivji MS (2006a) Identification of shark species composition and proportion in the Hong Kong shark fin market based on molecular genetics and trade records. Conserv Biol 20:201–211. https://doi.org/10.1111/j.1523-1739.2005.00247.x
Clarke SC, McAllister MK, Milner-Gulland EJ, Kirkwood GP, Michielsens CG, Agnew DJ, Pikitch EK, Nakano H, Shivji MS (2006b) Global estimates of shark catches using trade records from commercial markets. Ecol Lett 9:1115–1126. https://doi.org/10.1111/j.1461-0248.2006.00968.x
Cortés E (2000) Life history patterns and correlations in Sharks. Rev Fish Sci 8:299–344. https://doi.org/10.1080/10641260008951115
Dent F, Clarke S (2015) State of the global market for shark products. FAO, Rome, p 187
Department of Fisheries (2014) The marine fisheries statistics 2012 base on the sample survey. Fishery Statistics Analysis and Research Group. Fisheries Development Policy and Strategy Division, p 1654
Department of Fisheries (2018) Fisheries statistics of thailand 2016. Fisheries development policy and strategy division, department of fisheries, p 165
Department of Fisheries (2020) Thailand National Plan of Action for the Conservation and Management of Sharks (NPOA-Sharks, Thailand: Plan 1, 2020–2024). Fisheries, Ministry of Agriculture and Cooperatives, Bangkok, Thailand, p 62
Department of Fisheries (2021) Marine capture production of commercial fishery 2020. Department of Fisheries, Ministry of Agriculture and Cooperatives, Fishery Statistics Analysis and Research Group. Fisheries Development Policy and Strategy Division, p 187
Domingues RR, Bunholi IV, Pinhal D, Antunes A, Mendonça FF (2021) From molecule to conservation: DNA-based methods to overcome frontiers in the shark and ray fin trade. Conserv Genet Resour 13:231–247. https://doi.org/10.1007/s12686-021-01194-8
Dulvy NK, Forrest RE (2010) Life histories, population dynamics, and extinction risks in chondrichthyans. In: Carrier JC, Musick JA, Heithaus MR (eds) Sharks and their relatives II: biodiversity, adaptive physiology, and conservation. CRC Press, Boca Raton, pp 635–676
Dulvy NK, Fowler SL, Musick JA, Cavanagh RD, Kyne PM, Harrison LR, Carlson JK, Davidson LN, Fordham SV, Francis MP et al (2014) Extinction risk and conservation of the world’s sharks and rays. Elife 3:e00590. https://doi.org/10.7554/eLife.00590
Fahmi SK (2007) Size, sex and length at maturity of four common sharks caught from western indonesia. Mar Res Indonesia 32:7–19
Feitosa LM, Martins APB, Giarrizzo T, Macedo W, Monteiro IL, Gemaque R, Nunes JLS, Gomes F, Schneider H, Sampaio I et al (2018) DNA-based identification reveals illegal trade of threatened shark species in a global elasmobranch conservation hotspot. Sci Rep 8:3347. https://doi.org/10.1038/s41598-018-21683-5
Ferrette B, Domingues RR, Rotundo MM, Miranda MP, Bunholi IV, De Biasi JB, Oliveira C, Foresti F, Mendonca FF (2019) DNA barcode reveals the Bycatch of Endangered Batoids Species in the Southwest Atlantic: implications for sustainable Fisheries Management and Conservation efforts. Genes (Basel) 10:1–15. https://doi.org/10.3390/genes10040304
Fields AT, Abercrombie DL, Eng R, Feldheim K, Chapman DD (2015) A novel mini-DNA barcoding assay to identify processed fins from internationally protected shark species. PLoS ONE 10:e0114844. https://doi.org/10.1371/journal.pone.0114844
Fields AT, Fischer GA, Shea SKH, Zhang H, Abercrombie DL, Feldheim KA, Babcock EA, Chapman DD (2018) Species composition of the international shark fin trade assessed through a retail-market survey in Hong Kong. Conserv Biol 32:376–389. https://doi.org/10.1111/cobi.13043
Giles JL, Ovenden JR, Dharmadi, AlMojil D, Garvilles E, Khampetch KO, Manjebrayakath H, Riginos C (2014) Extensive genetic population structure in the Indo-West Pacific spot-tail shark, Carcharhinus sorrah. Bull Mar Sci 90:427–454. https://doi.org/10.5343/bms.2013.1009
Hall T (2011) Bioedit: an important software for molecular biology. GERF Bull Biosci 2:60–66
Hoenig JM, Gruber SH (1990) Life-history patterns in the elasmobranchs: implications for fisheries management. Department of Commerce, Washington, DC, p 16
Krajangdara T (2021) Species diversity and conservation status of shark in Thai Water. Fish Gaz 4:101–111 (in Thai)
Krajangdara T, Ali A, Vidthayanon C, Rodpradit S, Chansue N (2019) Guidebook to cartilaginous fishes of Thailand and adjacent water. Phuket Offset Printing, Phuket, Thailand, p 146. (in Thai)
Lam L, Sadovy de Mitcheson Y (2011) The sharks of South East Asia–unknown, unmonitored and unmanaged. Fish Fish 12:51–74. https://doi.org/10.1111/j.1467-2979.2010.00383.x
Marshall LJ, Barone M (2016) Shark Fin Guide: identifying sharks for their fins. food and agriculture organization of the nited nations, Rome, Italy, pp 144
Moore AB, McCarthy ID, Carvalho GR, Peirce R (2012) Species, sex, size and male maturity composition of previously unreported elasmobranch landings in Kuwait, Qatar and Abu Dhabi Emirate. J Fish Biol 80:1619–1642. https://doi.org/10.1111/j.1095-8649.2011.03210.x
Oktaviyani S, Kurniawan W, Fahmi F (2020) Fin Length and Total Length Relationships of Silky Shark Carcharhinus falciformis Landed at Tanjung Luar Fish Landing Port, West Nusa Tenggara, Indonesia. E3S Web of Conferences 147:02011. https://doi.org/10.1051/e3sconf/202014702011
Pacoureau N, Rigby CL, Kyne PM, Sherley RB, Winker H, Carlson JK, Fordham SV, Barreto R, Fernando D, Francis MP et al (2021) Half a century of global decline in oceanic sharks and rays. Nature 589:567–571. https://doi.org/10.1038/s41586-020-03173-9
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2 edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., p 2222
Stevens JD, Bonfil R, Dulvy NK, Walker PA (2000) The effects of fishing on sharks, rays, and chimaeras (chondrichthyans), and the implications for marine ecosystems. ICES J Mar Sci 57:476–494
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
The Customs Department (2021) https://www.customs.go.th/. Accessed 16, June 2021
Van Houtan KS, Gagne TO, Reygondeau G, Tanaka KR, Palumbi SR, Jorgensen SJ (2020) Coastal sharks supply the global shark fin trade. Biol Lett 16:20200609. https://doi.org/10.1098/rsbl.2020.0609
Villate-Moreno M, Pollerspöck J, Kremer‐Obrock F, Straube N (2021) Molecular analyses of confiscated shark fins reveal shortcomings of CITES implementations in Germany. Conserv Sci Pract. https://doi.org/10.1111/csp2.398
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B Biol Sci 360:1847–1857. https://doi.org/10.1098/rstb.2005.1716
Wong EH, Shivji MS, Hanner RH (2009) Identifying sharks with DNA barcodes: assessing the utility of a nucleotide diagnostic approach. Mol Ecol Resour 9:243–256. https://doi.org/10.1111/j.1755-0998.2009.02653.x
Acknowledgements
The authors would like to thank Department of Fisheries, Thailand for dedicating their time and facilitating collection of samples for the study. We would also like to thank Sutthirat Panchakhan and Nilubon Pirin for their assistance and hard work in collecting samples from various sites. This study was funded and supported by WildAid, a U.S. based non-profit organization with a mission to reduce demand for wildlife products.
Funding
This work was funded by WildAid.
Author information
Authors and Affiliations
Contributions
WK and SA contributed to the study conception and design. WK was responsible for samples collection, conducted the molecular work and analyses, designed all figures and tables, and wrote the first draft of the manuscript. All authors commented on and edited subsequent versions of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that have no competing or any conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Klangnurak, W., Arunrugstichai, S., Manopawitr, P. et al. DNA-based species identification of shark fins traded in thai markets. Conserv Genet 24, 537–546 (2023). https://doi.org/10.1007/s10592-023-01519-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01519-0