Abstract
The use of commercially produced seed material is a common practice in restoration. However, the impact of sowing on genetic variation of natural populations is still unclear. Aim of this study was, therefore, to test if genetic variation within and among populations restored with local seed material corresponds to the genetic variation of neighboring natural populations. We investigated each ten natural and restored populations of three common grassland species (Knautia arvensis, Silene vulgaris and Plantago lanceolata), situated in five study regions in southeastern Germany. Our study revealed significant genetic differentiation between natural and restored populations of the insect-pollinated K. arvensis and S. vulgaris although differentiation was much stronger for K. arvensis since most restored populations contained another ploidy level than natural populations. For the wind-pollinated P. lanceolata, genetic differentiation between natural and restored populations was comparable to the genetic differentiation between its natural populations. Genetic diversity within restored populations of each species was equivalent or even higher than within natural populations. Our study provides evidence that the local genetic structure especially of common insect-pollinated grassland species may be affected by the application of regional seed mixtures in restoration. Regional admixed provenancing in seed production is an important approach to preserve regional patterns and to provide seeds for the reestablishment of genetically variable populations. The method would however be an even more powerful tool in restoration when ploidy levels would be checked before seed production and seed transfer zones would be smaller.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ecological restoration of species-rich grasslands often depends on the availability of viable seeds in the soil seed bank of restoration sites or on native target species in the surrounding environment (Bakker et al. 1996). Landscape fragmentation can hamper seed dispersal between restoration sites and potential source populations (Münzbergova and Herben 2005; Hölzel et al. 2012). Therefore, the introduction of target species is a state-of-the-art method in conservation practice and especially sowing of local seed material has become a common tool in restoration ecology (Jongepierova et al. 2007; Török et al. 2010; Walker et al. 2015).
In forestry, guidelines for the use of local seed material have been established for several decades (FoVHgV 2003). Also across the world, the usage of local seeds and the implementation of seed transfer zones gain in importance for restoration purposes, for example in Australia (Krauss et al. 2013), Canada (Ukrainetz et al. 2011), the USA (Miller et al. 2011) and Europe (Malaval et al. 2010; Jørgensen et al. 2016). In Germany, a seed transfer zone concept including a seed transfer zone map and seed zone-specific species lists has been implemented since 2010 (Prasse et al. 2010). Seed transfer zones were determined on basis of the German system of 89 natural regions (Meynen et al. 1953-62), which were grouped together to 22 seed transfer zones within eight producing areas according to similar environmental conditions (Bucharova et al. 2018). Within a seed transfer zone, source seeds from several large populations have to be collected, mixed thoroughly, reproduced and can be transferred only within this zone. This seed sourcing strategy is called regional admixture provenancing and offers great advantages for restoration: The system provides almost unlimited amount of regionally adapted seed material for a huge number of species in every part of Germany. Generally, in restoration the use of local seeds or plant material is recommended (Mijnsbrugge et al. 2010) because plants are adapted to their surrounding environmental conditions. Ecological (isolation-by-environment) or geographical (isolation-by-distance) differences among habitats may cause the development of ecotypes and local adaptations (Joshi et al. 2001; Bischoff et al. 2006; Leimu and Fischer 2008). That is why blending genotypes originating from genetically differing seed sources may result in outbreeding depression (Hufford and Mazer 2003). Coadapted gene complexes can be destroyed and local adaptations get lost which leads to decreased fitness and performance of plant populations (Keller et al. 2000; Montalvo and Ellstrand 2001; Frankham et al. 2002). This may be avoided, when seeds reflect the gene pool of the naturally occurring individuals and populations near the restored areas (McKay et al. 2005).
However, genetic differentiation between populations does not only depend on ecological or geographic distances among populations, but also on life-history characteristics, such as mating system, pollination vector or dispersal unit (Hamrick and Godt 1996; Reisch and Bernhardt-Römermann 2014). For example, an outcrossing wind-pollinated plant species is likely to show lower genetic differentiation over large geographic distances as it is the case for an endemic outcrossing and insect-pollinated plant species. Considering the natural differentiation of plant populations due to abiotic and biotic factors the questions arise, how strong populations restored with local seed mixtures may vary from natural ones and if it is possible to ensure, that the genetic differentiation between natural and restored populations corresponds to the spatial genetic differentiation pattern of naturally occurring populations.
Furthermore, the production of seed material including sampling method of source seeds and propagation within a seed-farm may have major impacts on genetic diversity. This matters for example, when seeds were collected from small populations because these are less attractive to pollinators (Agren 1996; Kunin 1997). Reduced cross pollination increases mating with related individuals or even self-fertilization (Van Treuren et al. 1994), which may increase inbreeding and lead to reduced fitness and decreased genetic variation (Friar et al. 2000; Frankham et al. 2002). Furthermore, collecting seed material from a small number of source individuals in a large source population may cause genetic drift. A frequency shift of gene variants can reduce genetic diversity or local adaptations (Espeland et al. 2017) and lead to increased homozygosity and random loss or fixation of deleterious alleles (Ellstrand and Elam 1993; Young et al. 2005). Seed sampling is followed by cultivation of source seed and their reproduction. Stock individuals can be used several years for the production of local seeds. The multiple reproduction cycles may decrease genetic variation and increase the risk of inbreeding (Schoen and Brown 2001). Additionally, the plants are exposed to different environmental conditions than in their naturally habitat and unintended selection during the cultivation stage might be inavitable (Espeland et al. 2017; Nagel et al. 2018).
To avoid these negative effects caused by cultivation processes, there are procedural rules to follow for seed production. Therefore, commercially produced seed material is expected to exhibit a high genetic variability which is maintained by mixing source seeds of several large source populations. This procedure shall ensure the preservation of genetic variation. Finally, multiplying plant material only for a short period (for example 5 generations) should decrease the risk of unintended selection during the propagation and also the possibility of inbreeding depression and genetic erosion due to multiple reproduction cycles (Prasse et al. 2010; ErMiV 2011).
Although there are some genetic studies discussing seed origin and genetic differentiation among seed transfer zones (Michalski and Durka 2012; Bucharova et al. 2017; Durka et al. 2017; Listl et al. 2017a, b), the impact of sowing local seeds on the genetic variation of grassland species has much less frequently been studied. Only few studies directly compared genetic variation of plant populations from natural and restored grasslands (Aavik et al. 2012; Reiker et al. 2015).
Aim of this study was, therefore, to test if genetic variation within and among populations restored with local seed material corresponds to the variation of neighboring natural populations. We selected three widely distributed, outcrossing grassland species (Knautia arvensis, Silene vulgaris and Plantago lanceolata) and analyzed the genetic diversity and differentiation of natural and restored populations in a comparative approach using amplified fragment length polymorphisms (AFLPs). We asked the following questions
-
(i)
How strong is genetic differentiation among natural and restored populations?
-
(ii)
How large is the genetic diversity of natural and restored populations?
-
(iii)
Are local seed mixtures a promising tool to restore both species and genetic diversity of species-rich grasslands?
Methods
Study species and design
For our study, we selected three common outcrossing grassland species, occurring in natural and restored grasslands: the insect-pollinated Knautia arvensis (Coult.) and Silene vulgaris (Garcke) and the wind-pollinated Plantago lanceolata (L.).
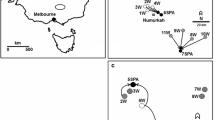
We sampled plant material for molecular analyses in populations of the study species from ten natural (N) and ten restored (R) species-rich grasslands. The populations were located in five study regions across Bavaria in Germany (Fig. 1; Table 1a–c). Study sites were not identical since not all species occurred at all sites simultaneously. However, we always compared ten natural populations and ten restored populations.
Geographic position of the study regions in south eastern Germany. Ten natural and ten restored populations of the study species Knautia arvensis, Silene vulgaris and Plantago lanceolata were investigated in five study regions. Populations of K. arvensis were situated in HE, NE, PF, RH and SC. Populations of S. vulgaris were situated within HA, NE, PF, RH and SC and the investigated populations of P. lanceolata were situated within HS, NE, PF, RE and RH. HE = Hetzmannsdorf, NE = Netzstall, PF = Pfatter; RH = Rannertshofen; SC = Schwaig; HA = Haag; HS = Herrnsaal; RE = Reichenau
Natural grasslands were historically old, which means that they have been continuously used as grassland since 19 th century and were identified using historical cadastral maps and recent maps from 2005 to 2015. Restored populations were located on former arable land. After topsoil removal ten to fifteen years ago, commercially produced local seed mixtures from a big, german seed farming company have been applied at these sites to restore species-rich grassland.
For molecular analysis, fresh leaf material was collected in situ from sixteen individuals per population. In total, material of 320 individuals was sampled and dried in teabags over silica gel.
Ploidy levels of the study species
As a first step, we applied Flow Cytometry (FCM) to identify potentially occurring different cytotypes, investigating the same plant material that was used for AFLP analysis. For each species, we tested one individual per population. Methodological details are attached in Appendix A.
Molecular analyses
DNA was isolated from 15 mg dried plant material applying the cetyltrimethylammonium bromide protocol by Rogers and Bendich (1994) with adaptions by Reisch (2007). All samples were standardized at a concentration of 7.8 ng/µL. The AFLP method was performed in accordance with the Beckman Coulter protocol as described before (Bylebyl et al. 2008).
Restriction-Ligation was performed in a reaction volume of 10 µL, containing genomic DNA, EcoRI (MBI Fermentas) and MseI (MWG Biotech) restriction enzymes and T4 DNA Ligase (MBI Fermentas). The samples were incubated for two hours at 37 °C.
PCRs were performed in a reaction volume of 5 µL. Preselective primers had one selective nucleotide (EcoRI-A; MseI-C). For selective amplification, a primer screening was conducted. For each species, 30 combinations were tested and then three combinations per species were selected for further analysis. The fluorescence labeled selective primers had three selective nucleotides (Appendix B).
The fluorescence labeled products were diluted with 5 µL (D2) and with 20 µL (D4) 1:10 TE buffer for DNA and then according to their size, separated by capillary gel electrophoresis on an automated sequencer (GeXP, Beckmann Coulter). Results were examined using the software Bionumerics 4.6 (Applied Maths, Kortrijk, Belgium). For quality control of the AFLP procedure a genotyping error rate was calculated (Bonin et al. 2004), which was 3.1% for K. arvensis, 2.9% for S. vulgaris and 4.7% for P. lanceolata.
Statistical analysis
For band detection, each strong and clearly defined fragment was taken either into account as present (1) or absent (0). The generated binary (0/1) matrix was used for further statistical analyses.
Bayesian cluster analyses were calculated with Structure, version 2.3.4 (Pritchard et al. 2000) to infer population structure in the data set and assign individuals into groups. The potential number of groups was calculated using 10000 Markov Chain Monte Carlo (MCMC) simulations with a burn-in-period of 100000 iterations. Analyses for the predefined value of K were run 20 times per K = 1–22 (Falush et al. 2003). Results were summarized by employing the program Structure Harvester (Earl and Vonholdt 2012). Group assignment was an ad hoc quantity procedure calculating ΔK (Evanno et al. 2005). According to the model, which gave the consistent results for multiple runs and the highest probability of the data, the best estimate of K for the data set was determined.
Patterns of genetic similarities between individuals were analyzed in the software GenAlEx 6 (Peakall and Smouse 2006) using principal coordinate analyses (PCoA) based on a squared Euclidean distance matrix.
Hierarchical analyses of molecular variance, AMOVA (Excoffier et al. 1992), were also conducted with the software GenAlEx 6. Thus genetic differentiation within and among populations was investigated in two- and three-level AMOVAs.
Correlation between genetic distances (ΦPT values calculated in the AMOVA) and geographic distances among populations was tested in a Mantel test with 999 permutations (Mantel 1967).
Gene diversity H was calculated using AFLPsurv (Vekemans 2002). A Wilcoxon-test was used to test for significant differences in genetic diversity between natural and restored populations applying the software IBM Statistics 24 for Windows (IBM Corp).
Results
Ploidy levels of the study species
FCM revealed different ploidy levels for Knautia arvensis. All natural populations and the restored populations from Schwaig (SC) were tetraploid. The restored populations of Hetzmannsdorf (HE), Netzstall (NE), Pfatter (PF) and Rannertshofen (RH) were diploid. We detected no different ploidy levels for Silene vulgaris and Plantago lanceolata.
Genetic differentiation among natural and restored populations
The Bayesian cluster analysis resulted in two groups for all study species. For K .arvensis (ΔK = 282.2), the first group included all natural and the restored populations SC_R1 and SC_R2 (tetraploid populations). The remaining restored populations (diploid populations) formed the second group (Fig. 2a). For S. vulgaris (ΔK = 153) all natural populations and most individuals of the restored populations from the regions RH and SC formed the first group. The restored populations belonging to the regions HA, NE and PF built the second group (Fig. 2b). Although the individuals of P. lanceolata (ΔK = 3.4) were assigned in two groups we detected no population grouping according to the grassland type or study region (Fig. 2c). For every species, K = 2 outputs of the 20 iterations were identical.
Bayesian Cluster Analysis for aKnautia arvensis based on 137 AFLP fragments, bSilene vulgaris based on 121 AFLP fragments and cPlantago lanceolata based on 127 AFLP fragments. Populations of K. arvensis (ΔKKa = 282.2; 2c) were assigned to two groups according to their ploidy level (light grey: diploid individuals; dark grey: tetraploid individuals). Individuals of S. vulgaris (ΔK = 153; 3c) were also assigned to two groups. Individuals of P. lanceolata (2 groups, ΔK = 3.4) were admixed. HE = Hetzmannsdorf, NE = Netzstall, PF = Pfatter; RH = Rannertshofen; SC = Schwaig; HA = Haag; HS = Herrnsaal; RE = Reichenau; N = natural populations; R = restored populations
The PCoA resulted in a strong separation of natural and restored populations for K. arvensis. According to the findings of FCM analysis, diploid restored populations from the study regions HE, NE, PF and RH built the first group. The tetraploid restored populations SC_R1 an SC_R2 were grouped with the tetraploid natural populations (Fig. 3a). Natural and restored populations of S. vulgaris showed stronger admixture and we identified two clusters according to the findings of the Bayesian cluster analysis (Fig. 3b). The natural and restored populations of P. lanceolata were admixed and no groups were distinguishable (Fig. 3c).
Results of the principal coordinates analysis (PCoA) based on AFLP data of the study species. Black circles: individuals of natural populations. Hollow circles: individuals of restored populations. aKnautia arvensis: Axis 1 explained 78.79% of variance; axis 2 explained 5.74% of variance in the data set. Individuals were separated clearly into two groups, according to their level of ploidy: diploid, restored individuals (first group, hollow circles) or tetraploid natural and restored individuals (second group, hollow and black circles). bSilene vulgaris: Axis 1 explained 78.79% of variance and axis 2 explained 5.74% of variance in the data set. PCoA resulted in two slightly mixed groups. Group 1: restored individuals. Group 2: natural and restored individuals. cPlantago lanceolata: Individuals of natural and restored populations were admixed. 21.48% of variance in the data set were explained by axis 1 and 15.28% by axis 2
The AMOVA (Table 2) revealed low genetic differentiation between the study regions for all study species. However, we observed very strong genetic differentiation for K. arvensis (Table 2a) among all populations (ΦPT = 0.49). With respect to the results of FCM, the ploidy levels were taken into count for further analysis: the genetic differentiation between tetraploid restored and natural populations was with a ΦPT value of 0.19 comparable low. However, we found lower levels of genetic differentiation among all natural populations (tetraploid populations; ΦPT = 0.13). Additionally, we conducted separate AMOVAs among all diploid and among all tetraploid populations and and found a genetic differentiation comparable to the differentiation between the natural populations (for both ΦPT = 0.14). For S. vulgaris (Table 2b), the AMOVA resulted in low genetic differentiation among all populations (ΦPT = 0.13) and slightly stronger differentiation between natural and restored populations (ΦPT = 0.16). Moreover, a low genetic differentiation among natural (ΦPT = 0.09) and restored (ΦPT = 0.09) populations was observed. The conducted AMOVAs for P. lanceolata (Table 2c) between all populations, natural and restored populations as well as among natural and among restored populations revealed low genetic differentiation (for all ΦPT = 0.03).
According to the FCM results, three Mantel-tests for K. arvensis were conducted, for all populations, for the diploid populations and for all tetraploid populations. We found no correlation between pairwise genetic and geographic distances for the species (rKa = 0.01, pKa = 0.62; rKa_diploid = 0.04, pKa_diploid = 0.34; rKa_tetraploid = 0.03, pKa_tetraploid = 0.39). The Mantel-tests for S. vulgaris and P. lanceolata also revealed no correlation between pairwise genetic distances and geographic distances (rSv = 0.05, 0.18; rPl = − 0.05, pPl = 0.30).
Genetic diversity of natural and restored populations
For K. arvensis, 82.48% of the fragments were polymorphic. In natural populations, Nei’s Gene Diversity (H) ranged from 0.14 to 0.21 (mean 0.19; Table 1a). In restored populations, H values were significantly higher than in natural populations and ranged from 0.17 to 0.23 (mean 0.21;, p = 0.04).
For S. vulgaris, 89.26% of the fragments were polymorphic. H ranged from 0.29 to 0.37 (mean 0.35) for natural populations and from 0.34 to 0.38 (mean 0.36) for restored populations. No significant difference could be detected between natural and restored populations (p = 0.39; Table 1b).
For P. lanceolata the percentage of polymorphic fragments was 83.46%. Nei’s gene diversity ranged from 0.29 to 0.35 (mean 0.32) for natural populations and from 0.28 to 0.34 (mean 0.31) for restored populations (Table 1c). Between natural and restored populations, no significant difference could be detected b (p = 0.80).
Discussion
Genetic differentiation between natural and restored populations
Gene flow and genetic drift strongly affect genetic differentiation (Slatkin 1987). The exchange of pollen, seeds or plant material among populations should reduce genetic differences between populations (Slatkin 1987) and result in comparatively low levels of differentiation especially among populations of widespread and outcrossing grassland species. It is, therefore, assumed that local seed mixtures originating from delineated seed transfer zones reflect the spatial genetic structure of common grassland species (Hufford and Mazer 2003).
However, our study revealed varying degrees of differentiation between natural and restored populations for the three investigated plant species. The differentiation was stronger between natural and restored populations of the two insect-pollinated species than between natural and restored populations of the wind-pollinated species, which can be attributed to the large-scale dispersal of pollen via wind, reducing the degree of differentiation.
For Knautia arvensis the genetic differentiation between natural and restored populations (ΦPT = 0.59) was very strong. This is mainly because different ploidy levels occurred in natural and restored populations. It has already been shown before that K. arvensis exhibits various cytotypes which do not interbreed (Kolar et al. 2009; Durka et al. 2017) and therefore, function as effective breeding barriers (Kohler et al. 2010). Consequently, the two ploidy levels can be regarded as separate taxonomic units.
Considering only tetraploid populations, genetic differentiation between natural and restored populations was with a ΦPT value of 0.19 slightly higher than the genetic variation between natural populations (ΦPT = 0.13). Genetic differentiation among diploid or tetraploid populations was comparable. This provides evidence that the natural genetic structure of the species seems not to be strongly affected by applying local seed material, when the correct ploidy level is used. In our study, all natural populations of K. arvensis were tetraploid, may be because of the limited population number. However, according to previous investigations, in our study region located in the Danube region both diploid and tetraploid populations of K. arvensis may occur (Kolar et al. 2009; Durka et al. 2017). The restoration of grassland with diploid populations closely located to tetraploid natural populations may therefore be acceptable, although not being optimal since the local genetic pattern is clearly affected.
Our study revealed also a significant differentiation between natural and restored populations of Silene vulgaris (ΦPT = 0.16), although populations were more strongly admixed than observed for K. arvensis. However, genetic differentiation between natural and restored populations was twice as high as among natural populations. Thus, the local seed material did not match the natural spatial genetic pattern of the species exactly. This observation goes in line with findings of a former study by Aavik et al. (2012) who also detected significant genetic differentiation between natural and restored populations of the widespread, outcrossing plant species Lychnis flos-cuculi L. in grasslands.
For P. lanceolata it has been reported in former studies that genetic differentiation between populations may depend on geographic distances between populations and on environmental distances between habitats (Bischoff et al. 2006; Crémieux et al. 2010). In our study genetic differentiation between natural and restored populations was, however, comparable to the genetic differentiation between natural populations and even lower than previously reported for other wind-pollinated species Reisch and Bernhardt-Römermann (2014). Therefore, the application of local seed material did not distort the natural spatial genetic structure of the species in our study area.
Summing up, our investigation revealed a slight but significant genetic differentiation between natural and restored populations of insect pollinated study species, which means that commercially produced seed mixtures did not fully reflect the local genetic structure of the species. This means not necessarily that the concept for the production of local seed mixtures failed. Mixing the seed material from several source populations within the seed transfer zone is supposed to guarantee high levels of genetic variation within populations but it is clear that this approach must cause genetic differentiation at the same time.
Thus, commercially produced seed material reflects the genetic potential of the entire seed transfer zone, but matches not exactly the local genetic pattern. Nevertheless, seed material from a commercially produced seed mixture will still be genetically closer to natural populations than seed material from anywhere.
Genetic diversity of natural and restored populations
In the context of using local seeds for restoration, it is often questioned whether commercially produced seed material is variable enough to establish vital populations (Espeland et al. 2017; Nagel et al. 2018). For example, the source populations of collected stock seeds maybe had been inbred due to small population size, isolation or fragmentation (Aavik et al. 2012). Genetic diversity can also be reduced, when only a few source individuals are sampled, which may cause bottleneck effects and enhance genetic drift (Friar et al. 2000). Furthermore, the seed stock for several reproduction cycles, which may lead to inbreeding and reduced genetic diversity (Schoen and Brown 2001). Studies showed, that genetic diversity of populations can be negatively affected by bottlenecks, isolation or small population size (Ellstrand and Elam 1993) and that fragmentation can have a negative impact on genetic diversity of common plant species as well as on rare ones (Honnay and Jacquemyn 2007).
However, we detected no reduced genetic diversity in the restored populations of our three study species. In contrast to the apprehensions, the observed genetic diversity of the restored populations was equal or even higher compared to the genetic diversity of the investigated natural populations. Our results support the few existing previous studies, where the authors also reported no decreased levels of genetic diversity in restored grassland plant populations (Aavik et al. 2012; Reiker et al. 2015). Furthermore, the genetic diversity observed in natural and restored populations of K. arvensis (mean H = 0.20), S. vulgaris (mean H = 0.36) and P. lanceolata (mean H = 0.32) was even higher than reported in literature (Reisch and Bernhardt-Römermann 2014).
The level of genetic diversity we observed in restored populations of the three study species support the system used for seed production in Germany. With the applied regional admixture provenancing (Bucharova et al. 2018) it seems to be possible to maintain high genetic diversity of common grassland species in local seed mixtures. The system is based on 89 natural regions across Germany, defined by Meynen et al. (1953-62). These 89 natural regions were summarized to 22 seed transfer zones. For the production of local seed mixtures stock seed from at least five large source populations distributed across a seed transfer zone are collected, mixed thoroughly and then be propagated for up to five generations (Prasse et al. 2010), which seems to be an suitable time span to avoid decreased genetic diversity.
Conclusions
The use of local seed mixtures is a frequently applied and effective practice in ecological restoration of species-rich grasslands (Zahlheimer 2009; Prasse et al. 2010; Kiehl et al. 2014). Nevertheless, such a general procedure may raise concerns about the quality of commercially produced seed material. It seems to be questionable whether the natural spatial genetic pattern of common plant species can be maintained while producing local seed material with a sufficient level of genetic diversity.
In our study, the seed material used for restoration reflected the natural genetic structure of the species to a very different degree. In the case of K. arvensis restored populations in four of five study regions differed in ploidy level from the corresponding natural populations. In our study area both ploidy levels of K. arvensis may occur (Kolar et al. 2009). Differing ploidy levels between natural and restored populations may therefore be acceptable. Nevertheless, it would be better to use the same ploidy level for restoration to preserve the local genetic pattern of the species. The distribution pattern of the cytotypes needs, therefore, to be investigated more precisely and should be carefully considered when sampling source populations in future. The difference between natural and restored populations was smaller in S. vulgaris than in K. arvensis. However, genetic differentiation between natural and restored populations was also nearly twice as large as between natural populations of the species, indicating that the natural genetic structure of S. vulgaris is affected at the local scale by the application of commercially produced seed material for restoration. For the wind-pollinated P. lanceolata genetic differentiation between natural and restored populations was within the range of the natural populations. The use of local seed material for restoration has therefore no impact on the local genetic structure of this species.
The application of local seed material is a big step forward in restoration practice and with the system of seed production and seed transfer zones an almost unlimited amount of regionally specific seeds for restoration is provided for a wide range of plant species. Our study clearly shows that the local genetic structure especially of insect-pollinated study species may be affected by the use of commercially produced seed material. It is clear that the system of regional admixed provenancing is not designed to match exactly the genetic structure of plant populations at a very small local scale, but rather to protect the broader patterns of genetic variation. Furthermore, commercially produced seed material may match the ecological conditions within a seed transfer zone. Finally, using regional seed mixtures for restoration is still better than using seeds from far away. However, there are possibilities to improve the system, for example by including different habitat types in the seed collection process or by minimizing the size of seed transfer zones, although we are aware that the size of zones has to be large enough to allow profit for the seed producers. Further genetic analyses are needed to better understand the patterns of genetic variation in common grassland species, which may then contribute to optimize the system of regional admixed provenancing.
Whereas natural and restored populations often differed genetically in our study, genetic diversity was comparable within both grassland types. The results presented here clearly support the assumption that highly diverse populations of grassland species can be created using commercially produced local seed material. The implemented regional admixture provenancing strategy (Bucharova et al. 2018) seems, therefore, to be an appropriate method to produce genetically diverse local seed material and with further genetic research and some adjustments in sampling and multiplying strategy of source seeds, the procedure will become an even more powerful tool in conservation management.
References
Aavik T, Edwards PJ, Holderegger R, Graf R, Billeter R (2012) Genetic consequences of using seed mixtures in restoration: a case study of a wetland plant Lychnis flos-cuculi. Biol Conserv 145:195–204
Agren J (1996) Population size, pollinator limitation, and seed set in the self-incompatible herb Lythrum salicaria. Ecology 77:1779–1790
Bakker JP, Poschlod P, Strykstra RJ, Bekker RM, Thompson K (1996) Seed banks and seed dispersal: important topics in restoration ecology. Acta Bot Neerl 45:461–490
Bischoff A, Cremieux L, Smilauerova M, Lawson CS, Mortimer SR, Dolezal J, Lanta V, Edwards AR, Brook AJ, Macel M, Leps J, Steinger T, Muller-Scharer H (2006) Detecting local adaptation in widespread grassland species—the importance of scale and local plant community. J Ecol 94:1130–1142
Bonin A, Bellemain E, Eidesen PB, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273
Bucharova A, Michalski S, Hermann JM, Heveling K, Durka W, Holzel N, Kollmann J, Bossdorf O (2017) Genetic differentiation and regional adaptation among seed origins used for grassland restoration: lessons from a multispecies transplant experiment. J Appl Ecol 54:127–136
Bucharova A, Bossdorf O, Hölzel N, Kollmann J, Prasse R, Durka W (2018) Mix and match: regional admixture provenancing strikes a balance among different seed-sourcing strategies for ecological restoration. Conserv Genet 1–11
Bylebyl K, Poschlod P, Reisch C (2008) Genetic variation of Eryngium campestre L. (Apiaceae) in Central Europe. Mol Ecol 17:3379–3388
Crémieux L, Bischoff A, Müller-Schärer H, Steinger T (2010) Gene flow from foreign provenances into local plant populations: fitness consequences and implications for biodiversity restoration. Am J Bot 97:94–100
Durka W, Michalski SG, Berendzen KW, Bossdorf O, Bucharova A, Hermann JM, Holzel N, Kollmann J (2017) Genetic differentiation within multiple common grassland plants supports seed transfer zones for ecological restoration. J Appl Ecol 54:116–126
Earl DA, Vonholdt BM (2012) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population-size—implications for plant conservation. Annu Rev Ecol Syst 24:217–242
ErMiV (2011) Verordung über das Inverkehrbringen von Saatgut von Erhaltungsmischungen (Erhaltungsmischungsverordnung). (ed. Bundesgesetzblatt), pp. 2641–2646
Espeland EK, Emery NC, Mercer KL, Woolbright SA, Kettenring KM, Gepts P, Etterson JR (2017) Evolution of plant materials for ecological restoration: insights from the applied and basic literature. J Appl Ecol 54:102–115
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation studie. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131:479–491
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164:1567–1587
FoVHgV (2003) Verordnung über Herkunftsgebiete für forstliches Vermehrungsgut (Forstvermehrungsgut-Herkunftsgebietsverordung - FoVHgV). (ed. Bundesgesetzblatt), p. 238
Frankham R, Ballou JD, Biscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Friar EA, Ladoux T, Roaldson EH, Robichaux RH (2000) Microsatellite analysis of a population crash and bottleneck in the Maune Kea silversword, Argyroxiphium sandwicense ssp. sandwicense (Asteraceae), and its implications for reintroduction. Mol Ecol 9:2027–2034
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans Roy Soc B 351:1291–1298
Hölzel N, Buisson E, Dutoit T (2012) Species introduction—a major topic in vegetation restoration. Appl Veg Sci 15:161–165
Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv Biol 21:823–831
Hufford KM, Mazer SJ (2003) Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends Ecol Evol 18:147–155
Jongepierova I, Mitchley J, Tzanopoulos J (2007) A field experiment to recreate species rich hay meadows using regional seed mixtures. Biol Conserv 139:297–305
Jørgensen M, Elameen A, Hofman N, Klemsdal S, Malaval S, Fjellheim S (2016) What’s the meaning of local? Using molecular markers to define seed transfer zones for ecological restoration in Norway. Evol Appl 9:673–684
Joshi J, Schmid B, Caldeira MC, Dimitrakopoulos PG, Good J, Harris R, Hector A, Huss-Danell K, Jumpponen A, Minns A, Mulder CPH, Pereira JS, Prinz A, Scherer-Lorenzen M, Siamantziouras ASD, Terry AC, Troumbis AY, Lawton JH (2001) Local adaptation enhances performance of common plant species. Ecol Lett 4:536–544
Keller M, Kollmann J, Edwards PJ (2000) Genetic introgression from distant provenances reduces fitness in local weed populations. J Appl Ecol 37:647–659
Kiehl K, Kirmer A, Shaw N, Tischew S (2014) Guidelines for native seed production and grassland restoration. Cambridge Scholars Publishing, Cambridge
Kohler C, Mittelsten Scheid O, Erilova A (2010) The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet 26:142–148
Kolar F, Stech M, Travnicek P, Rauchova J, Urfus T, Vit P, Kubesova M, Suda J (2009) Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales. Ann Bot 103:963–974
Krauss SL, Sinclair EA, Bussell JD, Hobbs RJ (2013) An ecological genetic delineation of local seed-source provenance for ecological restoration. Ecol Evol 3:2138–2149
Kunin WE (1997) Population size and density effects in pollination: pollinator foraging and plant reproductive success in experimental arrays of Brassica kaber. J Ecol 85:225–234
Leimu R, Fischer M (2008) A meta-analysis of local adaptation in plants. PLoS ONE 3:e4010
Listl D, Poschlod P, Reisch C (2017a) Do seed transfer zones for ecological restoration reflect the spatial genetic variation of the common grassland species Lathyrus pratensis?. Restoration Ecology 26:667–676
Listl D, Poschlod P, Reisch C (2017b) Genetic variation of liverleaf (Hepatica nobilis Schreb.) in Bavaria against the background of seed transfer guidelines in forestry and restoration. Biochem Syst Ecol 71:32–41
Malaval S, Lauga B, Regnault-Roger C, Largier G (2010) Combined definition of seed transfer guidelines for ecological restoration in the French Pyrenees. Appl Veg Sci 13:113–124
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
McKay JK, Christian CE, Harrison S, J. RK (2005) How local is local?a review of practical and conceptual issues in the genetics of restoration. Restor Ecol 13:432–440
Meynen E, Schmithüsen J, Gellert J, Neef E, Müller-Miny H, Schulze JH (1953) Handbuch der naturräumlichen Gliederung Deutschlands. Bundesanstalt fur Landeskunde, Remagen
Michalski SG, Durka W (2012) Assessment of provenance delineation by genetic differentiation patterns and estimates of gene flow in the common grassland plant Geranium pratense. Conserv Genet 13:581–592
Mijnsbrugge KV, Bischoff A, Smith B (2010) A question of origin: where and how to collect seed for ecological restoration. Basic Appl Ecol 11:300–311
Miller S, Bartow A, Gisler M, Ward K, Young A, Kaye TN (2011) Can an ecoregion serve as a seed transfer zone? evidence from a common garden study with five native species. Restor Ecol 19:268–276
Montalvo AM, Ellstrand NC (2001) Nonlocal transplantation and outbreeding depression in the subshrub Lotus scoparius (Fabaceae). Am J Bot 88:258–269
Münzbergova Z, Herben T (2005) Seed, dispersal, microsite, habitat and recruitment limitation: identification of terms and concepts in studies of limitations. Oecologia 145:1–8
Nagel R, Durka W, Bossdorf O, Bucharova A (2018) Rapid evolution in native plants cultivated for ecological restoration: not a general pattern. Plant Biol
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Prasse R, Kunzmann D, Schröder R (2010) Entwicklung und praktische Umsetzung naturschutzfachlicher Mindestanforderungen an einen Herkunftsnachweis für gebietseigenes Wildpflanzensaatgut krautiger Pflanzen. In: Abschlussbericht DBU-Projekt: AZ 23931 (ed. Umweltplanung If). Universität Hannover
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Reiker J, Schulz B, Wissemann V, Gemeinholzer B (2015) Does origin always matter? evaluating the influence of nonlocal seed provenances for ecological restoration purposes in a widespread and outcrossing plant species. Ecol Evol 5:5642–5651
Reisch C (2007) Genetic structure of Saxifraga tridactylites (Saxifragaceae) from natural and man-made habitats. Conserv Genet 8:893–902
Reisch C, Bernhardt-Römermann M (2014) The impact of study design and life history traits on genetic variation of plants determined with AFLPs. Plant Ecol 215:1493–1511
Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. In: G SB (ed) Plant Molecular Biology Manual. Springer, Dordrecht, pp 183–190 R.A. S,
Schoen D, Brown A (2001) The conservation of wild plant species in seed banks: attention to both taxonomic coverage and population biology will improve the role of seed banks as conservation tools. AIBS Bull 51:960–966
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236:787–792
Török P, Deák B, Vida E, Valkó O, Lengyel S, Tóthmérész B (2010) Restoring grassland biodiversity: Sowing low-diversity seed mixtures can lead to rapid favourable changes. Biol Conserv 143:806–812
Ukrainetz NK, O’Neill GA, Jaquish B (2011) Comparison of fixed and focal point seed transfer systems for reforestation and assisted migration: a case study for interior spruce in British Columbia. Can J Forest Res 41:1452–1464
Van Treuren R, Bijlsma R, Ouborg NJ, Kwak MM (1994) Relationships between plant density, outcrossing rates and seed set in natural and experimental populations of Scabiosa columbaria. J Evol Biol 7:287–302
Vekemans X (2002) AFLP-SURV version 1.0. In: Distributed by the author. Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles, Belgium
Walker EA, Hermann JM, Kollmann J (2015) Grassland restoration by seeding: seed source and growth form matter more than density. Appl Veg Sci 18:368–378
Young TP, Petersen DA, Clary JJ (2005) The ecology of restoration: historical links, emerging issues and unexplored realms. Ecol Lett 8:662–673
Zahlheimer W (2009) Autochthone begrünung: grundsätzliches und aktuelles. Naturschutz Niederbayern 6:81–91
Acknowledgements
We are obliged to the ALE for great commitment and financial support. Furthermore, we would like to thank Petra Schitko, Michaela Powolny and Christina Fischer for their help in the Lab, Anja Metko and Florian Wagner for their help with FCM, Sabine Fischer for assistance with the maps and Peter Poschlod for his generous support and many discussions. Two anonymous reviewers provided many helpful comments to improve our manuscript.
Author information
Authors and Affiliations
Contributions
C.R. conceived and designed the study. F.K. collected the data and performed the analyses. Both authors contributed to manuscript writing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaulfuß, F., Reisch, C. Restoration of grasslands using commercially produced seed mixtures: genetic variation within and among natural and restored populations of three common grassland species. Conserv Genet 20, 373–384 (2019). https://doi.org/10.1007/s10592-018-01138-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-018-01138-0