Abstract
Technological advancements have allowed geneticists to exploit an increasing array of molecular markers, many of which have different properties and may provide contrasting insights into the evolutionary history and structure of populations. This has important consequences for conservation managers attempting to identify units at which to conserve intraspecific diversity. In this study we compared the inferences derived from nuclear microsatellites and restriction-site associated DNA (RADseq) data for a threatened freshwater fish, the bull trout Salvelinus confluentus. For both marker types we generated data for the same suite of individuals collected from 24 populations distributed across the species range. The RADseq data were low coverage (mean site coverage < 3X), so we implemented a probabilistic genotyping approach. We performed a comparable suite of analyses for both datasets. Both datasets revealed similar broad patterns of subdivision that reflected primary evolutionary lineages (Coastal and Interior clades). However, the RADseq more clearly and consistently identified the hierarchical phylogenetic structure. Some populations had varying assignments to these lineages depending on the dataset. RADseq data also suggested admixture has shaped the genomic character of several populations. Such a signal was not apparent with the microsatellites, suggesting that the datasets are revealing different aspects of population history. Our study provides a valuable case study in how advances in molecular technology can enhance our understanding of a relatively well-studied species. It also underscores the importance of framing findings generated with high-throughput sequencing technology within the context of past research to enhance conservation decision making.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Characterizing patterns of intraspecific diversity and population history is one of the fundamental goals of population and conservation genetics. Within a given species, a multitude of evolutionary events and processes can generate complex patterns of variation and differentiation. Many species display hierarchical structure in which populations are nested within metapopulations and broader phylogenetic lineages (Excoffier et al. 1992; Unger et al. 2013; Pisa et al. 2015). Rarely are the relationships between these hierarchies simple, due to events such as secondary mixing between lineages, isolation by distance, and asymmetrical colonization (Excoffier et al. 2009; Martin et al. 2015; Gompert and Buerkle 2016). Assessing the distinctiveness of a population or lineage can be subjective (Ramey II et al. 2007) and the patterns of diversity we observe sometimes deviate from preconceived definitions of units we wish to conserve (McDevitt et al. 2009; Jensen et al. 2013; Wayne and Shaffer 2016; Groves et al. 2017).
An under-appreciated issue that complicates assessing intraspecific diversity is that we as a research community discern evolutionary relationships using imperfect systems of measurement. Although a true population history exists for a species, we are restricted to interpreting relationships using data, theory, models, and analytical techniques that may incompletely represent its history (Waples and Gaggiotti 2006). Until recently much of the knowledge in biodiversity genetics was built using a handful of loci for any given marker type, such as nuclear microsatellites, amplified fragment length polymorphism (AFLPs), restriction fragment length polymorphism (RFLPs), and mitochondrial genes. Although these data sources have been workhorses for molecular ecology and conservation genetics (Sunnucks 2000; DeYoung and Honeycutt 2008; Hodel et al. 2016), they have limitations (Putman and Carbone 2014). Now the proliferation of high-throughput sequencing technologies has made it possible to characterize significant portions of the genome for even non-model organisms. Genomic-level data will add to our knowledge of population structure, but may conflict with past findings and challenge existing notions of population relationships (Kohn et al. 2006; Twyford and Ennos 2012; Piccolo 2016). This is particularly important from a natural resource management perspective because decisions based on findings generated with traditional markers may require revisiting in light of new genomic data.
Within this context, we present a case study comparing the inferences of genetic structure derived from two different types of markers generated from the same dataset. Our target species was the bull trout Salvelinus confluentus, a freshwater salmonid native to the Pacific Northwest of the United States. Bull trout provide an interesting case study because there has been a large body of genetic and ecological research describing population relationships. This freshwater salmonid exhibits a variety of life history strategies including both resident fish that spend their entire life in small headwater streams and migratory fish that may travel over 100 km, even through saltwater, to feeding and maturing sites between spawning events (Northcote 1997; Rieman and Dunham 2000; Mogen and Kaeding 2005). However, two critical requirements of the species are access to cold-water spawning habitat and intact migration corridors (Rieman and McIntyre 1993; McPhail and Baxter 1996). Combined with strong fidelity to natal spawning location, this creates a patchwork of genetically discrete populations across the species’ range restricted to watersheds with suitable habitat. Previous genetic work involving nuclear microsatellite markers has emphasized this pattern (Spruell et al. 2003; Ardren et al. 2011; DeHaan et al. 2011). Additionally, nuclear and mitochondrial sequence markers suggested populations can be further aggregated into broad phylogenetic groups (Taylor et al. 1999; Spruell et al. 2003; Ardren et al. 2011). The main evolutionary division exists between populations west of the Cascade Mountain Crest (Coastal lineage) and those found east of the Cascade Mountain Crest in the interior Columbia River Basin (Interior lineage).
Even though the bull trout has previously been characterized with genetics, there are lingering evolutionary questions for specific populations and the species overall. For example, the Deschutes River basin in Central Oregon is east of the Cascade Mountain Crest (geographically consistent with the Interior lineage) but bull trout in this system cluster with other Coastal populations using microsatellites (Ardren et al. 2011). Further, bull trout in the Klamath River basin in southern Oregon cluster with those in the Willamette River in northern Oregon even though the distance between these basins’ respective entrance into the Pacific Ocean is several hundred kilometers. Most perplexing is the bull trout population in the St. Mary River of northern Montana: it is the only population in the contiguous US east of the Continental Divide yet with microsatellite markers it clusters with the Coastal lineage instead of Interior populations located in adjacent watersheds (Spruell et al. 2003; Ardren et al. 2011). There are other broad questions, such as the level of similarity among Coastal populations, despite being separated by saltwater, and the assignment of populations to lineages within the broader Interior group.
These questions are relevant in part because bull trout are listed as a threatened species under the Endangered Species Act (ESA) across their range in the coterminous United States. Currently the species is listed as a single entity with six defined recovery units (U.S. Fish and Wildlife Service 2015). All populations representing the Coastal lineage were combined into a single recovery unit (except for the Klamath, which was given its own recovery designation). Interior lineage populations were divided into three recovery units and the St. Mary was classified as a sixth distinct recovery unit. Given some of the uncertainties described above, additional information to help clarify the delineation of recovery units may be warranted.
Genetic data, such as single-gene regions of the mitochondrial genome and a suite of nuclear microsatellite markers, were used in part to designate recovery units. However, in totality these markers covered a limited portion of the bull trout genome, potentially obscuring complex evolutionary patterns (Putman and Carbone 2014). Therefore, we generated a restriction site-associated DNA sequencing (RADseq) dataset for 24 bull trout populations from across the species range in the coterminous United States. We then compared these data to a 16 locus microsatellite dataset generated for the same exact individuals. The anticipation was that both datasets would highlight the same broad phylogenetic patterns (e.g. coastal vs. interior) and the RADseq data would provide enhanced clarity for previously uncertain evolutionary relationships (e.g. Deschutes and Coastal lineage, Klamath and Willamette; St. Mary and Coastal lineage). Our study presents a valuable opportunity to evaluate the implications of new genomic sequencing technologies for characterizing intraspecific diversity and evolutionary patterns.
Materials and methods
RADseq library preparation
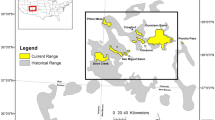
Our laboratories (Washington Department of Fish and Wildlife Molecular Genetics Laboratory [WDFW] and US Fish and Wildlife Service [USFWS] Abernathy Fish Technology Center) have repositories of bull trout samples collected as part of various research and management projects. Many of these samples were included in Ardren et al. (2011). The initial ascertainment library contained 380 individuals from 24 bull trout populations (Table 1; Fig. 1). We selected populations that provided sufficient geographic coverage and represented the distribution of previously known phylogenetic groups. Our dataset included eight populations west of the Cascades Mountains (including the Klamath River) and 16 populations east of the Cascades (including the St. Mary River). For much of the reporting we will reference populations according to relevant geographic groupings (Fig. 1). All samples were extracted for genomic DNA using Qiagen DNEasy ® kits (Qiagen Inc., Valencia, CA).
Map of the study area and the bull trout populations included in the RADseq libraries. Tributaries from which bull trout were collected are indicated by points, which are color-coded according to major drainage basins. The Columbia River Basin is shaded gray and the Klamath River Basin in purple. The solid red line indicates the Continental Divide and the solid blue line the highlights the Cascade Crest. Black lines indicate state and national boundaries
Restriction-site associated DNA (RAD) sequences were used (RADseq, Miller et al. 2007; Baird et al. 2008) to discover and genotype SNPs. DNA was quantitated using Quant-It™ BR assay kit (Life Technologies, Carlsbad, CA) and a QuantiFluor® ds DNA system (Promega, Madison, WI) to normalize DNA from all individuals at 1 µg/40 µL. Quantitated genomic DNA was digested using the enzyme Sbf I-HF® (New England Biolabs, Ipswich, MA) at 50 µL reaction volumes (400 U/mL SbfI-HF®, 1X Cutsmart™ buffer). Digests were conducted at 37 °C for 3 h followed by 65 °C for 20 min. The P1 adapters (Integrated DNA Technologies, San Diego, CA), which included a DNA barcode specific to each individual (96 unique barcodes in total), were ligated to digested DNA in 60 µL reaction volumes (8.3 nM P1 adapters, 0.17X NEBuffer 2 [New England Biolabs], 1 nM rATP [Promega], 16,666.7 U/mL T4 DNA Ligase [New England Bioloabs]). The reaction was incubated at room temperature for 1 h followed by 65 °C for 20 m, after which DNA from 95 individuals was pooled into a single reaction. A negative control was included in each library. Pooled DNA was sheared using a Bioruptor ® (Diagenode, Denville, NJ) for four to nine cycles of 30 s of shearing and 59 s resting, depending on DNA quality. Sheared DNA was purified and size selected using Agencourt® AMPure® XP PCR purification kits (Beckman Coulter Inc., Brea, CA), following manufacturers’ protocol. Genomic libraries were prepared, including the ligation of the P2 adapter (primer for the complimentary DNA strand), using the KAPA LTP Library Preparation Kit for Illumina® platforms (KAPA Biosystems, Cape Town, SA) following manufacturers’ protocol with the optional final PCR amplification step, annealing at 68 °C. Library DNA concentrations were evaluated using qPCR with the KAPA Library Quantification Kit for Illumina® platforms and an Applied Biosystems™ 7900 real-time PCR system (Life Technologies) following manufacturers’ protocol. Libraries were normalized to 10 nM and sent to University of Oregon’s Genomics and Cell Characterization Core Facility (UOGCF), where they were sequenced paired-end on an Illumina® HiSeq 2500 sequencer.
After the first round of sequencing the data were processed using the process_radtags module of Stacks 1.46 (Catchen et al. 2013) to evaluate average read count per individual. To increase total yield per individual and limit disparities in coverage, individual libraries were normalized again at the P1 ligation step based on read count; DNA was reduced for individuals with high read count and increased for those with low counts. RAD sequencing was then repeated. Library preparation proceeded as described above and the new libraries were submitted to the UOGCF for the second round of sequencing.
Bioinformatics
Amplification can introduce PCR clones into RADseq libraries, causing underestimates of heterozygosity and overestimates of coverage. Therefore, we applied the clone_filter module implemented by Stacks (Catchen et al. 2013) to our data. We performed a de novo assembly based on our RADseq data using the bioinformatic pipeline implemented by Stacks.
Certain parameters in the Stacks pipeline control the number of reads and the distance between them required to form ‘stacks’, which are then used to build contigs. The choice of parameter values can influence the number of contigs, number of SNPs, and genetic distance estimated with a RADseq dataset (Catchen et al. 2013; Mastretta-Yanes et al. 2015; Paris et al. 2017). We tested the impact of these parameters (m [stack depth], M [distance between stacks], n [mismatches between loci], and max_locus_stacks [stacks per locus]; see supplemental material) on contig discovery. Because we sequenced most of our samples twice in two separate HiSeq runs, we had independent replicate datasets to compare. For this experiment we selected sequencing data from ten individuals based on the smallest difference in the number of reads produced across the two sequencing runs, allowing at most only one individual from each population. Details on the parameters that were tested, the methodology, and results, are in the supplemental material.
Genotyping and population genetics
Based on the results of the Stacks pipeline experiment, we proceeded with the following parameter values for building loci for the entire bull trout dataset: m = 3, M = 2, max_locus_stacks = 3, and n = 1. Our sequence coverage was low (see “Results”) and selecting these parameters balanced the need of increasing mapping coverage while minimizing exclusion of reads from the dataset. To build our final catalog of contigs we incorporated the full suite of 344 individuals (see “Results”) that produced sufficient numbers of forward reads, combining data from the two replicates. We again ran the clone_filter function and then each of the individual Stacks core modules (ustacks, cstacks, and sstacks). After creating our catalog of contigs we removed any duplicates.
Because we had a large number of individuals per population and low sequencing coverage, we used the genotyping approach implemented in the program ANGSD (Korneliussen et al. 2014). Rather than directly calling genotypes at a particular genomic position for an individual, ANGSD relies on genotype likelihoods estimated using sequencing reads aligned to a reference genome. This method is advantageous for low coverage data and results in unbiased allele frequency estimates (Nielsen et al. 2011, 2012; Korneliussen et al. 2014). In this case, our constructed contigs from Stacks served as our ‘reference genome’. We aligned reads with Bowtie2 (Langmead and Salzberg 2012) using only the forward reads (i.e. reads originating from the restriction cut-site). The resulting sequence alignment/map (SAM) files produced by Bowtie were converted to binary alignment files (BAM) using SAMtools (Li et al. 2009). With ANGSD we measured per site coverage across our BAM files. To examine population structure we exploited several analyses integrated in the ANGSD framework. First we estimated the posterior genotype probabilities using the GATK method (McKenna et al. 2010) with the allele frequency prior and then used ngsCovar (Fumagalli et al. 2014) to conduct a principal component analysis (PCA). We also took the genotype likelihoods and conducted an admixture analysis with NGSadmix (Skotte et al. 2013). We ran ten iterations of every K value (i.e. number of genetic clusters) from one to 24. For both analyses we screened for base and mapping quality (see “Results”), identified variants across all individuals using a p-value threshold of 10− 6, only included sites for which reads were available from two or more individuals, removed tri-allelic sites, and set a minor allele frequency cut-off of 0.05.
We conducted an additional analysis with TreeMix (Pickrell and Pritchard 2012) to estimate a maximum-likelihood tree of population relationships and migration events. We added 1–17 migration edges, which reflect admixture events that improve the fit of the model, estimating the variance explained by the model with increasing number of edges. For TreeMix we generated SNP genotype calls for each individual based on the same ANGSD pipeline. We added RADseq data from brook trout Salvelinus fontinalis collected from Fishing Creek, Pennsylvania, USA to serve as an outgroup.
Microsatellites
We generated genotypes at 16 microsatellite loci following the protocol and procedures described in Ardren et al. (2011). All samples had been previously genotyped in Ardren et al. except for those from the Lewis and Clark Fork rivers, which were unique to this study. We constructed a PCA with these genotypes using the package adegenet 2.0 (Jombart 2008) for R 3.2 (R Core Team 2015). We performed a Bayesian clustering analysis of these genotypes using the program STRUCTURE (Pritchard et al. 2000) with both the uncorrelated and correlated allele frequency models (Falush et al. 2003). K ranged from one to 24 with five replicates per value and a 50,000 burn-in followed by 500,000 MCMC replicates per iteration. STRUCTURE runs were performed in parallel using the R package ParallelStructure (Besnier and Glover 2013). Along with the mean log-likelihood for each K value, we estimated the ΔK statistic (Evanno et al. 2005) to identify the optimal grouping of our populations.
Results
Sequencing results and stacks parameter testing
On average our initial set of four libraries produced ~ 37.9 million forward reads (SD 10.3 million) that were retained following removal of low quality reads based on the default process_radtags filter (e.g. -c and -d options selected). The average number of retained forward reads per individual following filtering and PCR clone removal was 401,824 (SD 397,535) with a median value of 256,218. Seventeen individuals were excluded in the second set of libraries because they produced a sufficient number of reads in the first normalization (between 1.2 and 1.86 million reads). Thirty-six individuals produced so few reads (all less than 30,000) that we excluded them from the analysis. The remaining individuals were re-sequenced. Our negative controls produced on average 6828 barcoded reads, with the highest value 7467 reads.
By far the parameter with the greatest impact on contig construction was stack depth (m): increasing this parameter value decreased the number of contigs in the catalog by nearly 20,000 for each incremental change (Fig. S1). Changing parameter values had little impact on contig error rates (i.e. proportion detected in one replicate but not in the other), although rates tended to decrease as m increased (Fig. S2). However, as m increased there were fewer contigs that were identical between the two replicates, suggesting different consensus contig sequences were produced between the replicates (Fig. S3). See the supplemental material for more detail.
Catalog construction
We then processed both sets of RADseq libraries together in the Stacks pipeline. Across the 344 individuals retained in the library, the average number of forward reads sequenced per individual was 602,924 (SD 343,403) and the median value 534,138. After removing PCR duplicates the average number of reads per individual was 513,240 (SD 272,329) and the median value 446,907. Our resulting Stacks catalog contained 165,847 de novo contigs: 37 were duplicates and were removed from the catalog. The remaining 165,810 contigs served as our reference genome. Aligning the forward reads to these contigs, our average within individual per site depth was 2.9X. This was variable across individuals: the maximum observed average coverage was 10.3X and the lowest was 0.8X. Nineteen individuals had an average coverage < 1X and another 110 had an average coverage of 1-2X. Our average per base quality score was 37.3 (out of maximum score of 40). There was a noticeable break in the distribution of base quality scores: 95.8% had a score of 27 or higher and the remainder had a score of 16 or lower. Thus, for subsequent analyses we filtered the data to include only bases with a quality score ≥ 27. Our average mapping quality score per individual was 29.5 with a range from 13.9 to 32.6. For subsequent analyses we removed reads with a mapping score below 10, which should remove reads aligned to multiple sequences (Urban 2014).
Population genetics
ANGSD identified 649,127 variable sites across individuals using the threshold parameters we selected. Of these 79,952 had a minor allele frequency greater than 0.05 and were included in the subsequent analyses. The first axis of the PCA produced by ngsCovar explained 8.34% of the variation in allele frequencies and the second axis explained 5.25%. When plotted, the first axis cleanly divided bull trout populations along the coastal and interior lineages (Fig. 2a). Populations from the Snake River basin, Upper Columbia, and St. Mary River all clustered among the Interior grouping; the Coastal grouping included the Deschutes, Lower Columbia, Klamath, and Puget Sound populations. The second axis split the Interior lineage between an Upper Columbia group (which included the St. Mary population) and a Snake River basin group. The population from the Yakima River basin in central Washington was intermediate to these clusters.
a Principal component plot of 344 bull trout based on allele frequencies estimated by ANGSD for sites with a minor allele frequency greater than 0.05. b Principal component plot of 322 bull trout based on microsatellite allele frequencies. The amount of variation in the data explained by the two axes is noted. Individuals are color coded to correspond with particular populations and regional groupings
The greatest increase in log likelihood estimates produced by NGSadmix occurred from K = 1 to K = 2 (Fig. S4, see Supplemental 2), which split the bull trout populations into groups corresponding to the Coastal and Interior lineages (Fig. 3). The Coastal cluster contained the Puget Sound, Klamath, and Willamette populations. The Interior cluster contained the Snake River, Upper Columbia, and St. Mary populations. Populations from the Lewis River (Lower Columbia) and Deschutes River had signatures of admixed ancestry (i.e. average q value for the two clusters both < 0.7) between the two lineages. The K = 3 split the Interior lineage into a group containing Snake River populations and another containing the Upper Columbia and St Mary populations. Several Upper Columbia populations, most notably the Yakima River basin, appeared to have admixed ancestry between these two interior groups. The K = 4 saw a division between populations from the Puget Sound and the Lower Columbia/Klamath. This was observed in nine out of the ten iterations of NGSadmix. This pattern was also observed in the ngsCovar PCA: the third PC, which explained 2.6% of the variation, separated populations from the Puget Sound and Lower Columbia/Klamath. Increasing values of K produced small increases in log-likelihood and greater inconsistency across runs, complicating assessment of hierarchical relationships (Fig. S4).
Plot of admixture coefficients estimated by NGSadmix for K = 2, 3, and 4. Populations are grouped into separate panels and the numbers beneath each panel correspond to the same numbers in Table 1 and Fig. 1. The black horizontal lines group populations by geographic region. Clustering patterns presented were observed in all ten iterations of NGSadmix for K = 2 and 3; nine out of ten iterations for K = 4
With no migration edges the maximum-likelihood tree produced by TreeMix had three broad clades that corresponded to the Coastal, Upper Columbia, and Snake River lineages (Fig. 4). The Willamette and Klamath populations grouped together and all populations from the Skagit River system (Upper Baker River, Illabot Creek, Ruby Creek) clustered together. The St. Mary population clustered with the Upper Columbia clade. Bull trout from the Yakima River were intermediate to the Upper Columbia and Snake River clades.
Maximum-likelihood graph of population relationships inferred using TreeMix based on RADseq data. Population labels match those in Table 1 and are color-coded to match geographic groupings described in the text. Brook trout RADseq data were used as an outgroup. The horizontal axis reflects the extent of genetic drift experienced by each branch in the graph, with longer branch lengths reflecting higher drift. The scale bar shows ten times the average standard error of the sample covariance matrix. This model assumed no migration events
Adding migration edges altered the position of some populations in the TreeMix tree but did not fundamentally change the primary clades (Fig. S9). The first added migration edge suggested admixture into the St. Mary population from the basal point of the entire Interior clade. Adding a second, third, and fourth migration edge suggested introgression from the St. Joe River population into the Lewis River, from the Interior lineage to the Elwha population, and from the Snake River clade into the Deschutes population, respectively. With no migration edges the model explained 97.08% of the covariance. Adding 13 (99.08%) and 15 migration edges (99.09%) resulted in models explaining the greatest proportion of covariance, but adding these edges began to alter tree topology. Also, few of these edges produced significant p-values with the Wald statistic, indicating that there was weak statistical support for their placement.
Microsatellite data
Twenty-two of the 344 bull trout included in the RADseq libraries failed to produce microsatellite genotypes. The first two dimensions of the PCA incorporating the microsatellite genotypes explained less variation (3.2% and 2.7%) than the RADseq data. Coastal and Inland lineages formed a rough divide along the first axis (Fig. 2b). St. Mary and Deschutes populations clustered intermediate to the two primary lineages. The second axis slightly separated the Upper Columbia and Snake River populations, although there was some overlap. Also along this axis the Klamath River population was highly divergent from those in the Lower Columbia.
With STRUCTURE the inference depended on the allele frequency model. Increasing K produced gradual increases in mean log-likelihood for the correlated model until K = 14: with higher values there were dramatic swings in log-likelihood scores (Fig. S5). This resulted in multiple values of K that had substantial support using the ΔK method (Fig. S6). The highest value was at K = 16, but there were other peaks at eight, ten, and 24. The K = 2 had the fifth highest ΔK score. At K = 2 STRUCTURE produced three different clustering patterns across our five replicates (Fig. S7). Three replicates produced a pattern that divided the Interior and Coastal lineages with the Deschutes and Klamath clustering with the Coastal and St. Mary with the Interior. One replicate clustered the Klamath with Interior populations and another clustered the Klamath and Willamette with the Interior and Warm Springs Creek with the Coastal. Regardless of the replicate, every population was virtually homogenous in ancestry for the cluster it was assigned; no population showed a pattern of introgression between the two clusters. When increased to K = 3, there were four different clustering patterns among the five replicates (Fig. S8). Although some of these patterns corresponded to geographic groupings, they were inconsistent.
The uncorrelated model produced a different pattern. Log-likelihood scores experienced the biggest leap from K = 2 to K = 3 with a gradual increase and plateauing of scores, although there were some large swings beyond K = 14 (Fig. S5). This meant K = 2 was by the far the most supported value using ΔK (Fig. S6). It again divided the Interior and Coastal lineages: for four of the replicates the St. Mary and an Upper Columbia population (Warm Springs Creek) clustered with the Coastal lineage (Fig. S7). Increasing K to three resulted in four different clustering patterns across the five replicates (Fig. S8).
Discussion
RADseq/microsatellite comparison
As conservation genetics moves into the genomic era there is increasing need to compare findings generated with traditional markers to high-throughput sequencing data. Although newer techniques may be attractive, many questions can still be adequately answered using traditional markers such as single-gene sequences or polymorphic microsatellites (Zink and Barrowclough 2008; Elbers et al. 2016; Hodel et al. 2016). Thus, it is important to weigh the benefits gained from using genomic data against the simplicity, cost, and efficiency of traditional markers (McMahon et al. 2014; Elbers et al. 2016; Puckett 2017).
Although the datasets produced similar overall findings, there were striking differences. In general the RADseq data produced sharper, more consistent patterns of genetic structure at broad phylogenetic scales. Similar findings have been observed in other studies comparing these marker types, with RADseq data revealing complex, previously unknown phylogenetic patterns within other species of fish (Bradbury et al. 2015; Jeffries et al. 2016). Comparably, though, the microsatellite data provided less resolution in identifying phylogenetic groups and was inconsistent in patterns of clustering. We believe these findings reflect the nature of microsatellite loci themselves rather than limitations of our specific dataset. Many of the microsatellite markers in this dataset were developed specifically for bull trout (DeHaan and Ardren 2005) or closely related species from the same genus (Angers et al. 1995; Crane et al. 2004), limiting potential ascertainment bias. Low sample sizes may have also affected the clustering patterns, but the broad patterns we observed with the microsatellites mirror those of Ardren et al. (2011) who had larger sample sizes per population. For example, they also found the St. Mary’s and Deschutes populations clustered with Coastal populations and did not observe admixture within populations.
RADseq-derived SNPs and microsatellite loci have different properties and reflect different aspects of an organism’s genomic history. Microsatellites often contain multiple alleles per locus, which can result in low individual frequencies of each allele. This makes microsatellites vulnerable to sudden shifts in allele frequencies due to genetic drift, especially bottlenecks (Luikart et al. 1998). Based on simulations, Haasl and Payseur (2010) suggested that microsatellites would detect recent divergence between populations more readily than SNPs.
Such properties likely explain the differing patterns of structure suggested by the markers used in this study. Bull trout populations are known for high genetic differentiation, even among neighboring tributaries (Spruell et al. 1999; Whiteley et al. 2006; Warnock et al. 2010; DeHaan et al. 2011). Ardren et al. (2011) found that although lower values of K discriminated the primary phylogenetic lineages, the highest supported K-value in their Bayesian clustering analysis equaled the total number of populations in the dataset. Every pairwise FST comparison between populations in their study was statistically significant. Many bull trout populations were founded after the retreat of the Pleistocene glaciers and/or are isolated by natural or anthropogenic barriers (Taylor et al. 1999; Costello et al. 2003; Spruell et al. 2003; Ardren et al. 2011); such recent divergence is likely to be reflected in the microsatellite data. Genome-wide SNPs, on the other hand, such as those generated with RADseq, can include heavily conserved and/or adaptive regions of the genome that are more likely to reflect deep divergences (Liu et al. 2005; DeFaveri et al. 2013). Thus, the RADseq was more likely to reveal phylogenetic divisions whereas the microsatellite data were obscured by more recent population processes.
Not only did we observe differences in clustering between the RADseq and microsatellite data, there were also inconsistencies in clustering patterns generated with the microsatellite data using STRUCTURE. Based on initial testing we ran STRUCTURE with both the uncorrelated and correlated allele frequency models. The correlated model accounts for the fact that closely related populations are likely to have non-independent allele frequencies while the uncorrelated model assumes populations have independent allele frequencies (Falush et al. 2003). It is difficult to predict which pattern fits any given biological system and selecting an ideal model is further complicated by hierarchical structure within the dataset. With the bull trout microsatellite data inferences of optimal K and overall clustering patterns were strongly influence by allele frequency model. This underscores the varying evolutionary signals and population histories that can be revealed by microsatellite data. We suggest using both models when investigating systems with strong hierarchical genetic structure.
Another strength of the genome-wide SNPs compared to microsatellites was their ability to detect admixture. Ardren et al. (2011) suspected admixture in some bull trout populations based on mtDNA and microsatellite incongruence, but did not observe admixed populations based solely on the microsatellites. We did not observe evidence of admixture with the microsatellites either. However, the RADseq data provided evidence that some populations have a history of admixture. Historical admixture is the more likely explanation for these patterns than contemporary hybridization based on the homogeneity of ancestry within populations and the overall lack of migrants detected in the dataset. Plus, many of the admixed populations are geographically located in potential contact zones between major phylogenetic lineages, a pattern that has been observed in other Pacific salmonids as well (Narum et al. 2010; Blankenship et al. 2011). Other studies have suggested that SNPs are superior to microsatellites for detecting admixture (Haasl and Payseur 2010; Väli et al. 2010; Bradbury et al. 2015). This is due to high numbers of SNPs that are fixed (i.e. homozygous) for a particular allele in populations and/or lineages: admixed individuals or populations would then display a heterozygous signal at these genomic regions.
Our study complements previous analyses that have compared findings generated with RADseq and microsatellite data (e.g. Corander et al. 2013; Bradbury et al. 2015; Jeffries et al. 2016; Thrasher et al. 2018). Previous studies typically approached RADseq data similarly, generating genotype calls for SNPs that were heavily filtered based on variables such as coverage and missing data. Approaching RADseq data in this way facilitates the use of similar analyses and software that have traditionally been used for microsatellite data. However, high-throughput sequencing data is fundamentally different from microsatellite data and can be processed in a variety of ways depending on the nature of the dataset and goals of the study.
Initial testing of our dataset suggested that the standard Stacks pipeline produced low genotyping rates due to our low coverage. Using the genotype likelihood approach implemented in ANGSD and ngsTools alleviated this issue and allowed us to identify a substantial number of potential SNPs. It also provided a way to avoid another issue: the high sample to cost ratio of RADseq compared to microsatellites. Low sample sizes (i.e. number of individuals per population) are often justified in RADseq analyses to balance the issue of sequencing coverage vs. cost of high-throughput sequencing (Elbers et al. 2016; Puckett 2017), resulting in lower sample sizes when compared to typical microsatellite datasets (Bradbury et al. 2015; Elbers et al. 2016; Jeffries et al. 2016). However, using a bioinformatics pipeline designed for low coverage data allowed us to directly compare the same suite of 300 individuals for both marker sets. Even though we had substantial amounts of missing data in terms of individual coverage per contig, adding low coverage contigs and/or variants may can also increase resolution by providing greater overall coverage of the genome (Hodel et al. 2017). Strict filtering of loci and variants based on arbitrary cut-offs may remove valuable information embedded within high-throughput sequencing data. Although using large samples sizes may result in lower coverage, this study and others demonstrate this approach can provide robust estimation of allele frequencies and subsequent assessment of genetic structure (Nielsen et al. 2012; Buerkle and Gompert 2013; Fumagalli et al. 2013).
Intra-specific diversity of bull trout
The RADseq analysis provided several important insights into bull trout evolutionary history, resolving some of the discrepancies noted by previous studies. Perhaps the most obvious finding is that the St. Mary population aligns with other populations from the Upper Columbia River basin instead of the Coastal lineage. Ardren et al. (2011) found that St. Mary’s bull trout clustered with the Coastal lineage with microsatellites but shared a mtDNA haplogroup with other Interior lineage populations. The congruence between mtDNA and RADseq data reflects biogeographic expectations, suggesting the microsatellite data provided misleading signals. This could have been due to random genetic drift producing similar allele frequencies as Coastal populations or homoplasy. Also, our results further corroborate previous studies documenting the similarity between the Klamath and Willamette populations. This is particularly interesting because the two watersheds are currently separated by the Umpqua and Rogue river basins in southern Oregon. In fact, the headwaters of the Deschutes basin are adjacent to those of the Klamath River basin, yet there was no evidence of recent shared ancestry between these populations. Further investigation involving additional species is needed to assess potential migration events between these two river basins. It also raises the question of whether bull trout were historically present in other Oregon Coastal Rivers with cold headwater systems found in the Cascade Mountains (e.g., Rogue and Umpqua Rivers).
A novel finding from our study was the ubiquity of admixture across the bull trout range. At the geographic scale covered by our populations, contemporary migration and gene flow between bull trout populations is very rare (Spruell et al. 2003; Ardren et al. 2011). These signatures of admixed ancestry likely reflect historical secondary contact between the primary biogeographical lineages. Our samples from the Deschutes River in central Oregon and the Lewis River in southwest Washington displayed ancestry from the Coastal and Interior lineages. This was hypothesized by Ardren et al. (2011): both clustered with Coastal populations using microsatellites, but a few populations in these basins had mtDNA haplotypes found in Interior populations. Our results support this hypothesis and further suggest these two populations possess admixture from different Interior lineages. Lewis River bull trout appeared to have higher admixture proportions from the Upper Columbia lineage whereas the Deschutes River bull trout had more from the Snake River. We also observed admixture within the Yakima River population, with ancestry from both of the Interior lineages (Upper Columbia and Snake River). Bull trout in this system also have mtDNA haplotypes from multiple lineages (Ardren et al. 2011).
The information from the RADseq analysis has implications for bull trout conservation. First, the assignment of populations to major lineages based on genetic data only partially aligns with their grouping into recovery units. The most obvious is the Mid-Columbia Bull Trout Recovery Unit, which encompasses populations such as the Yakima and Methow, and the Lower Snake River basin. This recovery unit includes populations from two distinct evolutionary lineages, but does not cover either lineage in totality. Also, combining all populations from the Coastal lineage into a single Coastal Recovery Unit does not represent the divergence between Puget Sound/Coastal Washington populations and those in the Lower Columbia River basin. As a more general trend, based solely on genetic relationships, many populations do not fit cleanly into simple dichotomies (e.g. coastal vs. interior). The Lewis River, Deschutes River, and Yakima River, for example, represent admixture between different lineages.
Our findings highlight a reoccurring theme in conservation genomics: the patterns of diversity being revealed with new genomic-level data do not always adhere to previous findings of population subdivision. Discrepancies inevitably cause confusion among the conservation community. Within this context it important for geneticists to emphasize that individual datasets are necessarily “right” or “wrong”, but instead can provide different windows into the genetic background of a species or population. Genetic marker type plays an important role in interpreting population history and great care should be given to selecting a marker that will adequately answer a given question. Also, no single genetic dataset exists in vacuum and should be complemented with previous genetic research and other biological information to provide a holistic perspective of population relationships. In the case of bull trout and many other species, additional types of biological data such as life history data, habitat availability, and connectivity may also be important for shaping conservation units. Designating management units can be further complicated when different types of data such as social beliefs and political boundaries are factored into the decisions (Polfus et al. 2016; Marin et al. 2017).
It is important to note that results presented here do not represent a comprehensive range-wide analysis of bull trout evolutionary history, but rather a comparison of results generated from different marker sets. Currently there are 187 subpopulations of bull trout distributed among 121 core habitat units identified by the US Fish and Wildlife Service (USFWS 2015). As has been noted in previous studies, gene flow between subpopulations is rare, even at very fine geographic scales (Costello et al. 2003; Ardren et al. 2011; DeHaan et al. 2011). Genetic similarities between populations that we observed likely reflect deep evolutionary divergence and past admixture, not contemporary gene flow. Differences in evolutionary patterns between this study and previous ones should be interpreted in light of the fact that this study contains a reduced number of populations relative to the range-wide distribution of bull trout.
Data availability
Data for this study will be submitted to the Dryad Digital Repository after the manuscript is accepted for publication.
References
Angers B, Bernatchez L, Angers A, Desgroseillers L (1995) Specific microsatellite loci for brook charr (Salvelinus fontinalis Mitchill) reveal strong population subdivision on a microgeographic scale. J Fish Biol 47:177–185
Ardren WR, DeHaan PW, Smith CT et al (2011) Genetic structure, evolutionary history, and conservation units of bull trout in the coterminous United States. Trans Am Fish Soc 140:506–525. https://doi.org/10.1080/00028487.2011.567875
Baird, NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoSOne 3:e3376
Besnier F, Glover KA (2013) Parallel structure: a R package to distribute parallel runs of the population genetics program STRUCTURE on multi-core computers. PLoS ONE 8:1–9. https://doi.org/10.1371/journal.pone.0070651
Blankenship SM, Campbell MR, Hess JE et al (2011) Major lineages and metapopulations in Columbia River Oncorhynchus mykiss are structured by dynamic landscape features and environments. Trans Am Fish Soc 140:665–684. https://doi.org/10.1080/00028487.2011.584487
Bradbury IR, Hamilton LC, Dempson B et al (2015) Transatlantic secondary contact in Atlantic Salmon, comparing microsatellites, a single nucleotide polymorphism array and restriction-site associated DNA sequencing for the resolution of complex spatial structure. Mol Ecol 24:5130–5144. https://doi.org/10.1111/mec.13395
Buerkle AC, Gompert Z (2013) Population genomics based on low coverage sequencing: how low should we go? Mol Ecol 22:3028–3035. https://doi.org/10.1111/mec.12105
Catchen J, Hohenlohe PA, Bassham S et al (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140. https://doi.org/10.1111/mec.12354
Corander J, Majander KK, Cheng L, Merilä J (2013) High degree of cryptic population differentiation in the Baltic Sea herring Clupea harengus. Mol Ecol 22:2931–2940. https://doi.org/10.1111/mec.12174
Costello AB, Down TE, Pollard SM et al (2003) Influence of history and contemporary stream hydrology on the evolution of genetic diversity within species: an examination of microsatellite DNA variation in bull trout, Salvelinus confluentus (Pisces: Salmonidae). Evolution 57:328. https://doi.org/10.1554/0014-3820(2003)057%5B0328:TIOHAC%5D2.0.CO;2
Crane PA, Lewis CJ, Kretschmer EJ, Miller SJ, Spearman WJ, DeCicco AL, Lisac MJ, Wenburg JK (2004) Characterization and inheritance of seven microsatellite loci from Dolly Varden, Salvelinus malma, and cross-species amplification in Arctic char, S. alpinus. Con Gen 5:737–741
DeFaveri J, Viitaniemi H, Leder E, Merilä J (2013) Characterizing genic and nongenic molecular markers: comparison of microsatellites and SNPs. Mol Ecol Resour 13:377–392. https://doi.org/10.1111/1755-0998.12071
DeHaan PW, Ardren WR (2005) Characterization of 20 highly variable tetranucleotide microsatellite loci for bull trout (Salvelinus confluentus) and cross-amplification in other Salvelinus species. Mol Ecol Notes 5:582–585. https://doi.org/10.1111/j.1471-8286.2005.00997.x
DeHaan PW, Bernall SR, Dossantos JM et al (2011) Use of genetic markers to aid in re-establishing migratory connectivity in a fragmented metapopulation of bull trout (Salvelinus confluentus). Can J Fish Aquat Sci 68:1952–1969. https://doi.org/10.1139/f2011-098
DeYoung RW, Honeycutt RL (2008) The molecular toolbox: genetic techniques in wildlife ecology and management. J Wildl Manage 69:1362–1384. https://doi.org/10.2193/0022-541X(2005)69%5B1362:TMTGTI%5D2.0.CO;2
Elbers JP, Clostio RW, Taylor SS (2016) Population genetic inferences using immune gene SNPs mirror patterns inferred by microsatellites. Mol Ecol Resour 17:481–491. https://doi.org/10.1111/1755-0998.12591
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes. Genetics 131:479–491. https://doi.org/10.1007/s00424-009-0730-7
Excoffier L, Foll M, Petit RJ (2009) Genetic consequences of range expansions. Annu Rev Ecol Evol Syst 40:481–501. https://doi.org/10.1146/annurev.ecolsys.39.110707.173414
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fumagalli M, Vieira FG, Korneliussen TS et al (2013) Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195:979–992. https://doi.org/10.1534/genetics.113.154740
Fumagalli M, Vieira FG, Linderoth T, Nielsen R (2014) NgsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics 30:1486–1487. https://doi.org/10.1093/bioinformatics/btu041
Gompert Z, Buerkle CA (2016) What, if anything, are hybrids: enduring truths and challenges associated with population structure and gene flow. Evol Appl 9:909–923. https://doi.org/10.1111/eva.12380
Groves CP, Cotterill FPD, Gippoliti S et al (2017) Species definitions and conservation: a review and case studies from African mammals. Conserv Genet 18:1247–1256. https://doi.org/10.1007/s10592-017-0976-0
Haasl RJ, Payseur B (2010) Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites. Heredity 106:158–171. https://doi.org/10.1038/hdy.2010.21
Hodel RGJ, Segovia-Salcedo MC, Landis JB et al (2016) The report of my death was an exaggeration: a review for researchers using microsatellites in the 21st Century. Appl Plant Sci 4:1600025. https://doi.org/10.3732/apps.1600025
Hodel RGJ, Chen S, Payton AC et al (2017) Adding loci improves phylogeographic resolution in red mangroves despite increased missing data: comparing microsatellites and RAD-Seq and investigating loci filtering. Sci Rep 7:17598. https://doi.org/10.1038/s41598-017-16810-7
Jeffries DL, Copp GH, Lawson Handley L et al (2016) Comparing RADseq and microsatellites to infer complex phylogeographic patterns, an empirical perspective in the Crucian carp, Carassius carassius, L. Mol Ecol 25:2997–3018. https://doi.org/10.1111/mec.13613
Jensen EL, Govindarajulu P, Russello M (2013) When the shoe doesn’t fit: applying conservation unit concepts to western painted turtles at their northern periphery. Conserv Genet 15:261–274. https://doi.org/10.1007/s10592-013-0535-2
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Kohn MH, Murphy WJ, Ostrander EA, Wayne RK (2006) Genomics and conservation genetics. Trends Ecol Evol 21:629–637. https://doi.org/10.1016/j.tree.2006.08.001
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinform 15:356. https://doi.org/10.1186/s12859-014-0356-4
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Liu N, Chen L, Wang S et al (2005) Comparison of single-nucleotide polymorphisms and microsatellites in inference of population structure. BMC Genet 6:26. https://doi.org/10.1186/1471-2156-6-S1-S26
Luikart G, Allendorf FW, Cornuet J-M, Sherwin WB (1998) Distortion of allele frequency distributions provides a test for recent population bottlenecks. J of Heredity 89:238–247
Marin K, Coon A, Fraser DJ (2017) Traditional ecological knowledge reveals the extent of sympatric lake trout diversity and habitat preferences. Ecol Soc 22:20. https://doi.org/10.5751/ES-09345-220220
Martin CH, Cutler JS, Friel JP et al (2015) Complex histories of repeated gene flow in Cameroon crater lake cichlids cast doubt on one of the clearest examples of sympatric speciation. Evolution 69:1406–1422. https://doi.org/10.1111/evo.12674
Mastretta-Yanes A, Arrigo N, Alvarez N et al (2015) Restriction site-associated DNA sequencing, genotyping error estimation and de novo assembly optimization for population genetic inference. Mol Ecol Resour 15:28–41. https://doi.org/10.1111/1755-0998.12291
McDevitt AD, Mariani S, Hebblewhite M et al (2009) Survival in the Rockies of an endangered hybrid swarm from diverged caribou (Rangifer tarandus) lineages. Mol Ecol 18:665–679. https://doi.org/10.1111/j.1365-294X.2008.04050.x
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110.20
McMahon BJ, Teeling EC, Höglund J (2014) How and why should we implement genomics into conservation? Evol Appl 7:999–1007. https://doi.org/10.1111/eva.12193
McPhail JD, Baxter JS (1996) A review of bull trout (Salvelinus confluentus) life-history and habitat use in relation to compensation and improvement opportunities. Department of Zoology, University of British Columbia, Vancouver
Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA (2007) Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 17:240–248
Mogen JT, Kaeding LR (2005) Identification and characterization of migratory and nonmigratory Bull Trout populations in the St. Mary River Drainage, Montana. Trans Amer Fish Soc 134:841–852
Narum SR, Hess JE, Matala AP (2010) Examining genetic lineages of Chinook salmon in the Columbia River Basin. Trans Am Fish Soc 139:1465–1477. https://doi.org/10.1577/T09-150.1
Nielsen R, Paul JS, Albrechtsen A, Song YS (2011) Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 12:443–451. https://doi.org/10.1038/nrg2986
Nielsen R, Korneliussen T, Albrechtsen A et al (2012) SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS ONE 7:e37558. https://doi.org/10.1371/journal.pone.0037558
Northcote TG (1997) Potamodromy in Salmonidae—living and moving in the fast lane. N Amer J Fish Manag 17:1029–1045
Paris JR, Stevens JR, Catchen JM (2017) Lost in parameter space: a road map for Stacks. Methods Ecol Evol 8:1360–1373. https://doi.org/10.1111/2041-210X.12775
Piccolo JJ (2016) Conservation genomics: coming to a salmonid near you. J Fish Biol 89:2735–2740. https://doi.org/10.1111/jfb.13172
Pickrell J, Pritchard J (2012) Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8:e1002967. https://doi.org/10.1371/journal.pgen.1002967
Pisa G, Orioli V, Spilotros G et al (2015) Detecting a hierarchical genetic population structure: the case study of the Fire Salamander (Salamandra salamandra) in Northern Italy. Ecol Evol 5:743–758. https://doi.org/10.1002/ece3.1335
Polfus JL, Manseau M, Simmons D et al (2016) Łeghágots’enetę (learning together): the importance of indigenous perspectives in the identification of biological variation
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puckett EE (2017) Variability in total project and per sample genotyping costs under varying study designs including with microsatellites or SNPs to answer conservation genetic questions. Conserv Genet Resour 9:289–304. https://doi.org/10.1007/s12686-016-0643-7
Putman AI, Carbone I (2014) Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol Evol 4:4399–4428. https://doi.org/10.1002/ece3.1305
Ramey IIRR, Wehausen JD, Liu H-P et al (2007) How King et al. (2006) define an “evolutionary distinction”. of a mouse subspecies: a response. Mol Ecol 16:3518–3521. https://doi.org/10.1111/j.1365-294X.2007.03397.x
R Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. https://www.R-project.org
Rieman BE, McIntyre JD (1993) Demographic and habitat requirements for conservation of bull trout. Ogen, Tampa
Rieman BE, Dunham JB (2000) Metapopulations and salmonids: a synthesis of life history patterns and empirical observations. Eco Fresh Fish 9:51–64
Skotte L, Korneliussen TS, Albrechtsen A (2013) Estimating individual admixture proportions from next generation sequencing data. Genetics 195:693–702. https://doi.org/10.1534/genetics.113.154138
Spruell P, Rieman BE, Knudsen KL et al (1999) Genetic population structure within streams: microsatellite analysis of bull trout populations. Ecol Freshw Fish 8:114–121. https://doi.org/10.1111/j.1600-0633.1999.tb00063.x
Spruell P, Hemmingsen AR, Howell PJ et al (2003) Conservation genetics of bull trout: geographic distribuition of variation at microsatellite loci. Conserv Genet 4:17–29
Sunnucks P (2000) Efficient genetic markers for population biology. Trends Ecol Evol 15:199–203. https://doi.org/10.1016/S0169-5347(00)01825-5
Taylor EB, Pollard S, Louie D (1999) Mitochondrial DNA variation in bull trout (Salvelinus confluentus) from northwestern North America: implications for zoogeography and conservation. Mol Ecol 8:1155–1170. https://doi.org/10.1046/j.1365-294X.1999.00674.x
Thrasher DJ, Butcher BG, Campagna L et al (2018) Double-digest RAD sequencing outperforms microsatellite loci at assigning paternity and estimating relatedness: a proof of concept in a highly promiscuous bird. Mol Ecol Resour. https://doi.org/10.1111/1755-0998.12771
Twyford AD, Ennos RA (2012) Next-generation hybridization and introgression. Heredity 108:179–189. https://doi.org/10.1038/hdy.2011.68
U.S. Fish and Wildlife Service (2015) Recovery plan for the coterminous United States population of bull trout. Portland, OR
Unger S Jr, Sutton OR, Williams T R (2013) Population genetics of the eastern hellbender (Cryptobranchus alleganiensis alleganiensis) across multiple spatial scales. PLoS ONE 8:1–14. https://doi.org/10.1371/journal.pone.0074180
Urban J (2014) How does bowtie2 assign MAPQ scores? [Blog] Biofinysics. URL: http://biofinysics.blogspot.com/2014/05/how-does-bowtie2-assign-mapq-scores.html
Väli Ü, Saag P, Dombrovski V et al (2010) Microsatellites and single nucleotide polymorphisms in avian hybrid identification: a comparative case study. J Avian Biol 41:34–49. https://doi.org/10.1111/j.1600-048X.2009.04730.x
Waples RS, Gaggiotti O (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439. https://doi.org/10.1111/j.1365-294X.2006.02890.x
Warnock WG, Rasmussen JB, Taylor EB (2010) Genetic clustering methods reveal bull trout (Salvelinus confluentus) fine-scale population structure as a spatially nested hierarchy. Conserv Genet 11:1421–1433. https://doi.org/10.1007/s10592-009-9969-y
Wayne RK, Shaffer HB (2016) Hybridization and endangered species protection in the molecular era. Mol Ecol 81:778–793. https://doi.org/10.1111/mec.13642
Whiteley AR, Spruell P, Rieman BE, Allendorf FW (2006) Fine-scale genetic structure of bull trout at the southern Llimit of their distribution. Trans Am Fish Soc 135:1238–1253. https://doi.org/10.1577/T05-166.1
Zink RM, Barrowclough GF (2008) Mitochondrial DNA under siege in avian phylogeography. Mol Ecol 17:2107–2121. https://doi.org/10.1111/j.1365-294X.2008.03737.x
Acknowledgements
Funding for this project was provided by the US Fish and Wildlife Service Fish and Aquatic Conservation Program and Washington State general funds. We sincerely thank the numerous individual biologists and technicians from the different federal, state, tribal, and non-governmental agencies who collected tissue samples used in these analyses. We also thank Sewall Young and Ken Warheit (WDFW) for sharing scripts for running Stacks. The findings and conclusions in this paper are those of the authors and do not necessarily represent the views of the US Fish and Wildlife Service.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bohling, J., Small, M., Von Bargen, J. et al. Comparing inferences derived from microsatellite and RADseq datasets: a case study involving threatened bull trout. Conserv Genet 20, 329–342 (2019). https://doi.org/10.1007/s10592-018-1134-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-018-1134-z