Abstract
The use of local provenances in restoration, agriculture and forestry has been identified and widely implemented as a measure for species and community conservation. In practice, provenances are often delineated based on climate, geomorphology and correlated spatial descriptors following the boundaries of larger natural regions. They are thought to comprise genetically homogenous plant material. Here we investigate genetic variation at AFLP loci in 26 natural populations of the regionally common grassland species Geranium pratense, which is often used in seed mixtures. Most studied populations are located in one previously delineated provenance in Germany. We assess within- and among provenance differentiation patterns and aspects of gene flow by investigating the mating system, the genetic structure at regional and local scale, gene dispersal and potential selective mechanisms that may have contributed to differentiation patterns found. Compared to other herbaceous, insect-pollinated grassland species and despite being outcrossed (mean t m = 0.88), G. pratense showed low genetic diversity (mean H E = 0.15), considerable genetic differentiation among populations within provenances (mean pairwise F ST = 0.20) and a pronounced within-population spatial genetic structure (mean Sp = 0.064). A genome scan approach identified three potentially adaptive loci. However, their allelic frequencies were only weakly related to climatic parameters thus providing little evidence for adaptive divergence. Nevertheless, the distribution of genetic diversity and derived gene dispersal estimates indicate limited dispersal ability, suggesting that gene flow at distances larger than 10 km is negligible. Our findings may question the approach of delineating provenances by general criteria, and highlight the importance of species-specific studies on differentiation and adaptation patterns.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The introduction of seed and plant material from different provenances into the local landscape by activities like compensatory measures, sowing of grassland areas or plantings in the course of restoration has been recognized as a potential risk to regional genetic and species diversity for several reasons (Vander Mijnsbrugge et al. 2010). First, maladaption of alien genotypes to the local environment may result in lower fitness compared to local genotypes (Hereford 2009). Second, intraspecific hybridization between alien and local genotypes can lead to outbreeding depression and hence, lower fitness in subsequent generations (Edmands 2007; Leimu and Fischer 2010). Third, alien genotypes can become invasive if they are superior to the locals (Saltonstall 2002; Hufford and Mazer 2003). Lastly, biotic interactions within a community might be negatively affected by introduced plant material. For example, differences in time of flushing or flowering between alien and local genotypes could affect insect species that are reproductively synchronized to these phenological events (Yukawa 2000; Vitou et al. 2008). As a consequence and fundamental for species and community conservation, it has been suggested to use locally adapted plant material for e.g. ecological restoration activities (Rogers and Montalvo 2004; Vander Mijnsbrugge et al. 2010). The implementation of this suggestion into practice needs the clear delineation of local provenances as a framework for commercial seed collectors, breeders and suppliers.
The delineation of provenances has been done for a number of countries using rather general criteria such as climate or geomorphology and correlated spatial descriptors and thus following the boundaries of larger natural regions (e.g. Mortlock 2000; Ying and Yanchuk 2006). However, whether a local population will suffer from introduced plant material is likely to depend on species-specific patterns of genetic differentiation and adaptation. These, in turn, depend on species-specific trait combinations and responses to selection, as well as to migration history and habitat characteristics. For example, neutral differentiation patterns are expected to be more pronounced in species with a restricted ability for genetic exchange among individuals and populations. Indeed, it is well known that selfing species or species with short range seed dispersal are in general more differentiated than outcrossing species or species with wind dispersed seeds (Hamrick and Godt 1996; Nybom 2004). However, strong differentiation among populations can also be the result of stochastic effects, i.e. drift because of fragmentation and isolation of habitats (e.g. Aegisdottir et al. 2009). Lastly, but of particular importance for the delineation of local provenances, populations with differing environmental conditions might be genetically differentiated because of divergent selection regimes, which in turn can lead quite rapidly to locally adapted genotypes (Jump et al. 2008). As a consequence, the spatial genetic structure observed at different scales, i.e. within or among population, can be the result of different causal effects or of the same set of causes acting with different intensities. Hence, the delineation of local provenances using species-specific spatiogenetic differentiation and adaptation patterns is challenging and requires studies which provide in-depth knowledge about single species (Kleinschmit et al. 2004; McKay et al. 2005).
AFLP markers (amplified fragment lengths polymorphisms) have helped to identify large scale differentiation patterns (Michalski et al. 2010), to describe the spatial genetic structure within populations (Eckstein et al. 2006), to infer the mating system (Peters et al. 2009) and dispersal distances (Hardy et al. 2006), and to demonstrate adaptive differentiation (Herrera and Bazaga 2008). Hence, AFLPs are a versatile tool for studying relevant patterns for the delineation of local provenances (Krauss and Koch 2004; Bussell et al. 2006).
Here we study genetic variation at AFLP loci in a regional set of populations of Geranium pratense L., a perennial, diploid herb (2n = 28) common in grasslands of continental Eurasia. G. pratense, the Meadow cranesbill, reaches its western limit in Western Europe. In central Europe it favors groundwater free, nutrient-rich clay-soils of hilly countries and forelands of higher mountains (Hundt 1975). According to Hundt (1975) the distribution area of the species in central Europe is currently in expansion, but limited by the intensification of grassland management. This expansion may have a starting point with the general increase in grassland area in the second half of the nineteenth century, but suitable habitats can be traced back in the literature to 1500 BC and were likely to occur even before that time (Kauter 2002). The species is large flowered, protandrous (Proctor et al. 1996) and highly attractive for bumble bees and honey bees (Dlussky et al. 2000) suggesting an outcrossing mating system (cf. Fryxell 1957). However, no formal assessment of the mating system is known to the authors. The species is frequently included in seed mixtures used for ornamental or restoration purposes. In Germany, seeds of G. pratense are commercially available from regionalized production, based on 22 provenances that have been delineated based on natural regions, climatic and geological criteria (http://regionalisierte-pflanzenproduktion.de). Two larger suppliers for localized seeds in Germany have a disposal of 30–70 and 20–50 kg seeds per year (Rieger-Hofman GmbH and Saaten-Zeller e.K., respectively, pers. comm.) which can amount to a sown area of up to 300 ha annually indicating a considerable impact of regionalized seeds.
To test whether an already delineated provenance in Germany reflects a natural pattern in a single species and to provide a basis for a more detailed delineation in G. pratense we investigate different aspects of gene flow within and among a regional set of natural populations. In particular we (1) infer the mating system, (2) ask at which spatial scale populations are genetically structured and how within-provenance differentiation differs from among provenance differentiation, (3) indirectly assess gene dispersal from spatial genetic structure within and among populations, and (4) ask whether selective mechanisms have contributed to differentiation patterns found.
Materials and methods
Sampling and genotyping
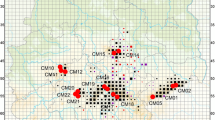
A population was defined as spatially delimited location with more or less continuously distributed individuals of the study species. Leaf tissue of individual plants was sampled from 22 natural populations in Central Germany (Table 1, Fig. 1). Sampled populations were all situated in one (‘Lowlands and Downs of Central Germany’) of the 22 local provenances in Germany (http://regionalisierte-pflanzenproduktion.de). Mean spatial distance among populations was 54 km, with minimal and maximal distances of 0.8 and 116 km, respectively. Sampling was done arbitrarily across patches. Plant density was estimated by averaging the number of individuals found in multiple surveys of a one square meter area scattered across the population randomly. To allow comparison between within- and among- provenance genetic differentiation we additionally sampled three populations from the provenance ‘Swabian Alb’ and one from the provenance ‘Southwestern German Highlands’ (Table 1), both located in Southern Germany. Mean spatial distance of these populations to the populations in Central Germany is 333 km, with minimal and maximal distances of 201 and 393 km, respectively. Sampling sites were managed grasslands or extensively managed road verges without evidence for introduction of seed material.
a Sampled populations of Geranium pratense in Central (blue, orange, green) and Southern Germany (red). Colours represent clusters found in the Bayesian structure analysis. Shades of gray represent altitude; solid lines indicate borders of German Federal States. b Hierarchical spatial genetic structure as identified by Structure
For the estimation of outcrossing rates from progeny arrays, open-pollinated seed families and maternal tissue were collected from three populations (ELB, VAT and CRA_I). Seeds were germinated and raised in a growth chamber for 2 months and leaf material of seedlings was sampled. For the analysis of small-scale spatial genetic structure exact coordinates of individual plants were recorded for populations BOES, ELB and GRO. For all samples, total genomic DNA was extracted using the DNeasy 96 Plant extraction kit (QIAGEN). AFLP analysis was done as described in Michalski et al. (2010). After primer screening the four following primer combinations were selected: 5′-FAM-GACTGCGTACCAATTCAAC-3′ and 5′-GATGAGTCCTGAGTAACAG-3′; 5′-VIC-GACTGCGTACCAATTCACA-3′ and 5′-GATGAGTCCTGAGTAACTT-3′; 5′-NED-GACTGCGTACCAATTCAAG-3′ and 5′-GATGAGTCCTGAGTAACAG-3′, and 5′-PET-GACTGCGTACCAATTCAGG-3′ and 5′-GATGAGTCCTGAGTAACAG-3′. Binning and scoring of fragments was done manually with GeneMapper version 3.7 (Applied Biosystems). The procedure was done first globally, including adult samples from all populations. For error analysis in the first data set, 66 (12%) of the samples were extracted and analyzed twice. Only loci with <8% error rate were retained in the data set resulting in an overall error rate of 2.6%. The four primer combinations resulted in a total of 122 loci, of which 118 (96.7%) were polymorphic and used for the analyses. Second, for the outcrossing rate analysis fragments for maternal and offspring samples were binned and scored for each population individually to increase the number of loci. Here, 5–19% of samples per population were included as replicates and only loci free of error were included in the final data sets, resulting in 36, 59 and 50 polymorphic loci for the three populations ELB, CRA_I and VAT, respectively.
Outcrossing rate estimates
Binary matrices with marker information for seedlings and mothers were analyzed with MLTR version 3.4 (Ritland 2002). Multilocus and single-locus outcrossing rates (t m and t s, respectively) as well as parental inbreeding coefficients (F) were computed by running the program with default options. We computed the difference between multilocus and single-locus outcrossing rates as a minimum estimate of biparental inbreeding (Shaw et al. 1981). Ninety-five per cent confidence intervals of mating system parameters were calculated based on 1,000 bootstrap replicates with families as units of resampling.
Genetic diversity and differentiation, isolation-by-distance (IBD)
Expected heterozygosity per population (H E) and pairwise genetic differentiation values (F ST) were calculated as implemented in AFLP-SURV (Vekemans 2002) using a Bayesian method with non-uniform prior distribution of allele frequencies to estimate allelic frequencies (Zhivotovsky 1999), following the treatment of Lynch and Milligan (1994). H E and F ST values were computed assuming an inbreeding coefficient of F = 0.209, based on the average value obtained from progeny arrays (see above). The effect of elevation, population density and the estimated total number of individuals in the population on H E was assessed by Pearson’s correlation. Sample sizes differed between populations due to population size restrictions and different sampling intensity (N = 8–32, mean = 20.5), however, estimates of genetic variation were not correlated to sample size (P = 0.35). The partitioning of genetic diversity according to the hierarchical sampling design of provenances and populations was evaluated by analysis of molecular variance (AMOVA) as implemented in Arlequin 3.5.1.2 (Excoffier et al. 2005).
To test whether populations are in gene-flow—drift equilibrium, pairwise genetic differentiation values (F ST/(1 − F ST), Rousset 1997) were correlated against log-transformed pairwise spatial distances. The null hypothesis that populations are at equilibrium can be rejected if the relationship fails to be monotonic and significantly positive over all distances (Hutchison and Templeton 1999). Furthermore, also the degree of scatter of the relationship is expected to increase with increasing spatial distances because of increased effects of genetic drift. To test these expectations, the significance of a Pearson’s correlations of genetic differentiation on spatial distances was evaluated using a Mantel test running 10,000 permutations. Similarly, absolute values of the residuals obtained from a standard linear regression were correlated with spatial distances and the relationship was tested for significance as just described.
Spatial genetic structure (SGS)
Population genetic structure and gene flow was assessed on different scales. First, genetic structure on the regional scale was inferred using a Bayesian approach implemented in Structure v2.3.3 (Pritchard et al. 2000; Falush et al. 2003; Falush et al. 2007). Using an admixture model with correlated allelic frequencies and allowing recessive alleles and without prior information on sampling origin, we estimated the most likely number of genetic clusters (K) for all sampled individuals. The log probability of data was calculated for K = 1 to K = N + 3, with N the number of populations sampled and 10 independent runs for each K. For each run the Burnin period was 10,000 replications and the log probability of data was calculated from additional 50,000 replications. As STRUCTURE detects only the upper hierarchical structure, we repeated the analysis for each cluster inferred in the first analysis. The most likely number of genetic clusters (K) was estimated using the method of Evanno et al. (2005). However, we took into account both, whether ∆K showed a local maximum (peak) at a particular K, and the absolute value of ∆K using ∆K = 20 as a lower threshold for significant sub-structuring.
Second, small-scale genetic structure was investigated for three populations (BOES, ELB and GRO). We tested for spatial autocorrelation in the three populations by computing kinship coefficients (Loiselle et al. 1995) between pairs of individuals within given distance classes. Upper limits for the distance classes were defined for all three populations equally as 10, 20, 30, 50, 200 and 445 m, with the last distance class for population GRO only. Mean pairwise kinship coefficients per distance class were tested against a null distribution by permuting individual locations among all individuals 1,000 times. We also computed the Sp statistic, a measure of spatial genetic structure (SGS) independent of the sampling scheme, which has been proposed for comparisons among species (Vekemans and Hardy 2004). The Sp statistic was calculated as Sp = −b/(1 − F (1)) where b is the slope of the linear regressions of pairwise kinship coefficients on log transformed pairwise spatial distances, and F (1) is the mean multilocus kinship coefficient for the first distance interval (Vekemans and Hardy 2004). Approximate confidence intervals of Sp were computed as ±two times the standard error of b estimated by jackknifing over loci. All these computations were done assuming inbreeding coefficients deviating from zero using either the direct estimate obtained from progeny arrays (ELB) or the mean parental F values among all populations studied for the outcrossing rate (F = 0.209 for BOES and GRO). All computations were performed with SPAGeDi 1.3c (Hardy and Vekemans 2002). To compare the SGS with that of other herbaceous species, we extended the compilation of Sp values collected by Vekemans and Hardy (2004) by a literature survey.
Indirect estimates of gene flow
Gene flow distances were estimated indirectly first on regional scale and second for the local scale in populations BOES, ELB and GRO. Both estimates assume that the observed spatial genetic structure is representative of an isolation-by-distance pattern at dispersal-drift equilibrium. First, according to Rousset (1997) and assuming the population genetic structure following a stepping stone model, the relationship between pairwise F ST/1 − F ST values among populations and the logarithm of pairwise spatial distances in the case of a two dimensional distribution of samples is expected to be approximately linear. The slope of the regression (b) can then be used to estimate the quantity 1/(4Dπσ2), where D is the effective population density and σ2 is half the average squared axial parent–offspring distance. To estimate σ from regional genetic structure, D was estimated as one-half to one-tenth the average density of individuals in the sampled populations (Frankham 1995). To evaluate the effect of sampling on the obtained estimate we report a 95% confidence interval obtained by jackknifing the sampled populations (N − 1) 500 times. All computations were done within the R environment (R Development Core Team 2010). Second, the dispersal parameter σ can be estimated from the slope of a regression of pairwise Kinship coefficients on spatial distances between individuals within a continuous population using an iterative approach (Vekemans and Hardy 2004). Here, 1/4Dπσ2 is estimated from the Sp value, computed as described above. D was estimated as one-half to one-tenth the average density of individuals across all sampled populations. Regression slopes were computed from a restricted distance range between σ and 20σ (Vekemans and Hardy 2004). Confidence intervals for σ were computed as ± two times the standard error of b estimated by jackknifing over loci. Intrapopulation estimates of gene flow were computed with SPAGeDi 1.3c (Hardy and Vekemans 2002).
Adaptive differentiation
To detect a possible adaptive genetic divergence among populations studied, environmental parameters were related to allelic frequencies at single marker loci by logistic regression analysis. Monthly sum of precipitation and the monthly average of air temperature for the years 1978–2008 were provided for each sampled populations from the nearest respective weather station by the German Meteorological Service (DWD, Offenbach Germany). From these data we extracted the average across years of sum, minimum and maximum monthly precipitation in the total growing season (March to September). Also, we extracted an average temperature value across years, and average minimum and maximum temperature. We also treated elevation as an environmental factor. To reduce dimensions and eliminate collinearity, we applied a principal component analysis (PCA) using the package ADE4 (Dray and Dufour 2007) for R. The two factors that accounted most for the variation in the environmental parameters (91%) were used subsequently in the regression analysis. Relating all AFLP loci individually to the two extracted factors might result in increased type I error and inflated number of significant outcomes. Hence, a preselection on all loci was accomplished by running the DFDIST program (http://www.rubic.rdg.ac.uk/~mab/stuff/), a modification for dominant markers of software developed by Beaumont and Nichols (1996). This software identifies loci that deviate from expectations of a neutral drift model. The software was run by an iterative procedure as described in Beaumont and Nichols (1996), i.e. the mean F ST to be met by the neutral simulation was adjusted after excluding outliers observed in a previous run. Loci outside a 95% confidence interval of F ST values obtained by the neutral simulation were considered to behave non-neutrally and were subjected to the regression analysis. For each preselected locus, AFLP allelic frequencies per population were explained by the two environmental factors (PCs) via separate logistic regression models, weighted by the number of included alleles per population, and with binomial error distributions. To account for overdispersion, a quasi-likelihood estimation approach was applied. Only those relationships were considered significant that remained so after Bonferroni correction for multiple tests.
The test for adaptive differentiation was done for the 22 populations from the provenance ‘Lowlands and Downs of Central Germany’ only, as the detection of outlier loci is not recommended for comparisons showing F ST values larger than 0.2 (see results; Pérez-Figueroa et al. 2010).
Results
Outcrossing rates
Multilocus outcrossing rates assessed by progeny array analyses were similarly high in all three populations (mean t m = 0.884, Table 2) suggesting a predominantly outcrossed mating system. Biparental inbreeding was substantial, exceeding 10% of the mating events. Parental inbreeding coefficients were significantly larger than zero (mean F = 0.209) indicating deviation from Hardy–Weinberg assumptions. None of the assessed parameters differed significantly among populations.
Genetic diversity and differentiation
Expected heterozygosity per population (H E) ranged from 0.108 to 0.202 (mean H E = 0.156, N = 26) and was unrelated to elevation, population density and log10-transformed total number of individuals in the population (P > 0.07). Within provenances genetic differentiation was well pronounced with pairwise F ST values ranging from 0.03 to 0.44 (mean F ST = 0.20). However, among provenance genetic differentiation was even higher with pairwise F ST values between 0.16 and 0.61 (mean F ST = 0.48). An AMOVA summarizing these results is presented in Table 3. We found significant isolation-by-distance patterns for all sampled populations (r = 0.71, Mantel P < 0.001) as well as for populations within the provenance ‘Lowlands and Downs of Central Germany’ only (r = 0.22, Mantel P = 0.009, Fig. 2). However, the degree of scatter of pairwise differentiation values was unrelated to spatial distances in both cases (P > 0.07).
Spatial genetic structure
According to the Bayesian cluster analysis, individuals were most likely to cluster in two groups, i.e. all individuals from Southern Germany in one and all individuals from the Central German provenance in a second cluster (K = 2, ∆K = 3764.5). In additional analyses the Southern German cluster split further apart (K = 2, ∆K = 2887.7), with populations being a mix of individuals from both clusters (Fig. 1). The Central German individuals were divided into two equally sized groups (K = 2, ∆K = 325.3), representing individuals of the western/northern and southern/eastern populations, respectively (Fig. 1). For the western/northern populations a strong further sub-structuring was evident, separating individuals in the West from those in the East (K = 2, ∆K = 191.1). However, for the southern/eastern populations we found only little evidence for further significant structuring (K = 5, ∆K = 15.5).
Within populations, spatial genetic autocorrelation was found to be significant in all three populations studied (Fig. 3). Mean kinship coefficients per distance intervals were very similar for populations ELB and BOES, whereas for population GRO mean Kinship coefficients were much higher for the first distance intervals. Nevertheless, the Sp statistic indicated a similar degree of spatial genetic structure for populations BOES and GRO (Sp = 0.097, CI 0.058–0.136 and 0.067, CI 0.054–0.081, respectively), whereas population ELB showed a significantly lower Sp value (Sp = 0.027 CI 0.015–0.040). For comparison, we compiled Sp values and estimates of the mating system (predominantly selfing, mixed mating, predominantly outcrossing) for in total 57 herbaceous species. Selfing, mixed mating and outcrossing species had average Sp values of 0.148 (SD 0.080, N = 11), 0.039 (0.032, N = 11) and 0.024 (0.026, N = 34), respectively (data available as Supplementary material).
Indirect estimates of gene flow
Indirect estimates of gene flow distances obtained from small scale and regional genetic structure are displayed in Table 4. Gene dispersal distances (σ) among populations BOES and ELB were similar in magnitude with partly overlapping confidence intervals, slightly varying depending on the estimate of the effective population size. Population GRO showed significantly larger σ values which is mainly based on the lowest census population density (0.1 individuals per m²) among all populations. σ values obtained from regional genetic structure based on Central German populations only approximated the values from small scale genetic structure within populations ELB and BOES (Table 4). Significant lower estimates of σ were derived from the genetic structure including all sampled populations.
Loci putatively under selection
Three out of 122 AFLP loci showed a putatively non-neutral differentiation pattern, i.e. they were more differentiated than expected (P > 0.984), and hence were included in the logistic regression analysis. Also, one marginally significant locus (AAC/CAG_239, P = 0.973) was included. Two loci showed less differentiation than expected (P < 0.025) putatively indicating balancing selection.
The first two PCA axes explained 91% of the variation in the environmental variables. Loadings of the environmental variables on the two axes and location of populations in the environmental space are displayed in Fig. 4. Of the four loci, only the allelic frequencies at locus AAC/CAG_228 showed a significant negative relationship with the second PCA factor after correction for multiple tests (Table 5).
Biplot of a principal component analysis on environmental parameters for the Central German populations of Geranium pratense studied. The first two axes account for 71 and 20% of the total variance, respectively. Arrows represent the loadings of the original parameters on the two axes (sum, minimum and maximum monthly precipitation in the total growing season—sumP, minP and maxP, respectively; average temperature, average minimum and maximum temperature—meanT, minT, maxT, respectively). Circles represent populations in the environmental space
Discussion
The main purpose of seed provenances for conservation and restoration is the conservation of genetic resources and the avoidance of harmful effects of introducing non-adapted gene pools into local communities. Therefore, the evaluation of spatial genetic differentiation patterns among and within provenances and the underlying processes, i.e. gene flow by pollen and seeds are fundamental. Our results on the mating system and the spatial genetic structure of G. pratense allow conclusions about the level of gene flow within and among natural occurring populations.
Outcrossing rates
Our results show that G. pratense is highly outcrossed. This is in concordance with findings on its pollinator spectrum (Dlussky et al. 2000) and its floral morphological features. For example, among congeneric species, the pollen/ovule ratio (P/O) of G. pratense is in the upper range of values reported by an extensive study on members of the Geraniaceae family (Fiz et al. 2008). Indeed, P/O ratios have been shown to correlate positively with the outcrossing rate (Cruden 1977; Michalski and Durka 2009), a relationship that has often more explanatory power within lineages of closely related species. However, outcrossing rates have not been quantified by molecular analysis of progeny arrays in species of Geranium, except for unpublished results for G. maculatum, a perennial species with large showy flowers similar to G. pratense, showing similarly high outcrossing rates (cited in Chang 2007).
Mean foraging distances of several hundreds meters, reported for the main pollinators of G. pratense, honey bees and bumble bees, should allow an effective pollen transfer within and even among populations (e.g. Walther-Hellwig and Frankl 2000; Wolf and Moritz 2008), which might result in a low population genetic structure. However, both the within-population spatial genetic structure (SGS), and the differentiation among populations indicate that gene pools are not effectively homogenized. The within-population SGS can also account for the substantial levels of biparental inbreeding (10–18%) found in our study (cf. Zhao et al. 2009).
Genetic diversity and differentiation
Generally, variation in the distribution of genetic diversity within and among populations can be well explained by life-history traits and historical processes affecting gene exchange among individuals (Hamrick and Godt 1996; Pannell and Dorken 2006). For example, outcrossing perennials tend to exhibit higher genetic diversity and larger genetic homogenization than selfing and short-lives species. However, genetic diversity for G. pratense at population level was low when compared to other species studied with dominant markers (cf. Nybom 2004). This becomes even more evident when genetic diversity for G. pratense (mean H E = 0.15) is directly compared to other species with similar life history traits. For example, a number of perennial, insect pollinated herbs, mostly with an outcrossing mating system and inhabiting grassland ecosystems such as Sanguisorba officinalis (Musche et al. 2008), Anthyllis vulneraria (Honnay et al. 2006), Ranunculus acris (Odat et al. 2004) or Trollius europaeus (Despres et al. 2002), exhibit higher values of within-population genetic diversity (H E = 0.19–0.30, mean H E across species = 0.27). Also, within sampled provenances, among population differentiation is unexpectedly high in G. pratense (mean pairwise F ST = 0.20) when compared to herbaceous grassland species studied at a similar spatial scale: Sanguisorba officinalis (Musche et al. 2008), Anthyllis vulneraria (Honnay et al. 2006), Ranunculus acris (Odat et al. 2004) or Cirsium dissectum and Succisa pratensis (Smulders et al. 2000), which all share quite low differentiation values (F ST (or analogues) <0.11, mean value across species = 0.07). Hence, the observed distribution of genetic diversity within and among populations of G. pratense in Central and Southern Germany seems to be shaped in particular by the local history of the species and its populations, e.g. range edge effects, which possibly masks the effects of life history traits.
Spatial genetic structure and gene flow
For G. pratense an overall increase of genetic differentiation between populations with increasing distance was found (Fig. 2). However, in a pattern of regional gene flow-drift equilibrium, also the degree of scatter should increase with distance (Hutchison and Templeton 1999), which was not the case. Rather, the regional population structure is affected by gene flow and drift depending on scale with populations still in genetic exchange at distances up to 10 km (Fig. 2). At larger distances the pattern seems to result from genetic drift only.
Within-population spatial genetic structure was also found to be very pronounced which is typical for isolation-by-distance patterns because of restricted dispersal ability. The Sp statistics obtained for the three populations (mean Sp = 0.064) are higher than the average value reported from other outcrossing herbs (mean Sp = 0.024) and rather comparable with values found for selfing and mixed mating species (mean Sp = 0.148 and 0.039, respectively) indicating a quite small genetic patch size in G. pratense. Thus, although the mating system analysis suggests that alleles might be homogenized within-populations because of outcrossing, the effective gene dispersal is indeed restricted.
This is further substantiated by the Structure analyses which showed that genetic diversity is strongly hierarchically structured. Even the two clusters identified for the subset of populations from the provenance ‘Lowlands and Downs of Central Germany’ (Fig. 1) still showed a pronounced within-cluster population differentiation (mean F ST = 0.154 and 0.191, for the southern/eastern cluster and the northern/western cluster, respectively). As no obvious geographical barriers between the well separated clusters are apparent, causes for the structuring of genetic diversity, e.g. historical land use differences or environmental gradients remain speculative and deserve further study.
Indirect estimates of gene dispersal for G. pratense (Table 4) are quite low, but well in the range reported for other herbaceous species (Van Rossum and Triest 2007; Llaurens et al. 2008; Rong et al. 2010). Low dispersal ranges would fit to the mode of seed dispersal in Geranium. Although seeds of Geranium species are dispersed by a ballistic mechanism it has been described as relatively inefficient for long distance dispersal with the majority of seeds remaining in close vicinity to the mother plant (<0.5 m, Hundt 1975; Herrera 1991).
Long distance dispersal events may occur by the anthropogenic spread of the species, e.g. following hay transports along roads (Hundt 1975), in the course of road construction (Brennenstuhl 2007) or by introduction of commercially available seed material. However, the significant differentiation among populations within provenances suggests that in our case anthropogenic dispersal did not contribute substantially to effective seed dispersal and genetic homogenization. To some extent this could be expected because our sampled populations were far from sites of restoration or ornamental activities.
Indirect estimates of gene flow critically depend on the effective population density used in the calculations (Vekemans and Hardy 2004). In low density populations the ratio of effective population size to census population size might be lower than in high density populations (Frankham 1995). Furthermore, pollinators have to move larger distances between flowering individuals. Both facts may explain the differences in within-population SGS found among the three studied populations. It is possible that the effective population density, the number of individuals that contribute genes to the next generation, is even smaller for G. pratense in Central Germany than estimated for our study. The grassland habitats of G. pratense are frequently mown for hay production before seed maturity. As a consequence, often only a fraction of flowering individuals is able to shed seeds which might affect within-population genetic structure.
Our indirect measures of gene dispersal based on the SGS within populations fitted quite well to the estimate obtained from the isolation-by-distance plots at the regional scales (Table 4). Hence, although a number of assumptions are needed to obtain these estimates (Rousset 1997; Vekemans and Hardy 2004) and which are not necessarily met in reality, the estimates can be seen as independent replicates supporting each other. This further suggests that the genetic structure at both spatial scales is the result of similar processes acting with similar intensity.
Adaptive differentiation
The occurrence and detection of outlier loci from genome scans (Beaumont and Nichols 1996; Foll and Gaggiotti 2008) is not necessarily a signature of selection, but can be caused also by neutral processes, expressed in form of a phylogeographical structure or by bottleneck effects (Holderegger et al. 2008). To allow more specific insights into a possible adaptive differentiation in G. pratense we asked whether allelic frequencies at outlier loci may correlate to climatic variables. However, with only one significant out of eight correlations we found little evidence for adaptive differentiation caused by climatic selection, which has been identified as a driving force of population differentiation on various scales (e.g. Michalski et al. 2010, Shi et al. 2011). However, it cannot be excluded that other environmental variables (e.g. soil type, pH-value) and thus other selective mechanisms have shaped the observed allelic frequency patterns at certain loci. Also, the overall strong degree of genetic differentiation (mean F ST = 0.20) may lower the efficiency of the method used for identifying outlier loci (Pérez-Figueroa et al. 2010). Whereas a combination of methods has been suggested recently to control the frequency of false positives found in such genome scans (Pérez-Figueroa et al. 2010), only selection experiments, e.g. a reciprocal transplant experiment, can provide a final proof for adaptive differentiation (Holderegger et al. 2008; Leimu and Fischer 2008).
Implications for conservation
Despite being an outcrossed, regionally common grassland species, G. pratense harbors relatively low levels of genetic diversity, is highly genetically differentiated at distances larger than 10 km and shows a pronounced within- and among population genetic structure, which partly reflects spatial distribution. We found that the already high population differentiation within provenances (maximum pairwise F ST = 0.44) can reach similar values as among provenance comparisons (mean pariwise F ST = 0.46). In contrast to common practice for species and community conservation in Germany, the results suggest spatially more narrowly defined provenances for G. pratense. However, it needs to be tested experimentally whether these large differentiation values can imply harmful effects on local communities because of outbreeding depression or maladaption when seed or plant material of G. pratense is transferred between populations and provenances. Our results differ from those for other herbaceous, insect-pollinated grassland species that display rather weak population differentiation which is rather concordant to the existing provenance delineation. However, our findings may question the approach of delineating provenances by general criteria, and highlight the importance of species-specific studies on differentiation and adaptation patterns (cf. Miller et al. 2011).
References
Aegisdottir HH, Kuss P, Stocklin J (2009) Isolated populations of a rare alpine plant show high genetic diversity and considerable population differentiation. Ann Bot 104:1313–1322
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B 263:1619–1626
Brennenstuhl G (2007) Bemerkenswerte Arten nach Strassenbaumaßnahmen in Salzwedel. Mitt Florist Kart Sachsen-Anhalt 12:95
Bussell JD, Hood P, Alacs EA, Dixon KW, Hobbs RJ, Krauss SL (2006) Rapid genetic delineation of local provenance seed-collection zones for effective rehabilitation of an urban bushland remnant. Austral Ecol 31:164–175
Chang SM (2007) Gender-specific inbreeding depression in a gynodioecious plant, Geranium maculatum (Geraniaceae). Am J Bot 94:1193–1204
Cruden RW (1977) Pollen-ovule ratios: a conservative indicator of breeding systems in flowering plants. Evolution 31:32–46
Despres L, Loriot S, Gaudeul M (2002) Geographic pattern of genetic variation in the European globeflower Trollius europaeus L. (Ranunculaceae) inferred from amplified fragment length polymorphism markers. Mol Ecol 11:2337–2347
Dlussky GM, Lavrova NV, Erofeeva EA (2000) Mechanisms of restiction of pollinator range in Chamaenerion angustifolium and two Geranium species (G. palustre and G. pratense). Zh Obshch Biol 61:181–197
Dray S, Dufour AB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20
Eckstein RL, O’Neill RA, Danihelka J, Otte A, Kohler W (2006) Genetic structure among and within peripheral and central populations of three endangered floodplain violets. Mol Ecol 15:2367–2379
Edmands S (2007) Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol 16:463–475
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fiz O, Vargas P, Alarcón M, Aedo C, García JL, Aldasoro JJ (2008) Phylogeny and historical biogeography of Geraniaceae in relation to climate changes and pollination ecology. Syst Bot 33:326–342
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a bayesian perspective. Genetics 180:977–993
Frankham R (1995) Effective population-size adult-population size ratios in wildlife—a review. Genet Res 66:95–107
Fryxell PA (1957) Mode of reproduction of higher plants. Bot Rev 23:135–233
Hamrick JL, Godt MJW (1996) Effects of life-history traits on genetic diversity in plant species. Phil Trans R Soc B 351:1291–1298
Hardy OJ, Vekemans X (2002) SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier MH, Doligez A, Dutech C, Kremer A, Latouche-Halle C, Troispoux V, Veron V, Degen B (2006) Fine-scale genetic structure and gene dispersal inferences in 10 neotropical tree species. Mol Ecol 15:559–571
Hereford J (2009) A quantitative survey of local adaptation and fitness trade-offs. Am Nat 173:579–588
Herrera J (1991) Herbivory, seed dispersal, and the distribution of a ruderal plant living in a natural habitat. Oikos 62:209–215
Herrera CM, Bazaga P (2008) Population-genomic approach reveals adaptive floral divergence in discrete populations of a hawk moth-pollinated violet. Mol Ecol 17:5378–5390
Holderegger R, Herrmann D, Poncet B, Gugerli F, Thuiller W, Taberlet P, Gielly L, Rioux D, Brodbeck S, Aubert S, Manel S (2008) Land ahead: using genome scans to identify molecular markers of adaptive relevance. Plant Ecol Divers 1:273–283
Honnay O, Coart E, Butaye J, Adriaens D, Van Glabeke S, Roldan-Ruiz I (2006) Low impact of present and historical landscape configuration on the genetics of fragmented Anthyllis vulneraria populations. Biol Conserv 127:411–419
Hufford KM, Mazer SJ (2003) Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends Ecol Evol 18:147–155
Hundt R (1975) Zur anthropogenen Verbreitung und Vergesellschaftung von Geranium pratense L. Vegetatio 31:23–32
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: Inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Jump AS, Penuelas J, Rico L, Ramallo E, Estiarte M, Martinez-Izquierdo JA, Lloret F (2008) Simulated climate change provokes rapid genetic change in the Mediterranean shrub Fumana thymifolia. Glob Change Biol 14:637–643
Kauter D (2002) »Sauergras« und »Wegbreit«?: Die Entwicklung der Wiesen in Mitteleuropa zwischen 1500 und 1900. Heimbach, Stuttgart
Kleinschmit JRG, Kownatzki D, Gegorius HR (2004) Adaptational characteristics of autochthonous populations—consequences for provenance delineation. For Ecol Manag 197:213–224
Krauss SL, Koch JM (2004) Rapid genetic delineation of provenance for plant community restoration. J Appl Ecol 41:1162–1173
Leimu R, Fischer M (2008) A meta-analysis of local adaptation in plants. PLoS ONE. doi:10.1371/journal.pone.0004010
Leimu R, Fischer M (2010) Between-population outbreeding affects plant defence. PLoS ONE. doi:10.1371/journal.pone.0012614
Llaurens V, Castric V, Austerlitz F, Vekemans X (2008) High paternal diversity in the self-incompatible herb Arabidopsis halleri despite clonal reproduction and spatially restricted pollen dispersal. Mol Ecol 17:1577–1588
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
McKay JK, Christian CE, Harrison S, Rice KJ (2005) “How local is local?”—A review of practical and conceptual issues in the genetics of restoration. Restor Ecol 13:432–440
Michalski SG, Durka W (2009) Pollination mode and life form strongly affect the relation between mating system and pollen to ovule ratios. New Phyt 183:470–479
Michalski S, Durka W, Jentsch A, Kreyling J, Pompe S, Schweiger O, Willner E, Beierkuhnlein C (2010) Evidence for genetic differentiation and divergent selection in an autotetraploid forage grass (Arrhenatherum elatius). Theor Appl Genet 120:1151–1162
Miller SA, Bartow A, Gisler M, Ward K, Young AS, Kaye TN (2011) Can an ecoregion serve as a seed transfer zone? Evidence from a common garden study with five native species. Restor Ecol 19:268–276
Mortlock BW (2000) Local seed for revegetation. Ecol Manag Restor 1:93–101
Musche M, Settele J, Durka W (2008) Genetic population structure and reproductive fitness in the plant Sanguisorba officinalis in populations supporting colonies of an endangered Maculinea butterfly. Int J Plant Sci 169:253–262
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Odat N, Jetschke G, Hellwig FH (2004) Genetic diversity of Ranunculus acris L. (Ranunculaceae) populations in relation to species diversity and habitat type in grassland communities. Mol Ecol 13:1251–1257
Pannell JR, Dorken ME (2006) Colonisation as a common denominator in plant metapopulations and range expansions: effects on genetic diversity and sexual systems. Landscape Ecol 21:837–848
Pérez-Figueroa A, Garcia-Pereira MJ, Saura M, Rolan-Alvarez E, Caballero A (2010) Comparing three different methods to detect selective loci using dominant markers. J Evol Biol 23:2267–2276
Peters MD, Xiang QY, Thomas DT, Stucky J, Whiteman NK (2009) Genetic analyses of the federally endangered Echinacea laevigata using amplified fragment length polymorphisms (AFLP)-inferences in population genetic structure and mating system. Conserv Genet 10:1–14
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Proctor M, Yeo P, Lack A (1996) The natural history of pollination. Timber Press, Portland
Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Heredity 88:221–228
Rogers DL, Montalvo AM (2004) Genetically appropriate choices for plant materials to maintain biologcial diversity. University of California. Report to the USDA Forest Service, Rocky Mountain Region, Lakewood, CO. http://www.fs.fed.us/r2/publications/botany/plantgenetics.pdf. Accessed 26 Feb 2011
Rong J, Janson S, Umehara M, Ono M, Vrieling K (2010) Historical and contemporary gene dispersal in wild carrot (Daucus carota ssp. carota) populations. Ann Bot 106:285–296
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Saltonstall K (2002) Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci USA 99:2445–2449
Shaw DV, Kahler AL, Allard RW (1981) A multilocus estimator of mating system parameters in plant populations. Proc Natl Acad Sci USA 78:1298–1302
Shi M–M, Michalski SG, Chen X-Y, Durka W (2011) Isolation by elevation: genetic structure at neutral and putatively non-neutral loci in a dominant tree of subtropical forests, Castanopsis eyrei. PLoS ONE 6:e21302
Smulders MJM, van der Schoot J, Geerts R, Antonisse-de Jong AG, Korevaar H, van der Werf A, Vosman B (2000) Genetic diversity and the reintroduction of meadow species. Plant Biol 2:447–454
R Develoment Core Team (2010) R: a language and environment for statistical computing. In: http://www.R-project.org. R Foundation for Statistical Computing, Vienna, Austria
Van Rossum F, Triest L (2007) Fine-scale spatial genetic structure of the distylous Primula veris in fragmented habitats. Plant Biol 9:374–382
Vander Mijnsbrugge K, Bischoff A, Smith B (2010) A question of origin: where and how to collect seed for ecological restoration. Basic Appl Ecol 11:300–311
Vekemans X (2002) AFLP-SURV version 1.0. Distributed by the autor, Laboratoir de Génétique et Ecologie, Végétale. Université Libre de Bruxelles, Belgium
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935
Vitou J, Skuhrava M, Skuhravy V, Scott JK, Sheppard AW (2008) The role of plant phenology in the host specificity of Gephyraulus raphanistri (Diptera: Cecidomyiidae) associated with Raphanus spp. (Brassicaceae). Eur J Entomol 105:113–119
Walther-Hellwig K, Frankl R (2000) Foraging habitats and foraging distances of bumblebees, Bombus spp. (Hym., apidae), in an agricultural landscape. J Appl Entomol 124:299–306
Wolf S, Moritz RFA (2008) Foraging distance in Bombus terrestris L. (Hymenoptera : Apidae). Apidologie 39:419–427
Ying CC, Yanchuk AD (2006) The development of British Columbia’s tree seed transfer guidelines: purpose, concept, methodology, and implementation. For Ecol Manag 227:1–13
Yukawa J (2000) Synchronization of gallers with host plant phenology. Popul Ecol 42:105–113
Zhao R, Xia H, Lu BR (2009) Fine-scale genetic structure enhances biparental inbreeding by promoting mating events between more related individuals in wild soybean (Glycine soja, Fabaceae) populations. Am J Bot 96:1138–1147
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Michalski, S.G., Durka, W. Assessment of provenance delineation by genetic differentiation patterns and estimates of gene flow in the common grassland plant Geranium pratense . Conserv Genet 13, 581–592 (2012). https://doi.org/10.1007/s10592-011-0309-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-011-0309-7